Abstract
A vancomycin-resistant Staphylococcus aureus mutant, COL-VR1 (MIC, 16 μg/ml), was isolated from methicillin-resistant S. aureus COL by exposure to vancomycin. COL-VR1 also showed decreased susceptibility to teicoplanin (8-fold), methicillin (2-fold), macarbomycin (8-fold), and moenomycin (16-fold). Macarbomycin and moenomycin are thought to directly inhibit transglycosylase activity. Characterization of the mutant revealed a thickened cell wall and suppression of penicillin-induced lysis, although the amounts of the five penicillin-binding proteins (PBPs 1, 2, 3, 4, and 2′) and the profiles of peptidoglycan hydrolases were not altered. Analysis of muropeptide profile and glycan chain length distribution by reversed-phase high-pressure liquid chromatography revealed slightly decreased peptide cross-linking and an increased average glycan chain length compared to those of the parent. These results together suggest that a transglycosylase activity was enhanced in the mutant. This may represent a novel mechanism of glycopeptide resistance in S. aureus.
The glycopeptide antibiotics vancomycin and teicoplanin are frequently used for the treatment of methicillin-resistant Staphylococcus aureus infections. Since the emergence of a clinically vancomycin-resistant S. aureus (VRSA) strain (strain Mu50) was reported in Japan (11), several reports have been conducted about similar glycopeptide-intermediate-susceptible S. aureus (GISA) strains in the United States and Europe (3, 27, 30), causing great concern about the possibility of losing the last resort for the treatment of multidrug-resistant staphylococci. The mechanism(s) of vancomycin and/or teicoplanin resistance has been the subject of intense research with clinical strains or laboratory mutants of GISA or glycopeptide-resistant S. aureus. On the basis of those investigations, it is generally thought that vancomycin resistance in S. aureus is mediated by the accumulation of surplus cell wall material, reduced peptidoglycan cross-linking, and/or other cell wall alterations, which lead to an increased but nonproductive level of drug binding to the d-alanyl-d-alanine of part of the peptidoglycan moiety, thereby decreasing the amount of glycopeptide capable of binding to the lipid-linked peptidoglycan precursors at the outside of the cytoplasmic membrane (8, 11, 26), which appears to provide the real glycopeptide antibiotic target (1), although the precise mechanism is not fully understood.
To date, several factors correlated with glycopeptide resistance in individual strains have been reported including PBP 2 overexpression (19, 25), pbpD inactivation (26), and the enhancement of glutamine synthetase and l-glutamine d-fructose-6-phosphate aminotransferase activities (8, 10). Furthermore, sigB (2), the tcaR-B operon (7), and ddh (5, 18) were genetically identified as factors involved in glycopeptide resistance. To investigate such factors involved in glycopeptide resistance, one approach is the isolation of glycopeptide-resistant mutants from sensitive strains by serial passage because clinical isolates of VRSA strains seem to possess multiple mutations, rendering them difficult to analyze. Such experimentally created mutants have the additional advantage that they are amenable for use for analysis of the mutation site(s) and the phenotypic consequence(s) of the mutation by direct comparison with the parent strain; a potential drawback of this strategy is, however, that such experimental mutations may not be relevant for the clinical situation.
Here, we describe the isolation of a glycopeptide-resistant mutant (MIC, 16 μg/ml) from methicillin-resistant S. aureus parent strain COL after exposure to vancomycin. Characterization of the mutant led us to propose a novel potential mechanism of glycopeptide resistance in S. aureus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in the present study are listed in Table 1. Escherichia coli and S. aureus were grown at 37°C in Luria-Bertani broth and Trypticase soy broth (TSB), respectively. When needed, ampicillin (100 μg/ml) was added to the medium.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Characteristics |
|---|---|
| S. aureus | |
| COL | Laboratory strain, Mcr |
| COL-VR1 | Vancomycin-resistant mutant from COL |
| E. coli | |
| HK4457 | E. coli BL21/pHK4457 |
| HK4386 | E. coli BL21/pHK4386 |
| HK4476 | E. coli M15(pREP4)/pHK4476 |
| Plasmids | |
| pHK4457 | pbpA gene cloned into pGEX-3T vector |
| pHK4386 | pbpD gene cloned into FLAG-MAC vector |
| pHK4476 | mecA gene cloned into pQE30 vector |
Isolation of vancomycin-resistant mutant.
MRSA strain COL was used to isolate spontaneous vancomycin-resistant mutants. An overnight culture of COL was plated on Trypticase soy agar (TSA) containing 8 μg of vancomycin per ml. Several colonies which grew on the plate were replated on TSA containing 8 μg of vancomycin per ml, and this procedure was repeated three times. Finally, 20 colonies growing on the TSA plate containing vancomycin (8 μg/ml) were picked and their vancomycin susceptibilities were measured. The stability of the mutants was checked by measuring the MIC of vancomycin after several passages without antibiotic selection pressure.
MIC determination and population analysis.
The MICs of various antibiotics were determined by the microdilution method as described elsewhere (15). Population analysis was also performed as described previously (14). Oxacillin, methicillin, bacitracin, and vancomycin (all from Sigma Chemical Co., St. Louis, Mo.) and fosfomycin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) were commercially available. Macarbomycin and moenomycin were provided by Meiji Seika, Ltd. (Tokyo, Japan) and Aventis Pharma (Frankfurt, Germany), respectively.
Electron microscopy.
Thin-section electron microscopy was performed for morphological characterization of the cell wall ultrastructure. Additionally, to detect and further characterize alterations of the cell surface, wheat germ agglutinin (WGA)-gold, which binds to peptidoglycan, was used to treat the cells before preparation for thin-section electron microscopy. WGA is a lectin which binds specifically to N-acetylglucosamine residues on the surfaces of S. aureus cells (35). S. aureus COL and the isogenic VRSA cells were harvested during the logarithmic growth phase, washed twice with Dulbecco’s phosphate-buffered saline (PBS), and suspended in PBS. The bacterial cell suspension was mixed with the suspension of WGA-gold (diameter, 5 nm; EY Laboratories, Inc., San Mateo, Calif.). The mixture was incubated for 90 min at room temperature and washed twice with PBS. After centrifugation, the pellet was doubly fixed with 2.5% glutaraldehyde and 1% OsO4. The samples were dehydrated in a series of ethanol concentrations and then embedded in Spurr’s Epon. Thin sections were cut on an ultramicrotome with a diamond knife and were examined in a JOEL JEM-2000 EXII electron microscope at 80 or 100 kV.
Cell wall analysis by HPLC. (i) Muropeptide analysis.
Preparation of murein and enzymatic digestion of muropeptides were carried out as described elsewhere (24, 31). The digested muropeptides were fractionated by reversed-phase high-pressure liquid chromatography (HPLC) as described elsewhere (24, 31).
(ii) Glycan chain analysis.
Glycan chain analysis was performed by the method described above for muropeptide analysis, except that the cell wall was digested with lysostaphin instead of mutanolysin, as described elsewhere (26). For control purposes, some samples were also digested with both enzymes.
Characterization of the mutant. (i) Growth curves.
Overnight cultures (100 μl) of COL or COL-VR1 were inoculated into 10 ml of TSB. When the cultures reached an optical density at 660 nm (OD660) of 0.3, various concentrations of vancomycin were added to the medium. The OD660 was monitored for 8 h after the addition of vancomycin.
(ii) Autolysis.
The autolysis rates of COL and COL-VR1 were monitored by a method described elsewhere (15).
(iii) Susceptibility to lysostaphin.
The susceptibility to lysostaphin was measured by a zymographic assay, as described elsewhere (21).
(iv) Zymography.
Using heat-killed S. aureus or Micrococcus luteus cells as a substrate, the profiles of bacteriolytic enzymes were analyzed by a method described elsewhere (32).
PBP analysis. (i) PBP fluorography.
Penicillin-binding proteins (PBPs) labeled with [3H]benzylpenicillin (10 to 30 Ci/mmol; Amersham Pharmacia, Tokyo, Japan) were detected as described elsewhere (13).
(ii) Western blotting analysis of PBPs 1, 2, 2′, 3, and 4.
Since several reports indicated distinct patterns of expression of PBP 2 or of the PBP 2 complex in GISA strains, we compared the levels of expression of PBPs 1, 2, 2′, 3, and 4 between COL and COL-VR1. Serum with anti-PBP 2 antibodies was kindly provided by K. Murakami (20), and serum with anti-PBP 2B antibodies was obtained as described elsewhere (13). Pinho et al. (23) demonstrated that PBP 2B was identical to PBP 3. For recombinant PBP 1 (rPBP 1) purification, a DNA fragment amplified with two primers (5′-GGTCAAGGATCCGTCATGAA-3′ and 5′-CAACAAGGATCCAATGAA-3′) with RN450 chromosomal DNA as a template was cloned into vector pGEX-2T (Amersham Pharmacia Biotech, Tokyo, Japan) and was then electroporated into E. coli BL21 to generate HK4457. For rPBP 4 purification, a DNA fragment amplified with two primers (5′-CAAAGCTTACTAACAGTGACGTAACC-3′ and 5′-GGAGATCTTTCCCACATACTTTTAGC-3′) from RN450 chromosomal DNA was cloned into the pFLAG-MAC vector (Eastman Kodak Company, New Haven, Conn.) and was then electroporated into BL21 to generate HK4386. For rPBP 2′ purification, a DNA fragment amplified with two primers (5′-ATGGATCCATTGATGCAATTGAAGAT-3′ and 5′-AAAAGCTTAAGCAACCATCGTTACGG-3′) from COL chromosomal DNA was cloned into vector pQE30 (QIAGEN, Tokyo, Japan) and was then electroporated into E. coli M15(pREP4) to generate HK4457. Glutathione S-transferase-rPBP 1, FLAG-rPBP4, and His-tag-rPBP 2′ were purified according to the protocols of the respective manufacturers. Serum with anti-PBP 1, anti-PBP 2′, and anti-PBP 4 antibodies were obtained by immunizing rabbits with each of the recombinant proteins by a previously described method (12).
RESULTS
Isolation of a mutant with reduced susceptibility to vancomycin.
By applying the selection procedure described in Materials and Methods, a strain for which the vancomycin MIC was elevated (16 μg/ml) compared to that for the parent strain (2 μg/ml) was isolated and was designated COL-VR1. The vancomycin MIC for COL-VR1 was stable after seven passages (one passage per day). The results of a population analysis of methicillin- or vancomycin-treated COL-VR1 and its parent are shown in Fig. 1. The population distribution curve of COL-VR1 was shifted to higher vancomycin concentrations compared to that of COL. Interestingly, the corresponding curve for methicillin showed a similar, albeit smaller shift to higher methicillin concentrations for COL-VR1. The MICs of various cell wall-active antibiotics for both strains are shown in Table 2. The MICs of oxacillin, methicillin, and imipenem for COL-VR1 were twofold higher than those for parent strain COL. The MICs of the glycopeptide antibiotics vancomycin and teicoplanin for COL-VR1 were fourfold and eightfold higher than those for COL, respectively. There were no differences in the MICs of bacitracin and fosfomycin for the mutant and the parent. It is noteworthy that the MICs of macarbomycin and moenomycin for COL-VR1 turned out to be 8- and 16-fold higher than those for COL, respectively.
FIG. 1.
Population analysis of COL (closed symbols) and COL-VR1 (open symbols). S. aureus cells cultured overnight were plated on TSA containing serial dilutions of methicillin (squares) or vancomycin (circles).
TABLE 2.
MICs of various antibiotics for COL and COL-VR1
| Strain | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MET | OXA | VAN | TEC | MM | MB | FOF | BC | |
| COL | 1,024 | 512 | 2 | 1 | 0.01 | 0.5 | 1,024 | 64 |
| COL-VR1 | 2,048 | 1,024 | 16 | 8 | 0.125 | 4 | 1,024 | 64 |
Abbreviations: MET; methicillin, OXA; oxacillin, VAN; vancomycin, TEC; teicoplanin, MM; moenomycin, MB; macarbomycin, FOF; fosfomycin, BC; bacitracin.
Growth curves.
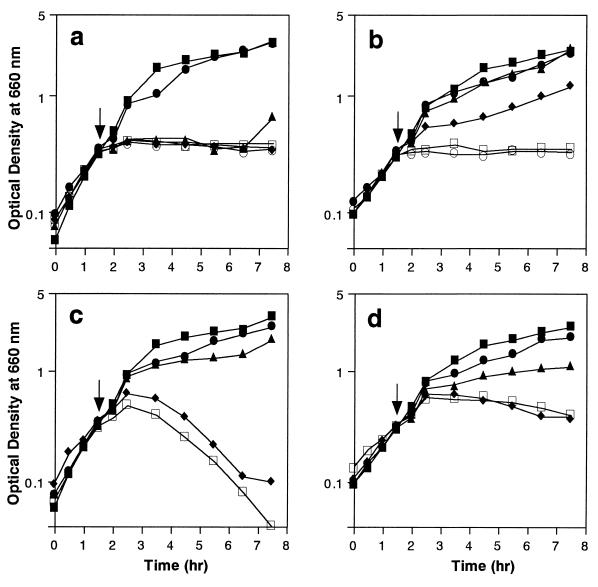
Growth curves are shown in Fig. 2. In the absence of antibiotics, the growth curves of COL and COL-VR1 were similar, although the growth rate was slightly lower for COL-VR1. In the presence of vancomycin, the growth of COL was inhibited at vancomycin concentrations above 4 μg/ml (two times the MIC for COL), while the growth of COL-VR1 was inhibited by concentrations above 16 μg/ml (the MIC for COL-VR1). However, vancomycin did not induce detectable cell lysis in either of the strains. In the presence of methicillin, the growth of both strains was inhibited only at concentrations above 1,024 μg/ml. Methicillin-induced lysis was clearly seen in the case of COL, but it was strongly reduced in the case of COL-VR1.
FIG. 2.
Growth curves of COL (a and c) and COL-VR1 (b and d) in the presence of methicillin (c and d) or vancomycin (a and b). A small portion of S. aureus cells cultured overnight was inoculated into fresh TSB and incubated at 37°C with shaking. Antibiotics were added at the times indicated by the arrows. The OD660 was monitored at 30-min intervals. Symbols: for panels a and b, ▪, control; •, vancomycin at 2 μg/ml; ▴, vancomycin at 4 μg/ml; ⧫, vancomycin at 8 μg/ml; □, vancomycin at 16 μg/ml; ○, vancomycin at 32 μg/ml; for panels c and d, ▪, control; •, methicillin at 10 μg/ml; ▴, methicillin at 100 μg/ml; ⧫, methicillin at 1,024 μg/ml; □, methicillin at 2,048 μg/ml.
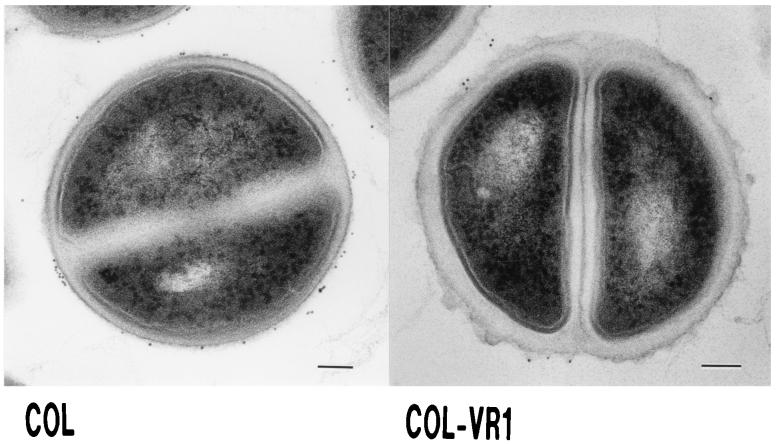
Electron microscopic features of the mutant.
The cell wall of COL-VR1 was clearly thickened compared with that of the parent (Fig. 3). COL-VR1 had a rough surface compared to that of the parent. Examination of specimens after WGA-gold binding to the cell wall revealed no significant differences between the parent and the mutant strains.
FIG. 3.
Thin sections of S. aureus COL and COL-VR1. Exponentially growing cultures of S. aureus cells were prepared for electron microscopy as described in Materials and Methods. Bars, 100 nm.
Peptidoglycan analysis.
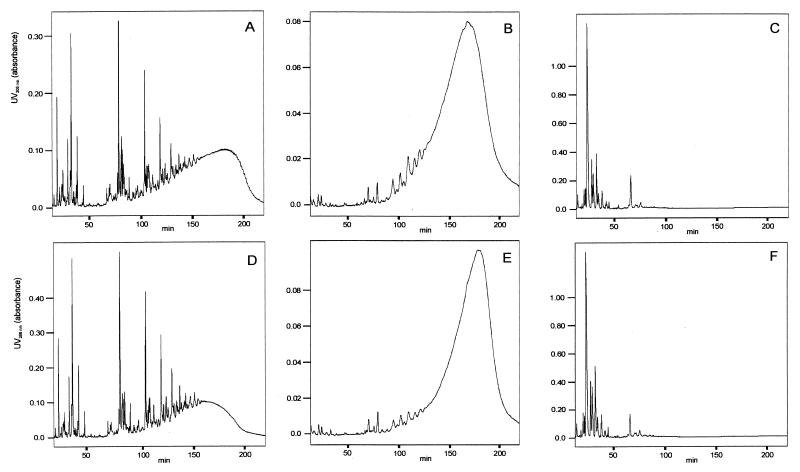
The muropeptide profile of COL obtained after digestion of isolated peptidoglycan with the muramidase mutanolysin was very similar to those of other wild-type strains described earlier (24). Comparison of the muropeptide profiles of COL and COL-VR1 revealed almost no differences (Fig. 4). Just the degree of peptide cross-linking was slightly lower for COL-VR1 than for COL, as can be judged from the bump of unresolved highly cross-linked muropeptides at long retention times. The HPLC elution profile of peptidoglycan digested with the endopeptidase lysostaphin instead of mutanolysin was recorded to analyze the peptidoglycan sugar chain length distribution, as described by Sieradzki et al. (26). The sugar chain length distribution obtained was clearly shifted to a longer average chain length for COL-VR1 compared to that for COL, as evident from the shift of the eluted broad peak toward longer retention times (Fig. 4). As a control, the HPLC profiles of peptidoglycan after double digestion with both enzymes, mutanolysin and lysostaphin, were recorded and no high-molecular-weight material was detected, indicating that the digestions were complete under our conditions.
FIG. 4.
HPLC analysis of peptidoglycan. Peptidoglycan was digested with mutanolysin (A and D) or lysostaphin (B and E), or both (C and F), and was subjected to HPLC. Data for COL are depicted in panels A, B, and C; and those for COL-VR1 are shown in panels D, E, and F.
Further characterization of the mutant.
Western blotting analysis of all PBPs with the respective antisera revealed no significant differences in the amounts of each of the PBPs between the two strains (Fig. 5a). Zymographic analysis with a gel with either M. luteus or S. aureus gave no hints about significant differences in autolysin quantity or specificity (Fig. 5b1 and b2); close inspection of the zymograms indicated that one of the bands had a somewhat lower lytic capacity in the case of COL-VR1, namely, the second lysis band, which is known to be related to the atl gene product. Other characterizations, including the autolysis rate, cell wall susceptibility to lysostaphin, and PBP fluorography, showed no significant differences between COL and COL-VR1 (data not shown).
FIG. 5.
Western blotting and zymographic analyses of S. aureus. (a) Membrane fractions of COL and COL-VR1 were analyzed by immunoblotting with serum with anti-PBP 1, anti-PBP 2, anti-PBP 3, anti-PBP 4, and anti-PBP 2′ antibodies. (b) Lysates of whole COL and COL-VR1 cells were subjected to zymography with a gel with M. luteus (b1) or a gel with S. aureus (b2). Protein marker bands are indicated at the left of the panel.
DISCUSSION
The vancomycin MIC (16 μg/ml) for COL-VR1, isolated from MRSA strain COL through three steps of exposure to vancomycin, was approximately eightfold higher than that for its parent strain. The data obtained in the present study suggest that the mechanism(s) of glycopeptide resistance for this isolate is at least partially distinct from those described previously for other clinical or laboratory derived isolates, adding to the complexity of the GISA and VRSA phenotypes observed previously (6). Several lines of evidence are in line with this proposal.
(i) While previous reports demonstrated that the levels of methicillin resistance of mutants selected by exposure to vancomycin or teicoplanin were strongly reduced, COL-VR1 not only retained its resistance to methicillin but also had an even slightly increased level of resistance to methicillin.
(ii) Alterations of PBP 2, PBP 4, or PBP 2′ expression were observed in several VRSA strains (19, 25, 26, 28). However, in the present study, such alterations were not found in our mutant either from penicillin binding data or by direct antibody detection experiments.
(iii) Cell wall structural alterations like a lack of amidation of the d-glutamic acid residue of the peptidoglycan stem peptide or a reduced level of cell wall peptide cross-linking, as has been found in several, although not all, GISA or VRSA mutants (6, 8, 10, 28), either were not observed at all (amidation) or were only slightly diminished (cross-linking). Instead, HPLC analysis of the products obtained after digestion of the isolated peptidoglycan with lysostaphin, an enzyme that cuts within the pentaglycine interpeptide bridge of the S. aureus peptidoglycan, thereby releasing soluble, non-cross-linked sugar chains with the peptides still attached, suggested an increase in the average sugar chain length for COL-VR1.
(iv) COL-VR1 also showed a peculiar pattern of susceptibility to cell wall-active antibiotics other than glycopeptides and β-lactams, in particular, drastically reduced sensitivities to the transglycosylase inhibitors macarbomycin and moenomycin (8- and 16-fold, respectively). To our knowledge, however, this feature has not been tested in other GISA or VRSA strains; it is therefore not possible to judge if this phenomenon might be common to several or even all of the mutants.
GISA and VRSA clinical isolates and laboratory mutants, including the mutant tested in the present study, have been shown to possess a characteristic, thickened cell wall (3, 8, 16, 29), which appears to be the most common phenotypic feature of these strains. Cui et al. (8) even reported that there is a direct correlation between cell wall thickness and susceptibility to vancomycin for Japanese clinical isolate Mu50 if it is grown under various culture conditions that allow differential expression of the cell wall thickness phenotype. They therefore proposed that the acceleration of cell wall synthesis contributed to the vancomycin resistance phenotype of Mu50. Whether this acceleration directly contributes to intrinsic resistance to vancomycin and cell wall thickness needs further investigation.
It is noteworthy that the average glycan chain length of peptidoglycan in COL-VR1 was longer than that in COL. A similar finding has been reported for highly glycopeptide-resistant mutants also derived from COL, which, however, differed in other aspects from COL-VR1 (26). Furthermore, Bischoff et al. (2) demonstrated that an S. aureus mutant with slightly decreased teicoplanin susceptibility due to inactivation of sigB expressed longer glycan chains compared to the lengths of the chains expressed by the parent, presumably due to less glucosaminidase activity. The peptidoglycan sugar chain length distribution is thought to be regulated by the interplay between synthetic and hydrolytic enzymes; in the case of S. aureus, these are transglycosylase and glucosaminidase. The S. aureus N-acetylglucosaminidase is known to be one of the atl gene products (22). Furthermore, Boneca et al. (4) described an additional endo-β-N-acetylglucosaminidase in atl mutants, although neither the gene nor the protein has yet been identified. For COL-VR1, however, the profiles of peptidoglycan hydrolases analyzed by zymography with S. aureus and M. luteus cells as a substrate showed no gross distinctions between COL and COL-VR1. The only hint to a potential difference between the mutant and the parent strain in this experiment was the slightly lower lytic activity of the second lysis band in the zymograms (Fig. 5), which is known to correspond to an atl gene product. However, since the other atl-related lysis bands in the same zymograms did not respond in the same manner and since this particular band is known to be somewhat unstable, this was deemed probably not significant. Also, the total autolysis rates were not different between COL and COL-VR1. These results indicate that a longer glycan chain length is not due to the reduced peptidoglycan hydrolase activity, although we cannot completely deny that possibility. On the other hand, we observed drastically increased macarbomycin and moenomycin MICs for COL-VR1 compared to those for COL (8- and 16-fold, respectively). Macarbomycin and moenomycin are believed to directly inhibit transglycosylase activity (12, 33, 34), while vancomycin and teicoplanin inhibit transglycosylase and transpeptidase activities indirectly by steric hindrance as a consequence of their binding to the d-Ala-d-Ala moiety of cell wall peptides (1). Since S. aureus PBP 2 belongs to class A PBPs, PBP 2 possesses transglycosylase domains in its N-terminal half (9, 19), but we could not demonstrate any difference in its level of expression between COL and COL-VR1 or alterations in its beta-lactam binding capacity. Matsuhashi and Park (17) demonstrated that, under their conditions, in S. aureus transglycosylase activity is not associated with PBP but is found in a membrane fraction separated from PBP, although they could not deny that the PBP(s) has transglycosylase activity. These data and our results taken together suggest that S. aureus uses multiple factors in providing transglycosylase activity and that in COL-VR1 increased transglycosylase activity plays an important role in the glycopeptide-intermediate susceptibility and vancomycin resistance phenotypes and possibly also in the thickened cell wall phenotype.
Acknowledgments
We thank Shin-ichiro Hiraoka and Karsten Servan for technical assistance. We also thank Alexander Tomasz for the generous gift of S. aureus strain COL.
This work was supported by a grant-in-aid for Encouragement of Young Scientists (grant 10770119) from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Barna, J. C. J., and D. H. Williams. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38:339–357. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bächi. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77–82. [DOI] [PubMed] [Google Scholar]
- 3.Bobin-Dubreux, S., M.-E. Reverdy, C. Nervi, M. Rougier, A. Bolmstrom, F. Vandenesch, and J. Etienne. 2001. Clinical isolate of vancomycin-heterointermediate Staphylococcus aureus susceptible to methicillin and in vitro selection of a vancomycin-resistant derivative. Antimicrob. Agents Chemother. 45:349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boneca, I. G., Z.-H. Huang, D. A. Gage, and A. Tomasz. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new endo-β-N-acetylglucosaminidase activity. J. Biol. Chem. 275:9910–9918. [DOI] [PubMed] [Google Scholar]
- 5.Boyle-Vavra, S., B. L. M. De Jonge, C. C. Ebert, and R. S. Daum. 1997. Cloning of the Staphylococcus aureus ddh gene encoding NAD+-dependent d-lactate dehydrogenase and insertional inactivation in a glycopeptide-resistant isolate. J. Bacteriol. 179:6756–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bächi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta 1523:135–139. [DOI] [PubMed] [Google Scholar]
- 8.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffin, C., and J.-M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu 50. J. Antimicrob. Chemother. 42:315–320. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673. [DOI] [PubMed] [Google Scholar]
- 12.Huber, G., and G. Nesemann. 1968. Moenomycin, an inhibitor of cell wall synthesis. Biochem. Biophys. Res. Commun. 30:7–13. [DOI] [PubMed] [Google Scholar]
- 13.Komatsuzawa, H., G. H. Choi, K. Ohta, M. Sugai, M. T. Tran, and H. Suginaka. 1999. Cloning and characterization of a gene, pbpF, encoding a new penicillin-binding protein, PBP 2B, in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komatsuzawa, H., M. Sugai, C. Shirai, J. Suzuki, K. Hiramatsu, and H. Suginaka. 1995. Triton X-100 alters the resistance level of methicillin-resistant Staphylococcus aureus to oxacillin. FEMS Microbiol. Lett. 134:209–212. [DOI] [PubMed] [Google Scholar]
- 15.Komatsuzawa, H., J. Suzuki, M. Sugai, Y. Miyake, and H. Suginaka. 1994. The effect of Triton X-100 on the in-vitro susceptibility of methicillin-resistant Staphylococcus aureus to oxacillin. J. Antimicrob. Chemother. 34:885–897. [DOI] [PubMed] [Google Scholar]
- 16.Marchese, A., G. Balistreri, E. Tonoli, E. A. Debbia, and G. C. Schito. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38:866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuhashi, M., and W. Park. 1984. Staphylococcus aureus and Micrococcus luteus peptidoglycan transglycosylases that are not penicillin-binding proteins. J. Bacteriol. 157:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milewski, W. M., S. Boyle-Vavra, B. Moreira, C. C. Ebert, and R. S. Daum. 1996. Overproduction of a 37-kilodalton cytoplasmic protein homologous to NAD+-linked d-lactate dehydrogenase associated with vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreira, B., S. Boyle-Vavra, B. L. M. DeJonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami, T., T. Fujimura, and M. Doi. 1994. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol. Lett. 117:131–136. [DOI] [PubMed] [Google Scholar]
- 21.Ohta, K., H. Komatsuzawa, M. Sugai, and H. Suginaka. 1998. Zymographic characterization of Staphylococcus aureus cell wall. Microbiol. Immunol. 42:231–235. [DOI] [PubMed] [Google Scholar]
- 22.Oshida, T., M. Sugai, H. Komatsuzawa, Y.-M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinho, M. G., H. De Lencastre, and A. Tomasz. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 182:1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roos, M., E. Pittenauer, E. Schmid, M. Beyer, B. Reinike, G. Allmaier, and H. Labischinski. 1998. Improved high-performance liquid chromatographic separation of peptidoglycan isolated from various Staphylococcus aureus strains for mass spectrometric characterization. J. Chromatogr. B 705:183–192. [DOI] [PubMed] [Google Scholar]
- 25.Shlaes, D. M., J. H. Shlaes, S. Vincent, L. Etter, P. D. Fey, and R. V. Goering. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942–18946. [DOI] [PubMed] [Google Scholar]
- 27.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517–523. [DOI] [PubMed] [Google Scholar]
- 28.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566–7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493–501. [DOI] [PubMed] [Google Scholar]
- 31.Stranden, A. M., K. Ehlert, H. Labischinski, and B. Berger-Bachi. 1997. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugai, M., T. Akiyama, H. Komatsuzawa, Y. Miyake, and H. Suginaka. 1990. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J. Bacteriol. 172:6494–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki, J., M. Hori, T. Saeki, and H. Umezawa. 1972. Macarbomycin, an inhibitor of peptidoglycan synthesis. J. Antibiot. 25:94–104. [DOI] [PubMed] [Google Scholar]
- 34.van Heijenoort, Y., M. Derrien, and J. van Heijenoort. 1978. Polymerization by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coli K12 and its inhibition by antibiotics. FEBS Lett. 89:141–144. [DOI] [PubMed] [Google Scholar]
- 35.Yamada, S., A. Matsumoto, K. Uehira, and T. Suda. 1994. Distribution of WGA-binding sites on the surface of clinical isolates of Staphylococcus aureus. J. Electron Microsc. 43:164–167. [PubMed] [Google Scholar]