Abstract
Abacavir is frequently used in antiretroviral combination therapies as a potent nucleoside reverse transcriptase inhibitor (NRTI). Four mutations are selected for by abacavir in vitro and in vivo: K65R, L74V, Y115F, and M184V. Abacavir resistance has also been observed in NRTI multidrug-resistant samples. Furthermore, abacavir resistance has been described in the context of zidovudine resistance. To evaluate the genetic basis of abacavir resistance, the viral genotype and phenotypic resistance were analyzed for 307 patient samples. Low- and high-level resistances were defined as 2.5- to 5.5-fold- and >5.5-fold-reduced susceptibility, respectively. If all samples with abacavir-selected and NRTI multidrug resistance-associated mutations were scored as resistant, 27.6% of the samples were misclassified, mainly due to samples falsely scored as susceptible. Therefore, the relative frequencies of other mutations were evaluated. Mutations at codons 44 and 118 were rarely detected in abacavir-susceptible samples but were overrepresented in resistant samples. Site-directed mutagenesis of E44D, V118I, and M184V resulted in low-level resistance for the double mutant 44/184 and the triple mutant. Low-level abacavir resistance was also detected for a viral clone carrying zidovudine mutations only. Additional insertion of M184V into the zidovudine background doubled the resistance, whereas 44/118 did not lead to a further increase. Incorporating combinations of zidovudine mutations and M184V into the scoring system markedly reduced the number of misclassified samples, whereas 44/118 did not improve the prediction. In conclusion, the combination of M184V with zidovudine mutations gives rise to high-level abacavir resistance, which may be clinically relevant. Thus, options for useful sequential combinations of NRTI are limited.
Since abacavir (1592U89) was approved as the sixth nucleoside reverse transcriptase inhibitor (NRTI) in 1998, it has frequently been used in antiretroviral combination therapies. Abacavir is at least as potent as the other NRTIs, has good oral bioavailability, and effectively penetrates into the central nervous system (1, 5). Furthermore, abacavir has been shown to be synergistic in combination with various antiretroviral drugs (11, 22). The combination with other NRTIs offers the opportunity of sparing other classes of antiretroviral drugs for subsequent use. In most cases, abacavir is well tolerated, with only minor side effects (9). A serious adverse event, a hypersensitivity reaction, has been reported to occur in 3 to 5% of patients, which precludes further use of abacavir in those patients (4).
Four drug resistance-associated mutations have been reported to be selected for by abacavir in vitro: K65R, L74V, Y115F, and M184V (23). These mutations were also selected in vivo, except that the codon Y115F mutation is only rarely observed in patient samples (6, 17). Additionally, abacavir resistance has been observed for two patterns selected by other nucleoside analogues, which are associated with NRTI multidrug resistance (MDR), the Q151 M complex (24) and the family of amino acid insertions between codons 67 and 70 of the reverse transcriptase (14). Furthermore, abacavir resistance has been described in the context of zidovudine and lamivudine resistance: M184V was selected after in vitro passage of a zidovudine-resistant viral clone in the presence of abacavir (23), and in some clinical isolates an increase in abacavir resistance was observed after M184V had developed in the context of zidovudine resistance (6).
The most frequent mutation implicated in abacavir resistance is M184V; however, controversial data exist concerning the relevance of this mutation for abacavir resistance. On one hand, the use of abacavir selects for M184V in vitro and in vivo (6, 17). On the other hand, the presence of M184V does not seem to preclude an antiviral response to abacavir in combination therapy (11). However, in heavily pretreated patients with a high prevalence of zidovudine and lamivudine resistance at baseline, the use of abacavir was associated with minor success (13).
Therefore, we wanted to investigate resistance to abacavir in more detail, using a database of corresponding genotypic and phenotypic results. The aims were to clarify the role of lamivudine resistance for abacavir resistance and to develop a model for more precise prediction of resistance to this drug.
(This material was presented in part at Progress in Clinical Virology IV of the European Society for Clinical Virology, Hamburg, Germany, September 1998 [abstr. 212]; the 3rd European Symposion on the Clinical Implications of HIV Drug Resistance, Frankfurt, Germany, February 2001 [abstr. 14]; and the 5th International Workshop on HIV Drug Resistance and Treatment Strategies, Scottsdale, Ariz., June 2001 [abstr. 70].)
MATERIALS AND METHODS
Selection of samples.
A total of 307 samples were obtained from 257 patients being treated at more than 20 outpatient centers and hospitals in Germany. The samples had been sent to the German National Reference Centre for Retroviruses for resistance testing between January 1998 and January 2000. Selection of samples was based on the availability of genotypic and phenotypic results only. In most cases, resistance testing was required because of permanently high viremia or rebound of viral load under antiretroviral therapy. Treatment histories were available for 229 of the 307 samples (74.6%). Of these, 81 samples (35.4%) were derived from abacavir-experienced patients.
Resistance testing.
Genotypic and phenotypic resistance tests were performed as described previously (20, 25). In brief, the genes for the protease and the relevant parts of the reverse transcriptase were amplified from patient plasma. Genotypic analysis was performed by direct sequencing of the product using dye terminators (Amersham, Cleveland, Ohio). Sequences were aligned with the Wisconsin Package version 10.0 (Genetics Computer Group, Madison, Wis.). The detection limit for minority species was about 30%. Resistance-associated mutations compiled by Schinazi et al. (19) were identified using the Stanford database (21). Phenotyping was performed by the recombinant-virus technique: the amplification product described above was cloned into a matched deletion mutant of the proviral human immunodeficiency virus type 1 clone NL4-3 (GenBank entry M19921). Viral stocks containing replication-competent recombinant viruses were obtained by transient transfection of 293T cells. A CEMx174-derived cell line containing the gene for the secreted alkaline phosphatase (SEAP) under the control of the simian immunodeficiency virus long terminal repeat (15) was used as an indicator cell line for titration of virus stocks and for drug susceptibility testing. Testing of abacavir resistance was performed in triplicate in 96-well plates containing 25,000 cells per well. SEAP activity was determined after 3 days. The inoculum was standardized to yield 10,000 relative light units/well after 3 days of culture without drugs. The resistance factor (RF) was calculated by dividing the 50% inhibitory concentration (IC50) for the respective recombinant virus by the IC50 for the nonresistant reference strain NL4-3, which was included in each independent assay.
Mutagenesis.
Viral clones were constructed by site-directed mutagenesis (10). For the insertion of E44D, V118I, and M184V into the pNL4-3 backbone, the following sense and antisense primers were used: E44D-s (5′-CAGAGATGGAAAAGGATGGGAA-3′), E44D-as (5′-TTTTCCCATCCTTTTCCATCTCT-3′), V118I-s (5′-TTTTTCAATTCCCTTAGATGAAGAC-3′), V118I-as (5′-GTCTTCATCTAAGGGAATTGAAAAA-3′), M184V-s (5′-TCTATCAATACGTGGATGATTTG-3′), and M184V-as (5′-CAAATCATCCACGTATTGATAGA-3′). For the construction of viral clones carrying zidovudine mutations, M41L was inserted with M41L-s (5′-TTGTACAGAGCTGGAAAAGGAA-3′) and M41L-as (5′-TCCTTTTCCAGCTCTGTACAAA-3′). Two strategies were used for the insertion of L210W and T215Y: viral clones resulting from mutagenesis with primers L210W/T215Y-s (5′-CTGTGGAGGTGGGGACTTTACACACC-3′)-and-L210W/T215Y-as(5′-GGTGTGTAAAGTCCCCACCTCCACAG-3′) contained 211R and 214L, whereas the use of primers L210W/R211K/T215Y-s (5′-CTGTGGAAGTGGGGATTTTACACACC-3′) and L210W/R211K/T215Y-as (5′-GGTGTGTAAAATCCCCACTTCCACAG-3′) resulted in viral clones with R211K and L214F. The drug susceptibilities of viral clones were determined as described above with at least four independent runs for each clone.
Algorithms.
Algorithms for the prediction of abacavir resistance from genotypic data were evaluated for sensitivity (percentage of phenotypically resistant viruses that were also predicted to be resistant by the algorithm) and specificity (percentage of phenotypically susceptible viruses that were also predicted to be susceptible by the algorithm).
RESULTS
A total of 307 samples were retrospectively analyzed for abacavir resistance. The IC50 of abacavir for the nonresistant reference virus NL4-3 was determined to be 4.58 ± 2.03 μM in 16 independent assays, a finding which is similar to previously published data (6, 23). The sample set was divided into three classes: samples with <2.5-fold resistance were defined as susceptible, whereas 2.5- to 5.5-fold and >5.5-fold resistances were used to distinguish between “low-level resistant” and “high-level resistant” samples, respectively. The lower limit of 2.5-fold was based on assay reproducibility. The upper limit of 5.5-fold was arbitrarily chosen. According to the classification presented above, 96 samples proved to be susceptible to abacavir, whereas 80 and 131 samples exhibited low- and high-level resistance, respectively.
In a first step, the frequencies of abacavir-selected mutations and the NRTI MDR patterns were determined. Mutations K65R (n = 2) and Y115F (n = 2) were detected in three samples only, whereas NRTI MDR (n = 14), L74V (n = 33), and M184V (n = 135) occurred more frequently. Comparing the relative frequencies of these mutational patterns with respect to the level of resistance revealed that NRTI MDR and L74V were predominantly present in highly resistant samples, whereas M184V was frequently detected in samples with low- and high-level resistance (Table 1).
TABLE 1.
Frequencies of mutations associated with resistance to abacavir in clinical isolates (n = 307), ranked with increasing resistance to abacavir
To evaluate whether these mutational patterns were sufficient to predict resistance to abacavir (RF > 2.5), samples were scored as resistant if they contained K65R, L74V, Y115F, NRTI MDR, or M184V. This resulted in a total of 85 misclassified samples (27.6%). While a specificity of 86.5% was achieved, sensitivity was considerably lower (65.9%). A low sensitivity means by definition that a high proportion of samples which are resistant in the phenotypic assay are predicted to be susceptible by this algorithm. This suggested that additional mutations were involved in abacavir resistance.
In a second step, the relative frequencies of other mutations were analyzed for abacavir-susceptible samples and for samples with low- and high-level resistance (Table 2). Mutations at codons 41, 67, 70, 210, 215, and 219 were detected in 9.4 to 24.0% of susceptible samples and two to three times more frequently in low-level resistant samples. In contrast, mutations at codons 44, 118, and 208 were present only in a very low percentage of abacavir-susceptible samples (2.1 to 5.2%) but were detected four to eight times more frequently in low-level resistant samples (Table 2).
TABLE 2.
Frequencies of mutations associated with resistance to zidovudine and lamivudine in clinical isolates (n = 307), ranked with increasing resistance to abacavir
| Fold reduced susceptibility to abacavira | Frequency (%) of mutations associated with resistance to zidovudine and lamivudine (19)
|
No. of samples | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M41L | E44D/A | D67N | K70R | V1181 | M184V/I | H208Y | L210W | R211K | L214F | T215Y/F/C | K219Q/E/R | ||
| <2.5 | 17.7 | 3.1 | 14.6 | 9.4 | 5.2 | 11.5 | 2.1 | 9.4 | 43.8 | 88.5 | 24.0 | 12.5 | 96 |
| 2.5–5.5 | 55.0 | 17.5 | 45.0 | 22.5 | 21.3 | 43.8 | 16.3 | 30.0 | 51.3 | 88.8 | 57.5 | 21.3 | 80 |
| >5.5 | 76.3 | 32.8 | 54.2 | 19.9 | 31.3 | 71.0 | 20.6 | 67.2 | 71.0 | 87.8 | 80.9 | 31.3 | 131 |
Fold increase in IC50 of the respective recombinant virus compared to the IC50 of the nonresistant reference virus NL4-3.
At that time, mutations at codons 44 and 118 had not been described as drug resistance associated. To investigate whether these “new” mutations, as well as M184V, contribute to abacavir resistance, the mutations were inserted into the nonresistant reference virus NL4-3. The single mutants exhibited less than twofold-reduced susceptibility to abacavir (MW01, MW02, and MW03) (Table 3). A 3.1-fold-reduced susceptibility to abacavir was detected for the double mutant 44/184 and the triple mutant 44/118/184 (MW05 and MW07).
TABLE 3.
Resistance profiles of viral clones constructed by site-directed mutagenesisa
| Mutation(s) at codons | Mutant | NRTI resistance profileb
|
|||||
|---|---|---|---|---|---|---|---|
| Zidovudine
|
Lamivudine
|
Abacavir
|
|||||
| RF | n | RF | n | RF | n | ||
| 44 | MW01 | 2.0 (1.2) | 5 | 1.3 (0.4) | 16 | 1.3 (0.5) | 15 |
| 118 | MW02 | 1.0 (0) | 5 | 1.9 (1.0) | 7 | 1.2 (0.3) | 6 |
| 184 | MW03 | 1.4 (0.5) | 5 | >100 (NA) | 16 | 1.5 (0.6) | 15 |
| 44/118 | MW04 | 1.2 (0.3) | 5 | 6.2 (2.6) | 5 | 2.2 (0.6) | 5 |
| 44/184 | MW05 | 1.2 (0.3) | 5 | >100 (NA) | 15 | 3.1 (1.2) | 14 |
| 118/184 | MW06 | 1.0 (0) | 5 | >100 (NA) | 7 | 1.8 (0.6) | 6 |
| 44/118/184 | MW07 | 1.0 (0) | 5 | >100 (NA) | 7 | 3.1 (1.3) | 7 |
| 41/210/211/215 | MW08 | >100 (NA) | 6 | 3.0 (0.6) | 7 | 3.8 (0.8) | 8 |
| 41/44/118/210/211/215 | MW09 | >100 (NA) | 4 | 6.2 (1.8) | 5 | 4.7 (1.9) | 7 |
| 41/184/210/211/215 | MW10 | 51.0 (24.0) | 4 | >100 (NA) | 5 | 8.0 (3.7) | 7 |
| 41/44/118/184/210/211/215 | MW11 | 32.2 (20.6) | 5 | >100 (NA) | 6 | 7.9 (2.3) | 8 |
Mutations that were inserted into the pNL4-3 backbone were M41L, E44D, V118I, M184V, L210W, R211K, and T215Y. Recombinant viruses from viral clones with 211R and 214L (44/118/210/215, 44/118/184/215, 41/44/118/184/215, and 44/118/184/210/215) replicated so poorly that resistance testing could not be performed.
RF, fold increase in IC50 compared with the nonresistant reference virus NL4-3; n, number of independent runs for each phenotypic resistance testing; NA, not applicable, because endpoint of resistance was not determined. Standard errors are included in parentheses.
After mutations E44D/A and V118I had been reported to be associated with moderate resistance to lamivudine in the presence of zidovudine mutations (7), we wanted to investigate whether a similar mechanism might be relevant for abacavir resistance. Thus, site-directed mutagenesis was extended to include the mutations M41L, L210W, and T215Y. These mutations were selected because their combination represented the most frequent pattern of zidovudine mutations in the clinical samples (107 of 237; 46.0%). In a first strategy, viral clones were constructed which contained zidovudine and lamivudine mutations in a background of 211R and 214L. However, recombinant viruses resulting from this strategy (44/118/210/215, 44/118/184/215, 41/44/118/184/210/215, and 44/118/184/210/215) replicated so poorly that resistance testing could not be performed. Mutations R211K and L214F have been reported to be associated with zidovudine and lamivudine dual resistance (19). Thus, in a second strategy, viral clones were constructed which contained R211K in addition to L214F as a natural polymorphism of the NL4-3 backbone. In this context, zidovudine mutations alone were sufficient to obtain a 3.8-fold-reduced susceptibility to abacavir (MW08) (Table 3). The additional presence of E44D and V118I added only a slight increase in abacavir resistance (MW09). An eightfold-reduced susceptibility to abacavir, however, was detected for M184V in addition to zidovudine mutations (MW10). In this context, E44D and V118I did not lead to any further increase of resistance (MW11).
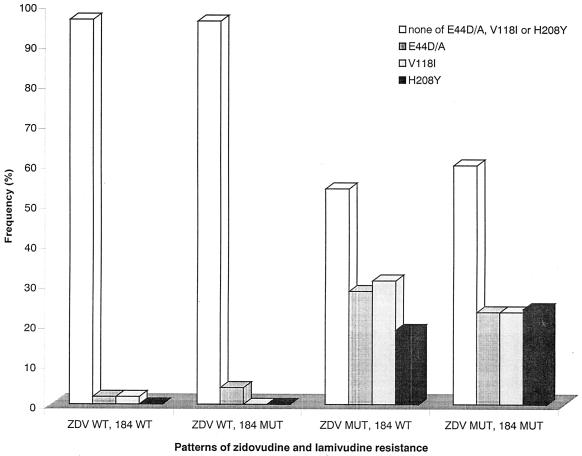
To clarify the discrepancy between the relative frequencies of mutations E44D/A and V118I in abacavir-resistant samples and their minor role in the clonal analyses, their relative frequencies were determined in clinical isolates with or without zidovudine resistance mutations (M41L, D67N, K70R, L210W, T215Y/F/C, and K219Q/E/R) and/or M184V/I. Both E44D/A and V118I were virtually absent in samples without any zidovudine resistance mutation, irrespective of the presence of M184V/I (Fig. 1). However, these mutations were detected in 18.6 to 31.0% of samples carrying at least one zidovudine mutation without M184V/I and with a similar frequency (23.1 to 23.9%) in samples carrying zidovudine mutations and M184V/I. The same distribution could be seen for H208Y, which has been linked to zidovudine and lamivudine dual resistance (16).
FIG. 1.
Relative frequencies of mutations E44D/A, V118I, and H208Y with respect to zidovudine resistance mutations and mutations at position 184 (n = 307). ZDV MUT is defined as the presence of one of the following mutations: M41L, D67N, K70R, L210W, T215Y/F/C, or K219Q/E/R. 184 MUT is defined as the presence of M184V/I. WT wild type, MUT mutant.
Based on the analysis of clinical samples and the results of the mutagenesis, different algorithms were evaluated for the prediction of abacavir resistance. Mutations K65R and Y115F were excluded from this evaluation, because they were too rare for a meaningful analysis. For the prediction of low-level abacavir resistance (RF > 2.5), the lowest number of misclassified samples (n = 43; 14.0%) was achieved by the algorithm “presence of L74V or NRTI MDR or M184V or any two mutations of 41/210/215,” which resulted in 91.9% sensitivity and 72.9% specificity. For the prediction of high-level resistance only (RF > 5.5), the best sensitivity (87.8%) and specificity (76.7%) were obtained for the algorithm “presence of L74V or NRTI MDR or any three mutations of 41/184/210/215,” resulting in 57 misclassified samples (18.6%). Inclusion of E44D/A and/or V118I did not improve the algorithms. This was also true for H208Y and for D67N and/or K70R, respectively.
DISCUSSION
This study confirmed that both M184V and zidovudine mutations contribute to abacavir resistance. In addition to the use of previously published data (6, 23), both a systematic analysis of clinical samples and site-directed mutagenesis were performed. Site-directed mutagenesis proved that the combination of both patterns had a synergistic effect on abacavir resistance, which is in contrast to the reduction of zidovudine resistance observed after the introduction of M184V. This was further supported by the evaluation of algorithms in clinical samples: low-level abacavir resistance could be predicted from the presence of either M184V or zidovudine mutations, whereas the presence of at least three mutations from a combination of zidovudine mutations and M184V was required for the prediction of high-level abacavir resistance. For both algorithms, most of the misclassified samples had resistance factors close to the chosen cutoffs (data not shown). Samples with resistance factors higher than two times the cutoff value were very rare (3 of 43 and 3 of 57 samples for the prediction of low-level and high-level resistance, respectively). The majority of misclassified samples were phenotypically susceptible but were classified as resistant, which is a conservative approach. Since these algorithms have been developed to predict the phenotype from the genotype, they still have to be evaluated for clinical relevance.
Some other mutations have been reported to develop in the context of zidovudine and lamivudine dual resistance: H208Y, R211K/L214F, and G333E/D (12, 16, 19). The role of mutations at codon 333 of the reverse transcriptase could not be evaluated, because the patient-derived part of the recombinant viruses only contained the first 300 amino acids of the reverse transcriptase. Mutations R211K and L214F were investigated by site-directed mutagenesis. Sufficiently replicating viruses were only obtained if these mutations were additionally present in a background of zidovudine mutations and M184V. This suggests that these mutations are important for the fitness of such viruses, facilitating zidovudine and lamivudine dual resistance. However, since viruses with zidovudine mutations and M184V without R211K and L214F were observed in patient samples, this is not an essential requirement.
A role for H208Y in abacavir resistance may be suspected from the overrepresentation of this mutation in resistant clinical samples. However, similar to E44D/A and V118I, mutation H208Y was virtually absent in isolates without other zidovudine resistance-associated mutations and seemed to appear independently of the presence of M184V/I (Fig. 1). For E44D and V118I, this distribution has already been described (3, 7). Additionally, none of these three mutations improved the prediction of abacavir resistance in clinical isolates. This supports the idea that these mutations are overrepresented in abacavir-resistant samples because they frequently develop in the context of zidovudine resistance. Thus, these mutations are good indicator mutations for abacavir resistance but do not necessarily contribute to abacavir resistance themselves.
The direct effect of mutations E44D and V118I was also assessed by site-directed mutagenesis. A slight increase in abacavir resistance was observed for viral clones carrying E44D compared to the respective clones without this mutation (MW05 versus MW03, MW07 versus MW06, and MW09 versus MW08), except for the highly resistant viruses (MW11 versus MW10). A similar effect could not be seen for the mutation V118I. Thus, the possibility that E44D may increase abacavir resistance in viruses with certain combinations of drug resistance-associated mutations cannot be excluded. Mutations E44D and V118I were originally reported to be associated with moderate resistance to lamivudine in the presence of zidovudine mutations (7). Thus, these mutations represent an alternative way for zidovudine-resistant viruses to acquire lamivudine resistance under simultaneous pressure of zidovudine and lamivudine. In contrast to M184V, resistance to lamivudine is only moderate, but it may be sufficient to enable replication under clinical conditions. However, the advantage compared to M184V is that zidovudine resistance is not reversed by E44D and V118I. Another aspect may be that E44D and V118I are possibly involved in resistance to other NRTIs. Thus, these mutations may be involved in abacavir resistance by a mechanism facilitating NRTI cross-resistance.
A problem in all studies of human immunodeficiency virus drug resistance is the definition of cutoff values to discriminate susceptible from resistant samples (2, 8, 18). In our study, the lower cutoff was defined according to technical restrictions, being based on assay reproducibility. The upper cutoff was arbitrarily chosen. Two cutoffs were chosen to address the point that drug resistance and consecutive treatment failure evolve as a continuum, i.e., the higher the resistance level, the more likely is therapy failure. Furthermore, the determination of meaningful cutoffs for phenotypic assays is still difficult. Recently, data on clinical cutoffs for abacavir resistance were presented (E. R. Lanier, N. Hellmann, J. Scott, M. Ait-Khaled, T. Melby, E. Paxinos, H. Werhane, C. Petropoulos, E. Kusaba, M. St. Clair, L. Smiley, and S. Lafon, 8th Conf. Retrovir. Opportunistic Infect., abstr. 254, 2001). In this study, response to abacavir in combination therapy was significantly worse if patients carried viruses with 4.5- to 6.5-fold-reduced susceptibility than if they carried viruses with less than 4.5-fold-reduced susceptibility. Therapy response was virtually absent if viruses with more than 6.5-fold-reduced susceptibility were present. Thus, the upper cutoff value that we have arbitrarily chosen is in the same range as these clinical cutoffs, although two different virus assays cannot be compared directly.
Finally, this study showed some interesting results concerning the role of M184V for abacavir resistance: the introduction of the single mutation into a wild-type background did not confer substantial abacavir resistance, which may explain why the presence of M184V alone does not preclude a therapy response to abacavir (11, 17). Nevertheless, since M184V is rapidly selected in the presence of abacavir in vitro and in vivo, this single-base mutation must confer some selective advantage (6). With M184V alone, this advantage may be very small, and possibly the slightly higher average resistance factor of 1.5 measured in the site-directed mutagenesis is an indicator of this advantage. If additional mutations are acquired, resistance increases to higher levels, which are more likely to be clinically relevant. These may be zidovudine resistance-associated mutations, which can be selected for by pretreatment with zidovudine and lamivudine as well as with stavudine and lamivudine. These combinations are frequent in the treatment history of heavily pretreated patients, which may explain the low success rate of abacavir in such patients (13).
In conclusion, our study supports the concept that cross-resistance is frequent not only within the classes of protease inhibitors and nonnucleoside reverse transcriptase inhibitors, but also within the class of NRTIs. Thus, the results of our study may have implications for daily clinical practice because they indicate that options for useful sequential NRTI combinations are limited.
Acknowledgments
We thank B. Fleckenstein for helpful discussion and continuous support. The indicator cell line was kindly provided by R. E. Means and R. C. Desrosiers.
We are also indebted to T. Harrer and M. Schmitt (Erlangen, Germany); M. Helm, G. Abelein, and W. Brockhaus (Nürnberg, Germany); L. Schneider (Fürth, Germany); S. Mauss, G. Schmutz, and T. Niehues (Düsseldorf, Germany); P. Braun and H. Knechten (Aachen, Germany); J. A. Rump (Freiburg, Germany); R. Billhardt (Offenburg, Germany); F. Schlote, E. Lauenroth-Mai, C. Schauenburg, F. Krauthausen, and M. Vocks-Hauck (Berlin, Germany); A. Groh and M. Scholz (Jena, Germany); U. Oesen and U. Bohr (Magdeburg, Germany); and C.-J. Reiβ (Halle, Germany) for providing clinical samples. Zidovudine, lamivudine, and abacavir were kindly provided by GlaxoSmithKline.
Financial support was provided by grants from the Bayerische Staatsministerium für Kultus, Erziehung und Wissenschaft (K. Korn) and the Robert Koch-Institute, Berlin, Germany (National Reference Centre for Retroviruses).
REFERENCES
- 1.Daluge, S. M., S. S. Good, M. B. Faletto, W. H. Miller, M. H. St. Clair, L. R. Boone, M. Tisdale, N. R. Parry, J. E. Reardon, R. E. Dornsife, D. R. Averett, and T. A. Krenitsky. 1997. 1592U89, a novel carbocyclic nucleoside analog with potent, selective anti-human immunodeficiency virus activity. Antimicrob. Agents Chemother. 41:1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeks, S. G., N. S. Hellmann, R. M. Grant, N. T. Parkin, C. J. Petropoulos, M. Becker, W. Symonds, M. Chesney, and P. A. Volberding. 1999. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J. Infect. Dis. 179:1375–1381. [DOI] [PubMed] [Google Scholar]
- 3.Delaugerre, C., M. Mouroux, A. Yvon-Groussin, J. M. Huraux, H. Agut, C. Katlama, and V. Calvez. 2000. Epidemiology and conditions of selection of 44D/A and/or 118I reverse transcriptase mutations in 344 patients. Antivir. Ther. 5(Suppl. 3):18. [Google Scholar]
- 4.Escaut, L., J. Y. Liotier, E. Albengres, N. Cheminot, and D. Vittecoq. 1999. Abacavir rechallenge has to be avoided in case of hypersensitivity reaction. AIDS 13:1419–1420. [DOI] [PubMed] [Google Scholar]
- 5.Foster, R. H., and D. Faulds. 1998. Abacavir. Drugs 55:729–736. [DOI] [PubMed] [Google Scholar]
- 6.Harrigan, P. R., C. Stone, P. Griffin, I. Najera, S. Bloor, S. Kemp, M. Tisdale, and B. Larder. 2000. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J. Infect. Dis. 181:912–920. [DOI] [PubMed] [Google Scholar]
- 7.Hertogs, K., S. Bloor, V. De Vroey, C. van Den Eynde, P. Dehertogh, A. van Cauwenberge, M. Sturmer, T. Alcorn, S. Wegner, M. van Houtte, V. Miller, and B. A. Larder. 2000. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob. Agents Chemother. 44:568–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertogs, K., M. P. de Béthune, V. Miller, T. Ivens, P. Schel, A. van Cauwenberge, C. van den Eynde, V. van Gerwein, H. Azijn, M. van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hervey, P. S., and C. M. Perry. 2000. Abacavir. A review of its clinical potential in patients with HIV infection. Drugs 60:447–479. [DOI] [PubMed] [Google Scholar]
- 10.Ho, S. N., H. D. Hunt, R. M. Horton, J. P. Pullen, and L. R. Pease. 1989. Site directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. [DOI] [PubMed] [Google Scholar]
- 11.Katlama, C., B. Clotet, A. Plettenberg, J. Jost, K. Arasteh, E. Bernasconi, V. Jeantils, A. Cutrell, C. Stone, M. Ait-Khaled, and S. Purdon. 2000. The role of abacavir (ABC, 1592) in antiretroviral therapy-experienced patients: results from a randomized, double-blind trial. CNA3002 European Study Team. AIDS 14:781–789. [DOI] [PubMed] [Google Scholar]
- 12.Kemp, S. D., C. Shi, S. Bloor, P. R. Harrigan, J. W. Mellors, and B. A. Larder. 1998. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and l-2′,3′-dideoxy-3′-thiacytidine. J. Virol. 72:5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khanna, N., T. Klimkait, V. Schiffer, J. Irigoyen, A. Telenti, B. Hirschel, and M. Battegay. 2000. Salvage therapy with abacavir plus a non-nucleoside reverse transcriptase inhibitor and a protease inhibitor in heavily pre-treated HIV-1 infected patients. Swiss HIV Cohort Study. AIDS 14:791–799. [DOI] [PubMed] [Google Scholar]
- 14.Larder, B. A., S. Bloor, S. D. Kemp, K. Hertogs, R. L. Desmet, V. Miller, M. Sturmer, S. Staszewski, J. Ren, D. K. Stammers, D. I. Stuart, and R. Pauwels. 1999. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleotide analog resistance. Antimicrob. Agents Chemother. 43:1961–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Means, R. E., T. Greenough, and R. C. Desrosiers. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 69:5431–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellors, J. W., H. Z. Bazmi, R. F. Schinazi, B. M. Roy, Y. Hsiou, E. Arnold, J. Weir, and D. L. Mayers. 1995. Novel mutations in reverse transcriptase of human immunodeficiency virus type 1 reduce susceptibility to foscarnet in laboratory and clinical isolates. Antimicrob. Agents Chemother. 39:1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, V., M. Ait-Khaled, C. Stone, P. Griffin, D. Mesogiti, A. Cutrell, R. Harrigan, S. Staszewski, C. Katlama, G. Pearce, and M. Tisdale. 2000. HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 14:163–171. [DOI] [PubMed] [Google Scholar]
- 18.Race, E., E. Dam, V. Obry, S. Paulous, and F. Clavel. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061–2068. [DOI] [PubMed] [Google Scholar]
- 19.Schinazi, R. F., B. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000–2001 update. Int. Antivir. News 8:65–92. [Google Scholar]
- 20.Schmidt, B., H. Walter, B. Moschik, C. Paatz, K. Van Vaerenbergh, A.-M. Vandamme, M. Schmitt, T. Harrer, K. Überla, and K. Korn. 2000. Simple algorithm derived from a geno-/phenotypic database to predict HIV-1 protease inhibitor resistance. AIDS 14:1731–1738. [DOI] [PubMed] [Google Scholar]
- 21.Shafer, R. W., D. R. Jung, and B. J. Betts. 2000. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat. Med. 6:1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staszewski, S., C. Katlama, T. Harrer, P. Massip, P. Yeni, A. Cutrell, S. M. Tortell, R. P. Harrigan, H. Steel, R. E. Lanier, and G. Pearce. 1998. A dose-ranging study to evaluate the safety and efficacy of abacavir alone or in combination with zidovudine and lamivudine in antiretroviral treatment-naive subjects. AIDS 12:F197–F202. [DOI] [PubMed] [Google Scholar]
- 23.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Laethem, K., M. Witvrouw, J. Balzarini, J. C. Schmit, S. Sprecher, P. Hermans, M. Leal, T. Harrer, L. Ruiz, B. Clotet, M. Van Ranst, J. Desmyter, E. De Clercq, and A. M. Vandamme. 2000. Patient HIV-1 strains carrying the multiple nucleoside resistance mutations are cross-resistant to abacavir. AIDS 14:469–471. [DOI] [PubMed] [Google Scholar]
- 25.Walter, H., B. Schmidt, K. Korn, A. M. Vandamme, T. Harrer, and K. überla. 1999. Rapid, phenotypic HIV-1 drug sensitivity assay for protease and reverse transcriptase inhibitors. J. Clin. Virol. 13:71–80. [DOI] [PubMed] [Google Scholar]