Abstract
An Italian nationwide survey was carried out to assess the prevalences and the antimicrobial susceptibilities of members of the family Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs). Over a 6-month period, 8,015 isolates were obtained from hospitalized patients and screened for resistance to extended-spectrum cephalosporins and monobactams. On the basis of a synergistic effect between clavulanate and selected β-lactams (ceftazidime, aztreonam, cefotaxime, cefepime, and ceftriaxone), 509 isolates were found to be ESBL positive (6.3%). Colony blot hybridization with blaTEM and blaSHV DNA probes allowed one to distinguish four different genotypes: TEM-positive, SHV-positive, TEM- and SHV-positive, and non-TEM, non-SHV ESBL types. MICs for each isolate (E-test) were obtained for widely used β-lactams, combinations of β-lactams with β-lactamase inhibitors, aminoglycosides, and fluoroquinolones. Among ESBL-positive strains, Klebsiella pneumoniae, Proteus mirabilis, and Escherichia coli accounted for 73.6% of isolates. Overall, TEM-type ESBLs were more prevalent than SHV-type enzymes (234 versus 173), whereas the prevalence of strains producing both TEM- and SHV-type ESBLs was similar to that of isolates producing non-TEM, non-SHV enzymes (55 and 38, respectively). In vitro, all but one of the ESBL-producing isolates remained susceptible to imipenem. Susceptibility to other drugs varied: piperacillin-tazobactam, 91%; amoxicillin-clavulanic acid, 85%; cefoxitin, 78%; amikacin, 76%; ampicillin-sulbactam, 61%; ciprofloxacin, 58%; and gentamicin, 56%. Associated resistance to aminoglycosides and ciprofloxacin was observed most frequently among TEM-positive strains. Since therapeutic options for multiresistant Enterobacteriaceae are limited, combinations of β-lactams and β-lactamase inhibitors appear to represent an important alternative for treating infections caused by ESBL-producing Enterobacteriaceae.
Among members of the family Enterobacteriaceae, the production of plasmid-mediated extended-spectrum β-lactamases (ESBLs) has emerged as an important mechanism of resistance to β-lactams, drugs that account for approximately 50% of antibiotic consumption (19). The vast majority of ESBLs are derivatives of TEM-1 and TEM-2 (common plasmid-mediated β-lactamases of Escherichia coli) or SHV-1 (the chromosomally encoded enzyme of Klebsiella pneumoniae) (6). These enzymes are capable of hydrolyzing a wide range of β-lactams, including most recently developed cephalosporins, but are not active against cephamycins and carbapenems (18). In interpreting the phenotype of ESBL-positive strains with regard to β-lactams, it should be borne in mind that drug resistance may also result from the combined activity of a specific ESBL together with other β-lactamases (e.g., the chromosomal AmpC) (18).
Among Enterobacteriaceae, ESBLs have been found mainly in Klebsiella spp. and E. coli but have been reported also in other genera, such as Citrobacter, Enterobacter, Morganella, Proteus, Providencia, Salmonella, and Serratia (6, 7, 11, 13, 36). Infections caused by ESBL-positive organisms often involve immunocompromised patients, making it difficult to eradicate these organisms in high-risk wards, such as intensive care units (2, 16, 20).
Microbiology laboratories play an important role in detecting and promptly reporting the isolation of ESBL-positive bacteria, since drug susceptibility data are of the utmost importance for the clinical management of patients infected by these organisms (32). From the laboratory standpoint, reduced susceptibility or resistance to extended-spectrum cephalosporins and/or monobactams represents the first indicator of ESBL production, but confirmatory tests of synergy between clavulanate and selected β-lactams are required (e.g., double-disk method, E-test special strips) (7, 10, 23, 34). The expression of an extended-spectrum enzyme does not always involve a phenotype that can be interpreted as resistant by the routine MICs and disk diffusion methods that follow current National Committee for Clinical Laboratory Standards (NCCLS) breakpoints (23). According to these criteria, ESBL-positive strains should be reported as resistant even if drug MICs are below breakpoints established for cephalosporins and aztreonam. This is defined for Klebsiella spp. and E. coli but has not been established for other Enterobacteriaceae.
In the case of ESBL-positive strains, microbiology laboratories should provide the clinician with reliable therapeutic options for successfully treating infected patients. Since ESBL distribution has been shown to differ among countries (13, 18, 29), monitoring of the prevalences and the types of extended-spectrum enzymes may contribute to defining the breadth of the problem in the geographic area of interest.
To this end, an Italian nationwide study was designed to assess the prevalences and distribution of ESBL-positive species among Enterobacteriaceae recovered from hospitalized patients. The second aim of this study was to evaluate the susceptibilities of ESBL-positive isolates to compounds useful in overcoming resistance traits that may be associated with ESBL production (21, 24, 33). Our results report the distribution of ESBLs in different species of Enterobacteriaceae and demonstrate that expression of these enzymes is often associated with resistance to nonrelated molecules, such as fluoroquinolones and aminoglycosides.
MATERIALS AND METHODS
Design of the study.
Ten microbiology laboratories distributed throughout Italy were enrolled in this study. Participating hospitals were the following: Ospedale Civile, Novara (G. Fortina and L. Soattini); Ospedale San Raffaele, Milan (R. Vaiani and G. Gesu); Ospedale Niguarda, Milan (E. Magliano and G. Ortisi); Ospedali Riuniti, Bergamo (A. Goglio and F. Vailati); Ospedale di Circolo, Varese (A. Toniolo and F. Luzzaro); Ospedale Careggi, Florence (P. Nicoletti and P. Pecile); Ospedale Monteluce, Perugia (F. Menichetti); Università Cattolica del Sacro Cuore, Rome (G. Fadda and T. Spanu); Università di Bari, Bari (G. Miragliotta); and Università di Catania, Catania (G. Nicoletti and G. Bonfiglio). Over a 6-month period (January to June 1999), the task of each laboratory was to evaluate epidemiological data and susceptibilities to β-lactams of Enterobacteriaceae recovered from hospitalized patients. To avoid duplicates, each laboratory included only one isolate per species from each patient, unless an isolate of the same species was subsequently obtained with clearly different resistance traits. Ampicillin-susceptible strains of E. coli, Proteus mirabilis, Salmonella spp., and Shigella spp. were not further tested. All other isolates were evaluated by the double-disk synergy test of interaction between clavulanate and selected β-lactams (14). Positive strains were subsequently assayed by two quantitative E-test special strips testing the synergistic effect of clavulanate with cephalosporins. At the molecular level, positivity was then confirmed by hybridization with blaTEM and blaSHV DNA probes. The profiles of susceptibilities of ESBL-positive strains to clinically relevant β-lactams and other antibiotics were evaluated by the quantitative E-test.
Testing synergy between clavulanate and β-lactams: double-disk assay and quantitative E-test.
The double-disk synergy test was performed as a standard disk diffusion assay on Mueller-Hinton agar (Oxoid, Milan, Italy). Disks containing aztreonam, ceftazidime, cefepime, ceftriaxone, and cefotaxime (30 μg each) were placed at variable distances depending on the species (20 to 30 mm center to center) around a disk containing amoxicillin (20 μg) plus clavulanic acid (10 μg). Enhancement of the inhibition zone toward the amoxicillin-plus-clavulanic acid disk, indicating a synergy between clavulanic acid and any one of the test antibiotics, was taken as presumptive evidence of ESBL production.
The synergistic activity of clavulanate with both ceftazidime and cefotaxime was confirmed by means of E-test special strips (AB Biodisk, Solna, Sweden) containing ceftazidime/ceftazidime plus clavulanate and cefotaxime/cefotaxime plus clavulanate. Production of ESBL was suggested if clavulanate caused at least a 3 twofold decrease of MICs of the above-mentioned drugs. Results of the E-test have been interpreted with particular caution in the case of the genera Citrobacter, Enterobacter, Providencia, Morganella, and Serratia, which may produce high levels of AmpC β-lactamase, because they can mask the synergistic effect of clavulanate with cephalosporins.
Molecular methods.
ESBL-producing isolates were studied by colony blot hybridizations performed on nylon membranes (Hybond-N; Amersham-Pharmacia Biotech, Milan, Italy) according to the manufacturer’s instructions, using random-primed 32P-labeled DNA probes. Amplicons generated by PCR, containing either the blaTEM-1 or the blaSHV-1 genes, were used as probes to detect the presence of blaTEM and blaSHV alleles, respectively. The primers used for amplification of the blaTEM-1 allele were MAb/F (5′-GGGGAGCTCATAAAATTCTTGAAGAC) and MAb/R (5′-GGGGGATCCTTACCAATGCTTAATCA). Cycling conditions were as follows: denaturation, 95°C for 30 s; annealing, 42°C for 1 min; extension, 72°C for 1 min; repeated for 30 cycles. The primers used for amplification of the blaSHV-1 allele were SHV/F (5′-GCCCGGGTTATTCTTATTTGTCGC) and SHV/R (5′-TCTTTCCGATGCCGCCGCCAGTCA). Cycling conditions were as follows: denaturation, 95°C for 30 s; annealing, 58°C for 1 min; extension, 72°C for 1 min; repeated for 30 cycles. All reactions were performed in a 100-μl volume using 2.5 U of the AmpliTaq Gold DNA polymerase (Perkin-Elmer, Monza, Italy) in the reaction buffer provided by the manufacturer containing 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 80 pmol of each primer, and 200 ng of plasmid DNA as a template. Methods have been described (26).
Measurement of MICs by the quantitative E-test.
MICs for each ESBL-producing isolate were obtained by the E-test method (AB Biodisk) for the following agents: piperacillin, cephalothin, ceftriaxone, cefotaxime, ceftazidime, cefepime, cefoxitin, aztreonam, imipenem, amoxicillin-clavulanate, ampicillin-sulbactam, piperacillin-tazobactam, gentamicin, amikacin, and ciprofloxacin. The E-test was performed in accordance with the manufacturer’s instructions using Mueller-Hinton agar plates (Oxoid). MICs were measured as the point of intersection of the inhibition ellipse with the E-test strip edge. Quality control strains (Escherichia coli ATCC 25922 and ATCC 35218, Pseudomonas aeruginosa ATCC 25783, and Klebsiella pneumoniae ATCC 700603) were included in each run.
RESULTS
Occurrence of ESBL-producing organisms.
A total of 8,015 isolates of the family Enterobacteriaceae were studied during a 6-month period (January to June 1999). The double-disk method showed that 509 out of 8,015 isolates (6.3%) were characterized by synergy between clavulanate and at least one of the tested β-lactams. As shown in Table 1, the most common ESBL-producing strain was K. pneumoniae (n = 189), followed by P. mirabilis (n = 131), E. coli (n = 55), and Enterobacter aerogenes (n = 31). However, when data were expressed as the prevalence of ESBLs within each species, Providencia stuartii appeared to harbor ESBLs at a frequency (28.1%) higher than that observed for other species. The above particularly high prevalence of ESBL-positive P. stuartii bacteria was mainly due to the contributions of 2 out of 10 centers participating in the study. The lowest intraspecies ESBL prevalence was observed for E. coli (1.2%).
TABLE 1.
Numbers of nonduplicated isolates of members of the family Enterobacteriaceae that produced ESBLa
| Species | No. of isolates | ESBL-producing isolates
|
|
|---|---|---|---|
| No. (%) | % ESBL positive | ||
| E. coli | 4,604 | 55 (1.2) | 10.8 |
| K. pneumoniae | 946 | 189 (20.0) | 37.1 |
| K. oxytoca | 166 | 25 (15.1) | 4.9 |
| P. mirabilis | 805 | 131 (16.3) | 25.7 |
| P. vulgarisb | 52 | 1 (1.9) | 0.2 |
| P. stuartii | 96 | 27 (28.1) | 5.3 |
| M. morganii | 191 | 9 (4.7) | 1.8 |
| E. aerogenes | 151 | 31 (20.5) | 6.0 |
| E. cloacae | 418 | 12 (2.9) | 2.4 |
| S. marcescens | 224 | 11 (4.9) | 2.2 |
| C. freundii | 256 | 12 (4.7) | 2.4 |
| C. koseri | 49 | 6 (12.2) | 1.2 |
| Other speciesc | 57 | 0 (0.0) | 0.0 |
| Total | 8,015 | 509 | 6.3 |
Isolates shown to produce ESBL by double-disk synergy test.
Proteus vulgaris.
Other species include Hafnia alvei, Salmonella spp., Proteus penneri, Providencia rettgeri, Enterobacter agglomerans, Enterobacter gergoviae, Enterobacter sakazakii, and Serratia liquefaciens.
The quantitative E-test (synergy between clavulanate and ceftazidime and/or cefotaxime) confirmed ESBL production in all isolates of the following species: E. coli, K. pneumoniae, Klebsiella oxytoca, P. mirabilis, and Citrobacter koseri. The E-test, however, was not able to detect all ESBL-positive isolates of Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, Morganella morganii, P. stuartii, and Serratia marcescens. On the whole, E-test strips evaluating synergy between clavulanate and ceftazidime proved more effective than strips containing clavulanate plus cefotaxime (data not shown).
Distribution of ESBL gene types in different members of the family Enterobacteriaceae.
Colony hybridization with blaTEM-1 and blaSHV-1 DNA probes could be performed for 500 out of 509 ESBL-positive isolates (Table 2). The assay showed that TEM-type ESBLs were more prevalent than SHV-type enzymes (234 versus 173) and that about 20% of ESBL-positive Enterobacteriaceae either hybridized with both TEM and SHV probes, or failed to hybridize with any of the above probes. The latter group (38 isolates) was supposed to produce non-TEM, non-SHV enzymes. On the basis of sequencing experiments, 7.5% of TEM-type β-lactamases were shown to be TEM-1 enzymes, whereas 3.2% of SHV-type enzymes were identified as SHV-1. All the other enzymes were found to be ESBLs (data not shown; M. Perilli, E. Dell’Amico, B. Segatore, M. R. de Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante, submitted for publication).
TABLE 2.
ESBL-producing Enterobacteriaceae: distribution of TEM- and SHV-derived genes as shown by colony blot hybridizationa
| Species | No. of ESBL-positive isolates | No. (%) with gene type within species
|
||||
|---|---|---|---|---|---|---|
| TEM type only | SHV type only | TEM and SHV | non-TEM/non-SHV | NAb | ||
| E. coli | 55 | 11 (20) | 28 (52) | 9 (17) | 6 (11) | 1 |
| K. pneumoniae | 189 | 32 (17) | 104 (55) | 34 (18) | 18 (10) | 1 |
| K. oxytoca | 25 | 9 (36) | 10 (40) | 1 (4) | 5 (20) | 0 |
| P. mirabilis | 131 | 127 (98) | 0 (0) | 0 (0) | 2 (2) | 2 |
| P. vulgarisc | 1 | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 |
| P. stuartii | 27 | 22 (88) | 0 (0) | 3 (12) | 0 (0) | 2 |
| M. morganii | 9 | 7 (78) | 1 (11) | 0 (0) | 1 (11) | 0 |
| E. aerogenes | 31 | 11 (35) | 14 (45) | 1 (3) | 5 (16) | 0 |
| E. cloacae | 12 | 3 (25) | 3 (25) | 6 (50) | 0 (0) | 0 |
| S. marcescens | 11 | 7 (64) | 4 (36) | 0 (0) | 0 (0) | 0 |
| C. freundii | 12 | 1 (11) | 7 (78) | 1 (11) | 0 (0) | 3 |
| C. koseri | 6 | 4 (67) | 2 (33) | 0 (0) | 0 (0) | 0 |
| Total | 509 | 234 (47) | 173 (35) | 55 (11) | 38 (8) | 9 |
Isolates shown to encode for TEM- and/or SHV-derived enzymes by hybridization with blaTEM and blaSHV DNA probes.
NA, number of isolates for which data were not available.
Proteus vulgaris.
TEM-type ESBLs appeared to be particularly prevalent (i.e., above 50%) for the following species: P. mirabilis, P. stuartii, M. morganii, C. koseri, and S. marcescens. SHV-type enzymes, in contrast, were widely diffused for C. freundii, K. pneumoniae, and E. coli. It is noteworthy that 50% of E. cloacae isolates expressed both TEM and SHV enzymes, whereas non-TEM, non-SHV enzymes were found in most species but were particularly frequent in K. pneumoniae.
MICs of β-lactams evaluated by the E-test.
Drug susceptibilities of ESBL-positive isolates were evaluated by the quantitative E-test. MICs of β-lactams have been listed separately for different species. As shown in Table 3, P. mirabilis and P. stuartii (both of which were found to produce almost exclusively TEM-derived enzymes) showed very similar resistance phenotypes, characterized by low MICs of aztreonam with elevated MICs of imipenem (presumably, a property of the involved species). However, for P. stuartii, the drug MICs at which 90% of the organisms were inhibited (MIC90s) were increased for ceftriaxone, ceftazidime, cefepime, and cefoxitin compared to results with P. mirabilis.
TABLE 3.
Susceptibilities to beta-lactams distributed by species and ESBL gene typea
| Species and ESBL type | No. of isolates | MIC50/MIC90 of beta-lactam (μg/ml)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Piperacillin | Cephalothin | Ceftriaxone | Cefotaxime | Ceftazidime | Cefepime | Cefoxitin | Aztreonam | Imipenem | ||
| P. mirabilis | ||||||||||
| TEM+ | 127 | >256/>256 | >256/>256 | 4/8 | 8/16 | 4/16 | 4/16 | 4/8 | 0.5/2 | 1/2 |
| P. stuartii | ||||||||||
| TEM+ | 22 | >256/>256 | >256/>256 | 4/32 | 8/16 | 32/>256 | 2/>32 | 2/128 | 4/16 | 1/2 |
| K. pneumoniae | ||||||||||
| TEM+ | 32 | >256/>256 | >256/>256 | 4/32 | 4/32 | >256/>256 | 2/8 | 2/16 | 32/>256 | 0.25/0.5 |
| SHV+ | 104 | >256/>256 | >256/>256 | 8/32 | 8/32 | >256/>256 | 2/16 | 2/16 | >256/>256 | 0.25/0.5 |
| TEM+ and SHV+ | 34 | >256/>256 | >256/>256 | 8/32 | 8/32 | >256/>256 | 2/16 | 2/16 | >256/>256 | 0.25/0.25 |
| Non-TEM/non-SHV | 18 | >256/>256 | >256/>256 | 8/>256 | 8/32 | >256/>256 | 2/8 | 4/16 | >256/>256 | 0.25/0.5 |
| E. coli | ||||||||||
| TEM+ | 11 | >256/>256 | >256/>256 | 4/32 | 8/16 | 16/128 | 4/16 | 2/4 | 4/8 | 0.25/0.25 |
| SHV+ | 28 | >256/>256 | >256/>256 | 4/32 | 4/32 | 32/>256 | 1/8 | 2/4 | 16/>256 | 0.25/0.25 |
| E. aerogenes | ||||||||||
| TEM+ | 11 | >256/>256 | >256/>256 | 8/16 | 8/16 | >256/>256 | 1/2 | >256/>256 | 16/32 | 0.25/0.25 |
| SHV+ | 14 | >256/>256 | >256/>256 | 16/64 | 16/32 | >256/>256 | 2/16 | >256/>256 | 64/>256 | 0.25/0.5 |
MICs were measured by the quantitative E-test assay. Values are not reported when fewer than 10 isolates were available.
Species that showed the most frequent production of SHV-derived enzymes (K. pneumoniae and E. coli) were characterized by elevated MICs of ceftazidime and aztreonam, with low MICs of imipenem. It is noteworthy that for TEM-producing E. coli isolates, MIC90s of aztreonam were reduced compared to results with isolates of the same species producing SHV-derived enzymes. In general, the resistance pattern of isolates producing TEM plus SHV or non-TEM, non-SHV enzymes was comparable to that of SHV-positive isolates.
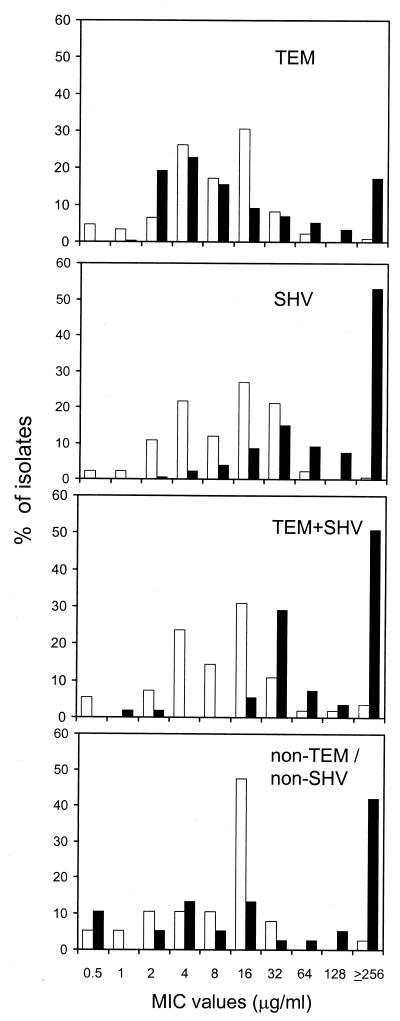
The frequency distribution for MICs of ceftazidime and cefotaxime was analyzed in more detail, due to their clinical relevance and to the fact that they represent good ESBL substrates. As shown in Fig. 1, most of the TEM-producing isolates (58%) were characterized by low MICs of both drugs (i.e., ≤8 μg/ml). In contrast, for isolates producing either SHV, TEM plus SHV, or non-TEM, non-SHV enzymes, drug MICs were high, particularly with ceftazidime.
FIG. 1.
ESBL-positive Enterobacteriaceae: frequency distribution of MICs of cefotaxime (□) and ceftazidime (▪). Results obtained from isolates carrying different ESBL gene types are shown in separate panels. Numbers of isolates were as follows: TEM type, n = 234; SHV type, n = 173; TEM and SHV type, n = 55; non-TEM-non-SHV, n = 38.
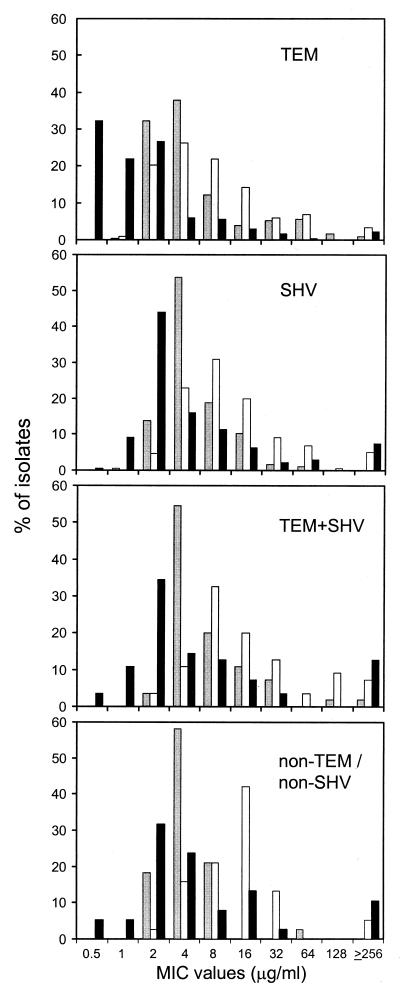
Figure 2 shows the frequency distribution of MICs of amoxicillin-clavulanate (susceptibility breakpoint, 8/4 μg/ml), ampicillin-sulbactam (breakpoint, 8/4 μg/ml), and piperacillin-tazobactam (breakpoint, 16/4 μg/ml). TEM-positive isolates were characterized by low MICs of β-lactam-β-lactamase inhibitor combinations, with 95.6% of them being susceptible to piperacillin-tazobactam. Amoxicillin-clavulanate and piperacillin-tazobactam had comparable MICs against isolates producing SHV, TEM plus SHV, and non-TEM, non-SHV enzymes. Ampicillin-sulbactam was the least active compound against ESBL-producing isolates (TEM+, 69.5%; SHV+, 58.4%; TEM+ SHV+, 47.2%; non-TEM, non-SHV, 39.5%).
FIG. 2.
ESBL-positive Enterobacteriaceae: frequency distribution of MICs of amoxicillin-clavulanate (gray bars), ampicillin-sulbactam (white bars), and piperacillin-tazobactam (black bars). Results obtained from isolates carrying different ESBL gene types are shown in separate panels. See Fig. 1 for numbers of isolates.
MICs of aminoglycosides and ciprofloxacin, evaluated by the E-test.
Susceptibility to clinically useful drugs other than β-lactams has been measured by the E-test. Results are listed by species and ESBL gene type (Table 4). For all species, MIC90s of ciprofloxacin were elevated. Interestingly, high MIC50s were found for P. mirabilis, P. stuartii, and E. aerogenes, indicating that in these species, resistance to fluoroquinolones is strongly associated with ESBL production.
TABLE 4.
Susceptibilities to drugs other than beta-lactams distributed by species and ESBL gene typea
| Species and ESBL type | No. of isolates | MIC50/MIC90 of drug (μg/ml)
|
||
|---|---|---|---|---|
| Gentamicin | Amikacin | Ciprofloxacin | ||
| P. mirabilis | ||||
| TEM+ | 127 | >256/>256 | 4/128 | 16/>32 |
| P. stuartii | ||||
| TEM+ | 22 | 4/64 | 16/64 | >32/>32 |
| K. pneumoniae | ||||
| TEM+ | 32 | 1/64 | 2/32 | ≤0,125/>32 |
| SHV+ | 104 | 2/16 | 2/32 | ≤0,125/2 |
| TEM+ and SHV+ | 34 | 2/32 | 4/32 | ≤0,125/>32 |
| Non-TEM/non-SHV | 18 | 4/32 | 1/4 | ≤0,125/4 |
| E. coli | ||||
| TEM+ | 11 | 4/>256 | 2/8 | 0,25/>32 |
| SHV+ | 28 | 2/16 | 2/32 | ≤0,125/16 |
| E. aerogenes | ||||
| TEM+ | 11 | 1/16 | 64/64 | >32/>32 |
| SHV+ | 14 | 2/64 | 8/16 | 2/>32 |
MICs were measured by the quantitative E-test assay. Values are not reported when less than 10 isolates were available.
On the whole, for all species, MIC90s of both gentamicin and amikacin were high, independently of the type of ESBL produced. In contrast, MIC50s were usually within the sensitivity range. Two main exceptions, however, occurred: 72% of P. mirabilis isolates were resistant to gentamicin, and more than 90% of E. coli isolates remained susceptible to amikacin.
Susceptibilities of ESBL-positive isolates to potentially active drugs.
Drugs potentially active against ESBL-positive Enterobacteriaceae include β-lactam-β-lactamase inhibitor combinations, cephamycins, carbapenems, aminoglycosides, and fluoroquinolones (Table 5). MICs have been interpreted according to the NCCLS (23). All isolates were susceptible to imipenem with the exception of a TEM-positive isolate; the above strain was identified as P. mirabilis, for which the MIC was 8 μg/ml, and was then classified as intermediate. Though cefoxitin is not hydrolyzed by ESBLs, in vitro it was active only against 78% of strains, possibly due to overexpression of chromosomal AmpC β-lactamase. Among combinations of β-lactams and β-lactamase inhibitors, amoxicillin-clavulanate and piperacillin-tazobactam were highly active (85 and 91%, respectively) against ESBL-producing Enterobacteriaceae, whereas only 61% of strains were susceptible to ampicillin-sulbactam. Interestingly, compared to strains producing only one type of ESBL, isolates producing both TEM- and SHV-derived enzymes were characterized by the highest degree of resistance to β-lactam-β-lactamase inhibitor combinations. Associated resistance to aminoglycosides and ciprofloxacin was observed most frequently among TEM-positive strains. Based on in vitro results, only 56 and 58% of ESBL-positive strains were susceptible to gentamicin and ciprofloxacin, respectively. Amikacin, however, remained active against 76% of ESBL-producing Enterobacteriaceae.
TABLE 5.
Susceptibilities to potentially active drugs of all isolates of ESBL-positive Enterobacteriaceaea
| ESBL type | No. of isolates | % Susceptibility to drug
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Cefoxitin | Amoxicillin plus clavulanate | Piperacillin plus tazobactam | Ampicillin plus sulbactam | Gentamicin | Amikacin | Ciprofloxacin | ||
| TEM+ | 234 | 99.6 | 79 | 82 | 95 | 69 | 45 | 68 | 32 |
| SHV+ | 173 | 100 | 79 | 88 | 88 | 59 | 66 | 86 | 85 |
| TEM+ and SHV+ | 55 | 100 | 75 | 78 | 84 | 47 | 71 | 69 | 76 |
| Non-TEM/non-SHV | 38 | 100 | 76 | 97 | 89 | 39 | 55 | 95 | 71 |
| Total | 500 | 99.8 | 78 | 85 | 91 | 61 | 56 | 76 | 58 |
MICs have been interpreted according to current NCCLS criteria (23).
DISCUSSION
Over the last decade, several studies assessed the occurrence of ESBLs among Enterobacteriaceae recovered from hospitalized patients. A recent survey in Brooklyn showed that ESBLs were produced by 17.2% of selected bacteria (K. pneumoniae, P. mirabilis, and E. coli) (31). In contrast, a wider French investigation revealed that the overall prevalence of ESBL-producing Enterobacteriaceae was 3.2% (8); these results were in agreement with Japanese data showing that ESBLs were expressed by less than 10% of isolates of E. coli and Klebsiella spp. (17).
The results of this wide Italian survey with 8,015 isolates obtained from 10 important medical centers located through the entire nation indicated that 6.3% of Enterobacteriaceae harbor ESBL genes. The prevalence and type of ESBLs varied according to species, with prevalences of 28.1% for P. stuartii, 20% for K. pneumoniae, 16.3% for P. mirabilis, and 1.2% for E. coli. However, it should be noted that while P. stuartii isolates derived almost exclusively from two centers, the other species were found at comparable frequencies in all centers.
Among ESBL-positive strains, the prevalence of TEM-type enzymes was higher than that of SHV-type enzymes; in particular, P. mirabilis coded almost exclusively for TEM-derived enzymes, but non-TEM, non-SHV genes were also found in two isolates. The observation that this species does express mainly TEM-type enzymes may have significant therapeutic implications, since P. mirabilis has been recognized as an important cause of morbidity in hospitalized patients (9).
Eight percent of strains harbored ESBLs other than TEM- or SHV-derived enzymes; among these, K. pneumoniae was particularly notable. This epidemiological finding confirms the relevance recently given to the emerging problem of non-TEM, non-SHV enzymes that are spreading worldwide (4, 5, 28). Though many of these unusual enzymes have been detected only in occasional isolates (SFO-1, TLA-1, VEB-1, and BES-1), enzymes of the CTX-M family have been reported from Europe, South America, and Asia (4), and PER-type enzymes have been found in Turkey, France, Italy, and Argentina (3, 25).
Two features of the present survey are worthy of note: (i) the expression of TEM- and/or SHV-type enzymes is frequently associated with resistance to aminoglycosides and fluoroquinolones; and (ii) most ESBL-positive strains remained susceptible in vitro to β-lactams combined with either clavulanate, sulbactam, or tazobactam.
Hospital outbreaks of ESBL-producing Enterobacteriaceae have been observed at an increasing frequency over the last few years (27, 29), with responsible strains often characterized by resistance to multiple drugs, including ciprofloxacin, gentamicin, and aminoglycosides (12, 21, 24). Our survey confirms and extends the above-cited reports, showing a marked association between ESBL production and resistance to ciprofloxacin, especially for P. mirabilis, P. stuartii, and E. aerogenes. On the whole, resistance to aminoglycosides did not appear to be associated with the type of produced enzyme(s). Association, however, depends on the location of ESBL genes on integrons containing promoters for the coordinated expression of multiple resistance gene cassettes (28).
From the clinical point of view, it should be considered that no precise association can be found between the ESBL type produced and susceptibility to different β-lactams, since susceptibility is multifactorial, depending on ESBL substrate specificity, production of additional β-lactamases, and changes of outer membrane permeability (6, 18, 22). In our study, ESBL-producing Enterobacteriaceae maintained susceptibility to imipenem; on the contrary, less than 80% were susceptible to cefoxitin. Our data indicate that a valuable option for treatment is represented by amikacin, a bactericidal drug effective against 76% of strains. In addition, β-lactam-β-lactamase inhibitor combinations remained quite active against most isolates, with the exception of ampicillin-sulbactam. The reduced activity of the latter combination against E. coli in hospitalized patients has already been reported (15).
The present survey on Enterobacteriaceae assesses, for the first time, the breadth of the ESBL problem in Italy by using classical bacteriological methods and molecular techniques. The finding of ESBL-associated resistance traits supports the notion that curing and eradicating infections caused by ESBL-producing Enterobacteriaceae cannot be done with disregard for laboratory data. As previously proposed (1), the results of our survey suggest the judicious use of piperacillin-tazobactam or amoxicillin-clavulanate either alone or together with amikacin. Systematic carbapenem therapy alone, in fact, may favor the selection of Stenotrophomonas maltophilia (a species naturally resistant to these drugs) and other nonfermenting gram-negative rods. In the Mediterranean area, strains of P. aeruginosa and P. putida encoding novel metallo-β-lactamases have already emerged (30, 35).
Acknowledgments
This work was partially supported by grants from the Italian Ministry for the University and Scientific Research (COFIN-1999 and FAR-1999) and the Italian Ministry of Health.
We thank Wyeth-Lederle Italia for its generous support.
REFERENCES
- 1.Amyes, S. G. B., and R. S. Miles. 1999. Extended-spectrum β-lactamases: the role of inhibitors in therapy. J. Antimicrob. Chemother. 42:415–417. [DOI] [PubMed] [Google Scholar]
- 2.Babini, G., and D. M. Livermore. 2000. Antimicrobial resistance amongst Klebsiella spp. from intensive care units in southern and western Europe in 1997–1998. J. Antimicrob. Chemother. 45:183–189. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, Ö. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R., J. L. M. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R., J. L. M. Sampaio, C. Chanal, D. Sirot, C. De Champs, J. L. Viallard, R. Labia, and J. Sirot. 2000. A novel class A extended-spectrum β-lactamase (BES-1) in Serratia marcescens isolated in Brazil. Antimicrob. Agents Chemother. 44:3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structures. Antimicrob. Agents Chemother. 39:1211–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coudron, P. E., E. S. Molland, and C. C. Sanders. 1997. Occurrence and detection of extended-spectrum β-lactamases in members of the family Enterobacteriaceae at a Veterans Medical Center: seek and you may find. J. Clin. Microbiol. 35:2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and The French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Champs, C., R. Bonnet, D. Sirot, C. Chanal, and J. Sirot. 2000. Clinical relevance of Proteus mirabilis in hospital patients: a two-year survey. J. Antimicrob. Chemother. 45:537–539. [DOI] [PubMed] [Google Scholar]
- 10.Emery, C. L., and L. A. Weymouth. 1997. Detection and clinical significance of extended-spectrum β-lactamases in a tertiary-care medical center. J. Clin. Microbiol. 35:2061–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franceschini, N., M. Perilli, B. Segatore, D. Setacci, G. Amicosante, A. Mazzariol, and G. Cornaglia. 1998. Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived extended spectrum beta-lactamase, TEM-60. Antimicrob. Agents Chemother. 42:1459–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grogan, J., H. Murphy, and K. Butler. 1998. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a Dublin paediatric hospital. Br. J. Biomed. Sci. 55:111–117. [PubMed] [Google Scholar]
- 13.Jacoby, G. A., and A. A. Medeiros. 1991. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:1697–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarlier, V., M.-H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867–878. [DOI] [PubMed] [Google Scholar]
- 15.Kaye, K. S., A. D. Harris, H. Gold, and Y. Carmeli. 2000. Risk factors for recovery of ampicillin-sulbactam-resistant Escherichia coli in hospitalized patients. Antimicrob. Agents Chemother. 44:1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaye, K. S., H. S. Fraimow, and E. Abrutyn. 2000. Pathogens resistant to antimicrobial agents. Epidemiology, molecular mechanisms, and clinical management. Infect. Dis. Clin. North Am. 14:293–319. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, M. T., K. Yamaguchi, D. J. Biedenbach, and R. N. Jones. 1999. In vitro evaluation of cefepime and other broad-spectrum β-lactams in 22 medical centers in Japan: a phase II trial comparing two annual organism samples. The Japan Antimicrobial Resistance Study Group. Diagn. Microbiol. Infect. Dis. 35:307–315. [DOI] [PubMed] [Google Scholar]
- 18.Livermore, D. M. 1995. β-lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore, D. M. 1998. β-lactamase-mediated resistance and opportunities for its control. J. Antimicrob. Chemother. 41(Suppl. D):25–41. [DOI] [PubMed] [Google Scholar]
- 20.Luzzaro, F., M. Perilli, R. Migliavacca, G. Lombardi, P. Micheletti, A. Agodi, S. Stefani, G. Amicosante, and L. Pagani. 1998. Repeated epidemics caused by extended-spectrum beta-lactamase-producing Serratia marcescens strains. Eur. J. Clin. Microbiol. Infect. Dis. 17:629–636. [DOI] [PubMed] [Google Scholar]
- 21.Luzzaro, F., M. Perilli, G. Amicosante, G. Lombardi, R. Belloni, A. Zollo, C. Bianchi, and A. Toniolo. 2001. Properties of multidrug-resistant, ESBL-producing Proteus mirabilis isolates and possible role of β-lactam/β-lactamase inhibitor combinations. Int. J. Antimicrob. Agents 17:131–135. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Martinez, L., A. Pascual, S. Hernandez-Alles, D. Alvarez-Diaz, A. I. Suarez, J. Tran, V. J. Benedi, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7–A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Paterson, D. L., L. Mulazimoglou, J. M. Casellas, W. Ko, H. Goossens, A. Von Gottberg, S. Mohapatra, G. M. Trenholme, K. P. Klugman, J. G. McCormack, and V. L. Yu. 2000. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin. Infect. Dis. 30:473–478. [DOI] [PubMed] [Google Scholar]
- 25.Pereira, M., M. Perilli, E. Mantengoli, F. Luzzaro, A. Toniolo, G. M. Rossolini, and G. Amicosante. 2000. PER-1 extended spectrum β-lactamase production in an Alcaligenes faecalis clinical isolate resistant to expanded-spectrum cephalosporins and monobactams from a hospital in northern Italy. Microb. Drug Res. 6:85–90. [DOI] [PubMed] [Google Scholar]
- 26.Perilli, M., B. Segatore, M. R. De Massis, M. L. Riccio, C. Bianchi, A. Zollo, G. M. Rossolini, and G. Amicosante. 2000. TEM-72, a new extended-spectrum beta-lactamase detected in Proteus mirabilis and Morganella morganii in Italy. Antimicrob. Agents Chemother. 44:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. 2Clin. Microbiol. Rev. 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel, L., I. Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analysis of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahal, J. J. 2000. Extended-spectrum β-lactamases: how big is the problem? Clin. Microbiol. Infect. 6(Suppl. 2):2–6. [DOI] [PubMed] [Google Scholar]
- 30.Rossolini, G. M., M. L. Riccio, G. Cornaglia, L. Pagani, C. Lagatolla, L. Selan, and R. Fontana. 2000. Carbapenem-resistant Pseudomonas aeruginosa with acquired blaVIM metallo-β-lactamase determinants, Italy. Emerg. Infect. Dis. 6:312–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saurina, G., J. M. Quale, V. M. Manikal, E. Oydna, and D. Landman. 2000. Antimicrobial resistance in Enterobacteriaceae in Brooklyn, NY: epidemiology and relation to antibiotic usage patterns. J. Antimicrob. Chemother. 45:895–898. [DOI] [PubMed] [Google Scholar]
- 32.Schiappa, D. A., M. K. Haiden, M. G. Matushek, F. N. Hashemi, J. Sullivan, K. Y. Miyashiro, J. P. Quinn, R. A. Weinstein, and G. M. Trenholme. 1996. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli bloodstream infection: a case-control and molecular epidemiologic investigation. J. Infect. Dis. 174:529–536. [DOI] [PubMed] [Google Scholar]
- 33.Shen, D., D. J. Biedenbach, P. L. Winokur, M. A. Pfaller, and R. N. Jones. 1999. Phenotypic and genotypic characterizations of Chinese strains of Escherichia coli producing extended-spectrum β-lactamases. Diagn. Microbiol. Infect. Dis. 34:159–164. [DOI] [PubMed] [Google Scholar]
- 34.Tenover, F. C., M. J. Mohammed, T. S. Gorton, and Z. F. Dembek. 1999. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J. Clin. Microbiol. 37:4065–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsakris, A., S. Pournaras, N. Woodford, M.-F. I. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzelepi, E., G. Panagiota, D. Sofianou, V. Loukova, A. Kemerglou, and A. Tsakris. 2000. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J. Clin. Microbiol. 38:542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]