Abstract
An open reading frame located 230 nucleotides downstream from the stop codon of vanSc and in the opposite direction relative to the other genes of the vanC cluster was identified in Enterococcus gallinarum BM4174. This gene (designated ddl2) encoded a protein of 343 amino acids that had significant predicted structural similarity to d-Ala:d-Ala ligases and displayed 33 and 35% amino acid identity to VanC-1 and the previously reported partial sequence of Ddl from E. gallinarum, respectively. Biochemical characterization by thin-layer chromatography confirmed that Ddl2 is a d-Ala:d-Ala ligase with no detectable d-Ala:d-Ser ligase activity. The vancomycin dependence of Enterococcus faecalis BM4320 (ddl mutant) was lost on electroporation of a plasmid construct expressing ddl2 constitutively. The latter strain was able to grow in the absence of vancomycin, and peptidoglycan precursor analysis under the same conditions indicated the synthesis of pentapeptide[d-Ala] as the main precursor, confirming the activity of Ddl2 in vivo. Expression of ddl and ddl2 in BM4174 was tested by reverse transcription-PCR: results suggested that both d-Ala:d-Ala ligases were expressed concomitantly. Our findings indicate that vancomycin-resistant E. gallinarum BM4174 is likely to express one d-Ala:d-Ser and two d-Ala:d-Ala ligase genes.
Resistance to glycopeptide antibiotics in enterococci is due to the synthesis of modified peptidoglycan precursors that display decreased affinity for vancomycin and teicoplanin (4). The alteration of the peptidoglycan biosynthetic pathway is the result of the expression of specific gene clusters. In VanA, VanB, and VanD isolates, UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Lac is synthesized as the main peptidoglycan precursor (4). In VanC and VanE isolates, UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ser (4, 15, 30) is used by the transglycosylase and transpeptidase enzymes to synthesize the mature peptidoglycan (20). The resistant biochemical pathway is completed with the destruction of “glycopeptide-susceptible” precursors (ending in d-Ala-d-Ala): all vancomycin-resistant strains encode enzymes with d,d-dipeptidase and/or d,d-carboxypeptidase activities that ensure that UDP-MurNAc-pentapeptide [d-Ala] is unavailable for peptidoglycan synthesis (29, 31).
The d-Ala:d-Ala ligases are ATP-dependent enzymes that operate by using two chemically identical substrate molecules of d-Ala in distinct ways. The enzyme activates the N-terminal d-Ala as an electrophilic acyl phosphate, by transfer of the γ-PO32− from ATP, and then activates the C-terminal d-Ala as a nucleophile to attack the bound acyl-P; this gives rise to a tetrahedral adduct as intermediate that then decomposes to yield the dipeptide product (35). Vancomycin-resistant enterococci have acquired genes that encode d-Ala:d-X ligase activities with altered substrate specificities: VanA, VanB, and VanD function as d-Ala:d-Lac ligases using a chemical mechanism that rejects the protonated form of d-Ala at subsite 2 of the active site (23). VanC and VanE synthesize the dipeptide d-Ala-d-Ser (6, 15). Using the three-dimensional structure of DdlB of Escherichia coli (13) and VanA from Enterococcus faecium (32), site-directed mutagenesis studies have defined the residues responsible for substrate specificity (13, 21, 23, 24, 35).
The existence of two d-Ala:d-Ala ligase genes has been previously reported in E. coli and Salmonella enterica serovar Typhimurium (41). In this work we provide evidence for the first time that vancomycin-resistant Enterococcus gallinarum possess at least three dipeptide ligase genes, two encoding d-Ala:d-Ala ligases and the third encoding a d-Ala:d-Ser ligase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are described in Table 1. Enterococci were grown in brain heart yeast extract (BHY) (Difco Laboratories, Detroit, Mich.) broth or on BHY agar. Vancomycin-dependent E. faecalis BM4320 (ddl mutant) (5) used for complementation studies was grown in BHY supplemented with vancomycin (10 μg/ml); 150 γg of gentamicin (Sigma, Steinheim, Germany) per ml was added to the medium for pAT392-containing derivatives of this strain. E. coli XL1-Blue (7) was grown in Luria-Bertani (LB) (Difco Laboratories) broth or agar supplemented with gentamicin (8 μg/ml) E. coli derivatives containing pAT392 (3). E. coli M15(pREP4) (Qiagen) was grown in LB agar or broth supplemented with kanamycin (50 μg/ml). Ampicillin (100 μg/ml) was added for pQE-30 (Qiagen)-containing derivatives.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac F [proAB+laclqlacZΔM15 Tn10(Tetr)] | 7 |
| E. coli M15(pREP4) | Nalr Strs Rifrlac ara gal mtl F−recA+uvr+lon+ (pREP4) | Qiagen |
| E. faecalis BM4320 | VanB, vancomycin-dependent strain harboring an S319→I mutation in Ddl | 5 |
| E. gallinarum BM4174 | Vanr strain, clinical isolate | 10 |
| Plasmids | ||
| pCA8 | 2.5-kb SacI-XbaI inverse PCR product containing the 3′ end of vanSc and ddl2 cloned into pUC18 | 1 |
| pAT392 | oriRpAMβ1 oriRpUC oriTRK2 spc lacZα P2 aac(6′)-aph(2") | 3 |
| pQE-30 | Plasmid expression vector with T5 promoter and 6His tag encoding sequence 5′ to the cloning region | Qiagen |
| pHA1 | 1,081-bp fragment containing ddl2, including the RBS, cloned into pAT392 | This work |
| pHA2 | 1,032-bp fragment containing the coding region of ddl2, cloned into pQE-30 | This work |
DNA manipulations, and plasmid construction and sequencing.
E. gallinarum BM4174 total DNA was extracted as described previously (28). Cloning, digestion with restriction endonucleases (Boehringer GmbH, Mannheim, Germany), isolation of plasmid DNA (Wizard Plus SV Minipreps; Promega), ligation, and transformation were carried out by standard methods (33). The sequence of ddl2 was obtained from an insert in plasmid pCA8 that has been described previously (1). Plasmid pHA1 was obtained after cloning a SacI-XbaI fragment containing the ddl2 gene including the putative ribosome-binding site (RBS) into the shuttle vector pAT392 under the control of the constitutive P2 promoter (3). This fragment was obtained after amplification with Pwo polymerase (Boehringer) using total DNA of E. gallinarum BM4174 with primers A (5′-GACTGAGCTCGCTAGCTATTTCTAAACCAAAAC) and B (5′-ACGCTCTAGACTATTTGTACCAGCTTTC) (SacI and XbaI sites underlined, respectively). pHA1 was purified from E. coli and electroporated into the vancomycin-dependent E. faecalis BM4320 (ddl) (5), as described previously (9). Transformants from electroporation experiments were isolated on BHY plates in the absence of vancomycin. No transformants were obtained when the vector (lacking the fragment encoding ddl2) was used or in the absence of plasmid DNA under the same conditions.
DNA containing the coding region of ddl2 was amplified by PCR with Pwo polymerase (Boehringer) using chromosomal DNA from BM4174 as a template with primers C (5′-GCTAGGATCCATGAATATTGTGGTATTA), which includes a BamHI site (underlined), and D (5′-CGGTAAGCTTCTATTTGTACCAGCTTTC), which includes a HindIII site (underlined) and the stop codon of ddl2 (italicized). The product was purified, digested, and cloned into pQE-30 under the control of the T5 promoter using BamHI and HindIII. The resultant plasmid was designated pHA2 and allowed the expression of a gene encoding a protein with the incorporation of six histidine residues (6His tag) at the N terminus to enable affinity chromatography to be carried out. Sequencing of the inserts in pCA8 (two independent clones), pHA1, and pHA2 was performed on both strands by the dideoxy-chain terminator method (34) using fluorescent cycle sequencing with dye-labeled terminators (ABI Prism Dye Terminator cycle-sequencing ready reaction kit; Perkin-Elmer) on a 373A automated DNA sequencer (Perkin-Elmer).
Purification of Ddl2.
E. coli M15 (pREP4) containing plasmid pHA2 encoding the His-tagged Ddl2 was grown at 37°C in LB broth (300 ml) until the absorbance at 600 nm was 0.5. The culture was cooled and incubated at 18°C for 20 h in the presence of 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacteria were harvested at 4°C, washed in lysis buffer (50 mM Bis-Tris propane [pH 7.5], 300 mM NaCl, 10 mM imidazole), resuspended in 5 ml of the same buffer, and sonicated. The broken-cell preparation was centrifuged at 40,000 × g for 20 min at 4°C, and the supernatant was collected. All the following steps were carried out at 4°C. The supernatant was applied to a 2-ml nickel-containing agarose column (Agarose Ni-NTA; Qiagen) equilibrated with the same buffer. The column was washed with at least 100 ml of 50 mM Bis-Tris propane (pH 7.5)-300 mM NaCl-30 mM imidazole buffer, and Ddl2 was eluted with 50 mM Bis-Tris propane (pH 7.5)-300 mM NaCl-250 mM imidazole buffer. The purified protein was dialyzed against the assay buffer (2× concentrated) containing 200 mM Tris-HCl (pH 8.6), 20 mM KCl, 20 mM MgCl2, and 1 mM dithiothreitol. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% polyacrylamide gel was carried out under denaturing conditions using the Laemmli buffer system (22) to determine the purity of Ddl2. For calibration, standard proteins in the range Mr 9,000 to 175,000 (New England Biolabs, Hitchin, United Kingdom) were used. Proteins were stained with 0.1% (wt/vol) Coomassie blue in 50% (vol/vol) methanol-10% (vol/vol) acetic acid for 30 min at 37°C and destained with 10% (vol/vol) methanol-10% (vol/vol) acetic acid at room temperature overnight.
TLC assays of Ddl2.
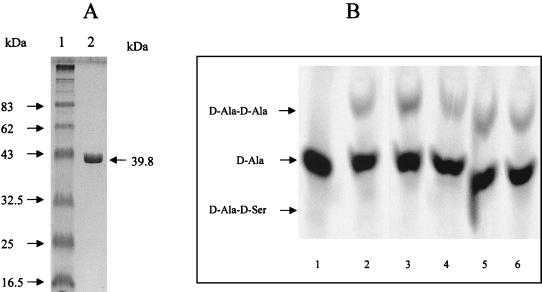
Assay mixtures (20 μl) contained 100 mM Tris-HCl (pH 8.6), 10 mM KCl, 10 mM MgCl2, 0.5 mM dithiothreitol, 6 mM ATP, 2 to 5 μg of enzyme, and combinations of different concentrations of [14C]d-Ala and d-Ser (see Fig. 2B). The samples were incubated at 37°C for 2 h, and the reaction was stopped by boiling the sample for 2 min. A 10-μl portion of the reaction mixture was spotted on a 0.1-mm microcrystaline cellulose thin-layer chromatography (TLC) plate (Polygram Cel 400; Macherey-Nagel) and developed for 4.5 h in butanol-glacial acetic acid-water (12:3:5 vol/vol/vol). The plate was dried for 5 min at 70°C, wrapped, and exposed to a Storage Phosphor-Screen (Molecular Dynamics, Sunnyvale, Calif.). Dipeptide formation was identified by comparing the spots (see Fig. 2B) with standards of d-Ala-d-Ala, d-Ala, d-Ala-d-Ser, and d-Ser after staining in 0.25% ninhydrin in acetone followed by drying for 5 min at 70°C. The radioactive spots were quantitated using image software (ImageQuant; Molecular Dynamics). The enzyme activity results were expressed in moles of product per minute per mole of enzyme.
FIG. 2.
(A) SDS-PAGE of purified His-tagged Ddl2. Lanes: 1, standard proteins; 2, sample containing His-tagged Ddl2 (Mr 39,828). (B) TLC of the products formed by Ddl2. Lanes: 1, 0.125 mM d-[14C]Ala standard; 2, 1 mM d-[14C]Ala incubated with purified His-tagged Ddl2; 3, as lane 2 but incubated with 10 mM d[14C]Ala; 4, Purified His-tagged Ddl2 incubated with 1 mM d[14C]Ala and 10 mM d-Ser; 5, as lane 4 but incubated with 10 mM d-[14C]Ala and 10 mM d-Ser; 6, as lane 4 but incubated with 1 mM d-[14C]Ala and 1 mM d-Ser.
RNA extraction.
E. gallinarum BM4174 was grown in 50 ml of BHY broth to an absorbance at 600 nm of 0.7 to 0.8. The bacteria were harvested, resuspended in 100 μl of TE (10 mM Tris HCl [pH 8.0], 1 mM EDTA) containing lysozyme (5 mg/ml), and incubated at 37°C for 30 min. Total RNA was extracted using a commercial kit (RNeasy mini kit; Qiagen). RNA was treated with RNase-free DNase I (DNA-free; Ambion, Austin, Tex.) to remove contaminating genomic DNA.
RT-PCR of ddl and ddl2.
Total RNA was reverse transcribed using a commercial kit (Omniscript reverse transcriptase; Qiagen) in a total volume of 20 μl containing 0.5 mM each dATP, dCTP, dGTP, and dTTP, 1 μM reverse primers (5′- TCAGGAATAATTCGTTTTTGTTCG for ddl; 5′-CTTTGTTCACTAATACGGTC for ddl2), 10 U of RNase inhibitor (SUPERase-IN; Ambion), 4 U of reverse transcriptase in corresponding buffer, and 2 μg of template RNA. The reverse transcription (RT) reaction was carried out at 37°C for 60 min. PCR was performed in a final volume of 25 μl which contained 2 μl of cDNA template, using Taq DNA polymerase (Pharmacia) and 0.5 μM primers specific for ddl (forward, 5′-TTGAAAATTATTTTATTATATGGCGG; reverse as above) and ddl2 (forward, 5′-CACCGAGCATTATTGTTGG; reverse as above). Amplification was carried out for 30 cycles (95°C for 1 min, 58°C for 1 min, and 72°C for 1 min). The sample was subjected to electrophoresis on a 1% agarose gel containing ethidium bromide. Non-reverse-transcribed PCR controls were used to indicate the absence of contaminating genomic DNA. To test for the specificity of the PCR targeting the ddl gene, primers used for ddl were included in a PCR assay with purified plasmid pHA1 as the template under the same conditions.
Extraction and analysis of peptidoglycan precursors from E. gallinarum and E. faecalis BM4320 and derivatives.
Extraction and analysis of peptidoglycan precursors was performed as described previously (30). In brief, E. gallinarum BM4174, E. faecalis BM4320, and derivatives were grown in BHY medium supplemented with vancomycin if needed. Ramoplanin (3 μg/ml) was added to inhibit peptidoglycan synthesis, and incubation was continued for 0.5 mean generation time (ca. 19 min) to allow the accumulation of peptidoglycan precursors. Bacteria were harvested, and cytoplasmic precursors were extracted, desalted on G10-Sephadex and analyzed by high-performance liquid chromatography.
Nucleotide sequence accession number.
The nucleotide sequence of ddl2 has been deposited in GenBank (accession number AF363615).
RESULTS AND DISCUSSION
Nucleotide sequence of ddl2 and amino acid comparisons.
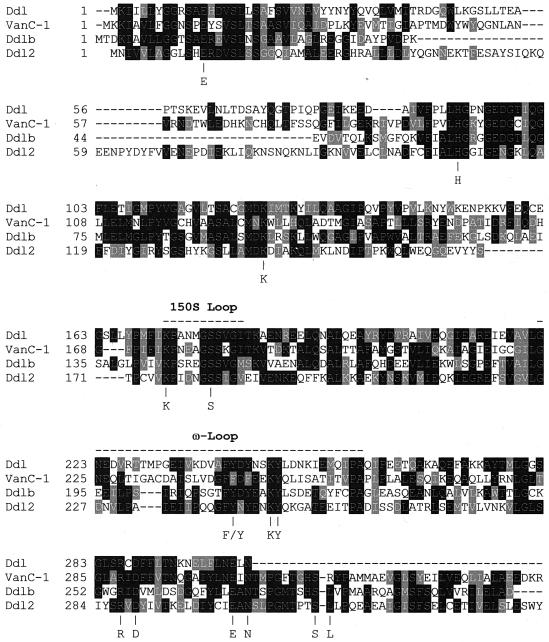
The sequence of ddl2 was obtained from sequencing the 3′ end of the insert of plasmid pCA8 (1). The putative open reading frame was located 230 nucleotides from the stop codon of vanSc (1) and in the opposite direction to the other genes of the vanC cluster. The gene was preceded by a putative RBS (5′-AGAAGGGATAN6ATG) that displayed similarity (underlined) to the 3-OH terminus of Bacillus subtilis 16S rRNA (3′-OH UCUUUCCUCC) (27). The putative product of ddl2 was a soluble protein of 343 amino acids with a calculated Mr of 38,430. Screening of the OWL protein sequence database showed similarity to d-Ala:d-X ligases from a variety of organisms. Ddl2 had 31 and 37% amino acid identity to DdlA and DdlB ligases, respectively, from E. coli. Compared with other d-Ala:d-X ligases previously described in E. gallinarum (10, 12) (Fig. 1), Ddl2 exhibited 33 and 35% identity to VanC-1 (a d-Ala:d-Ser ligase) and to a 303-amino-acid sequence of Ddl respectively, indicating the presence of three different ligase genes in this organism. Amino acid identity to the depsipeptide ligases VanA and VanB was 31 and 32%, respectively. Based on the crystal structure of DdlB from E. coli (13), all the important residues involved in binding ATP (K141, K176, K247, E302, and N304), d-Ala1 (E13, H107, S182, and R287), and d-Ala2 (Y248, D289, S313, and L314) were identified (Fig. 1). Additionally, by analogy to DdlB, residue 242 in Ddl2 should be involved in holding a loop of the protein over the nucleotide and the substrate-binding cavity: in d-Ala:d-Ala ligases this residue is invariably Tyr whereas in d-Ala:d-Ser ligases it is Phe (10, 15, 21, 26). The structure of DdlB also revealed two important amino acid loops: the first loop (the 150S loop) is part of an X-Gly-Ser-Ser-X-Gly motif that is conserved throughout all d-Ala:d-X ligases and contains the Ser150 residue, which is part of a hydrogen-bonding triad that orients Glu15 for binding the first d-Ala and which is also believed to hold the second loop in place (24). The second loop, called the ω-loop, forms a short helix, spanning residues 210 to 217 in DdlB, which closes over the bound ADP and reaction intermediates during catalysis, but it is believed to be flexible and exposed in the absence of substrates (13). All these residues were also conserved in Ddl2 (Fig. 1).
FIG. 1.
Alignment of the deduced amino acid sequence of Ddl2 with that of Ddl from E. gallinarum (303 amino acids) (12), Ddlb from E. coli (13), and VanC-1 (10). The alignment was performed with Clustalw (37). Predicted amino acids that interact with ATP, d-Ala1 and d-Ala2, are indicated below the alignment. The putative ω and 150S loops are shown above the alignment (13). Black and shaded boxes indicate amino acid identity and similarity, respectively.
Purification of Ddl2 and d-Ala:d-Ala ligase activity.
N-terminal His-tagged Ddl2 was purified from E. coli strain M15(pREP4) containing pHA2 (Fig. 2A). The ability of Ddl2 to synthesize a mixed dipeptide containing d-alanine was tested by incubation of the enzyme with different combinations of d-[14C]alanine (concentration range, 0.125 to 10 mM) and d-serine, followed by thin-layer chromatography and visualization in a Phosphor Imager. The results indicated that Ddl2 was able to synthesize d-Ala-d-Ala but not d-Ala-d-Ser (Fig. 2B). Maximum d-Ala:d-Ala ligase activity was 2.8 mol/min/mol of enzyme.
In vivo activity of Ddl2.
E. faecalis BM4320 is a vancomycin-dependent strain carrying the vanB cluster, with an S139→I mutation in Ddl which abolishes d-Ala:d-Ala ligase activity (5). The strain cannot grow in the absence of vancomycin, because the antibiotic is required for induction of the vanB gene cluster and consequent synthesis of d-Ala-d-Lac for peptidoglycan assembly (5). Analysis of peptidoglycan precursors of E. faecalis BM4320 grown in the presence of vancomycin (10 μg/ml) confirmed that the strain synthesized pentadepsipeptide as the main precursor (Table 2). No pentapeptide[d-Ala] was detected under these conditions.
TABLE 2.
Peptidoglycan precursors of E. faecalis BM4320 and derivatives and E. gallinarum BM4174 in the presence or absence of vancomycin
| Strain | Vancomycin (10 μg/ml) present | Presence of peptidoglycan precursor (%)a
|
||||
|---|---|---|---|---|---|---|
| UDP-MurNAc-tripeptide | UDP-MurNAc-tetrapeptide | UDP-MurNAc-pentapeptide[Ser] | UDP-MurNAc-pentapeptide[Ala] | UDP-MurNAc-pentadepsipeptide | ||
| E. faecalis BM4320 | Yes | 0.5 | 0.5 | 0 | 0 | 99 |
| E. faecalis BM4320/pAT392 | Yes | 2.5 | 0.5 | 0 | 0 | 97 |
| E. faecalis BM4320/pHA1(ddl2) | Yes | 0.5 | 0.5 | 0 | 0.5 | 98.5 |
| E. faecalis BM4320/pHA1(ddl2) | No | 33 | 0 | 0 | 67 | 0 |
| E. gallinarum BM4174 | No | 0 | 76 | 24 | 0 | 0 |
Determined by HPLC from the integrated peak areas.
Growth of E. faecalis BM4320 in the absence of vancomycin was restored by electroporation of plasmid pHA1, which encodes Ddl2 under the control of the constitutive P2 promoter. Control electroporation experiments carried out in the absence of plasmid DNA or with pAT392 lacking the ddl2 fragment yielded no colonies on BHY plates without vancomycin, confirming that growth restoration was not due to a spontaneous reversion of the dependent phenotype. The peptidoglycan precursor profile of this strain (analyzed in the absence of glycopeptide in the growth medium) indicated the presence of pentapeptide[d-Ala] and no pentadepsipeptide (Table 2). The amount of UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys (tripeptide) was found to be significant (33%) when E. faecalis BM4320/pHA1(ddl2) was grown in the absence of glycopeptides, suggesting that the rate of synthesis of pentapeptide[d-Ala] (and hence the synthesis of d-Ala-d-Ala) was low compared to the availability of the tripeptide precursor. When the same strain was grown in the presence of vancomycin, the amount of tripeptide was almost undetectable and pentadepsipeptide was virtually the sole peptidoglycan precursor synthesized (Table 2): the absence of pentapeptide indicates that VanXB (d,d-dipeptidase) is efficient in hydrolyzing d-Ala-d-Ala synthesized by Ddl2. Similarly, the high proportion of accumulated tetrapeptide in E. gallinarum BM4174 (76%) suggests that the bifunctional d,d-carboxypeptidase/d,d-dipeptidase (VanXYc) (29) prevents the utilization of pentapeptide[d-Ala] as the main precursor for wall synthesis: VanXYc effectively hydrolyzes pentapeptide that is likely to be made available by both ddl and ddl2. These findings confirmed that Ddl2 functioned as a d-Ala:d-Ala ligase in vivo.
Expression of ddl and ddl2 in E. gallinarum BM4174.
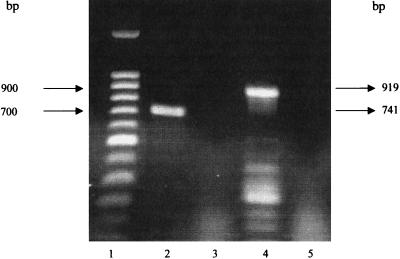
To investigate if both ddl and ddl2 were expressed at the same time, RT-PCR targeting both genes was performed with primers specific for each gene. To test such specificity, a PCR assay was performed with primers obtained from the ddl sequence but using plasmid pHA1(ddl2) as a template. No amplification product was observed under these conditions (data not shown), confirming the specificity of the assay. Expression of both genes was detected by RT-PCR (Fig. 3), indicating that both ddl1 and ddl2, encoding d-Ala:d-Ala ligases, are likely to be functional in E. gallinarum BM4174. The existence of two d-Ala:d-Ala ligase genes has been previously reported in E. coli and S. enterica serovar typhimurium (40), and our findings indicate for the first time that a gram-positive bacterium also possesses two ddl genes. The physiological relevance of this phenomenon is not clear. There are examples in bacteria and fungi of two genes encoding apparently similar enzymes: usually they are used for two separate metabolic pathways, e.g., two anthranilate synthases, trpE and phnA, in Pseudomonas aeruginosa (11) or two dehydroquinase activities of Neurospora crassa (18). On the other hand, both proteins are isoenzymes that are mostly regulated by feedback inhibition from different end products: the E. coli isoenzymes of acetolactate synthase, DAHP (3-deoxy-d-arabino-heptulosonate-7-phosphate) synthase, and aspartate kinase (38) are further examples of this phenomenon. In other situations the reasons are unclear, such as the two E. coli lysyl-tRNA synthetases (8). Two genes encoding similar enzymes involved in the synthesis of peptidoglycan precursors have also been reported: the alanine racemase alr and dadB genes (40). The constitutively expressed alanine racemase (Alr) is likely to be involved in the synthesis of peptidoglycan (17), and the inducible DadB racemase functions in the utilization of d-Ala as a sole carbon source. E. gallinarum BM4174 also harbors at least two racemases: an alanine racemase and a serine racemase (2). Due to the importance of the peptidoglycan synthetic pathway for bacteria, one could speculate that the two d-Ala:d-Ala ligases might be involved in different systems as well. Interestingly, gene homologues encoding the d-Ala-d-Ala-hydrolyzing enzyme VanX have been found in E. coli (ecovanX) associated with a putative dipeptide permease gene cluster. The genes form an operon under the control of the stationary-phase transcription factor RpoS. The consecutive action of EcoVanX and the membrane-bound d-amino acid dehydrogenase and pyruvate oxidase would allow the cell to utilize d-Ala-d-Ala released during the turnover of cell wall peptidoglycan as an energy source for cell survival during starvation (25).
FIG. 3.
RT-PCR of ddl and ddl2 using total RNA from E. gallinarum BM4174 as a template. Lanes: 1, Molecular size markers; 2, ddl2 RT-PCR; lane 3, as lane 2 but without reverse transcriptase; 4, ddl RT-PCR; 5, as lane 4 but without reverse transcriptase.
The presence of additional ligase genes in enterococci could also affect the emergence of the vancomycin-dependent phenotype (14, 16, 19, 36, 39) in two ways: (i) the presence of more than one functional ddl gene would decrease the probability of emergence of such a phenotype, or (ii) the presence of additional unexpressed ddl genes in vancomycin-dependant strains (due to specific regulatory mechanisms or repressors) would increase the possibility of restoring a previously mutated ligase gene by, for example, homologous recombination.
In conclusion, the characterization of a second d-Ala:d-Ala ligase gene indicates that vancomycin-resistant E. gallinarum BM4174 is likely to express three ligase genes: two encoding d-Ala:d-Ala ligases and one encoding a d-Ala:d-Ser ligase.
Acknowledgments
We are grateful to Tom Wilhelmsens Stiftelse for personal financial support to O.H.A. C.A.A. received personal financial assistance from COLCIENCIAS (Instituto Colombiano para el Desarrollo de la Ciencia y Tecnología, “Francisco José de Caldas”) and from the Overseas Research Scheme Award operated by the Committee of Vice-Chancellors and Principals of Universities in the United Kingdom. Part of this work was funded by an International Development Award from the Wellcome Trust.
We are indebted to Patrice Courvalin for the gift of plasmid pAT392 and E. faecalis BM4320. We thank Michel Arthur and Ireena Dutta for helpful discussions and J. Lester and C. Hill, Cambridge Center for Molecular Recognition, for DNA sequencing and synthesis of oligonucleotides, respectively.
REFERENCES
- 1.Arias, C. A., P. Courvalin, and P. E. Reynolds. 2000. vanC cluster of vancomycin resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 44:1660–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, C. A., M. Martin-Martinez, T. L. Blundell, M. Arthur, P. Courvalin, and P. E. Reynolds. 1999. Characterization and modelling of VanT, a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 31:1653–1664. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., F. Depardieu, H. A. Snaith, P. E. Reynolds, and P. Courvalin. 1994. Contribution of Van Y d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob. Agents Chemother. 38:1899–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, M., P. E. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401–407. [DOI] [PubMed] [Google Scholar]
- 5.Baptista, M., F. Depardieu, P. E. Reynolds, P. Courvalin, and M. Arthur. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25:93–105. [DOI] [PubMed] [Google Scholar]
- 6.Billot-Klein, D., L. Gutmann., S. Sable, E. Guittet, and J. van Heijenoort. 1994. Modification of peptidoglycan precursors is a common feature of the low-level vancomycin-resistant VANB-type Enterococcus D366 and of the naturally glycopeptide-resistant species Lactobacillus casei, Pediococcus pentosaceus, Leuconostoc mesenteroides, and Enterococcus gallinarum. J. Bacteriol. 176:2398–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue—a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376. [Google Scholar]
- 8.Clark, R. L., and F. C. Neidhardt. 1990. Roles of the two lysyl-tRNA synthetases of Escherichia coli: analysis of nucleotide sequences and mutant behavior. J. Bacteriol. 172:3237–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine-treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152–154. [DOI] [PubMed] [Google Scholar]
- 10.Dutka-Malen, S., C. Molinas, M. Arthur, and P. Courvalin. 1992. Sequence of the vanC gene of Enterococcus gallinarum BM4174 encoding a d-alanine:d-alanine ligase-related protein necessary for vancomycin resistance. Gene 112:53–58. [DOI] [PubMed] [Google Scholar]
- 11.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evers, S, B. Casadewall, M. Charles, S. Dutka-Malen, M. Galimand, and P. Courvalin. 1996. Evolution of structure and substrate specificity in d-alanine:d-alanine ligases and related enzymes. J. Mol. Evol. 42:706–712. [DOI] [PubMed] [Google Scholar]
- 13.Fan, C., P. C. Moews, C. T. Walsh, and J. R. Knox. 1994. Vancomycin resistance: structure of d-alanine:d-alanine ligase at 2.3 Å resolution. Science 266:439–443. [DOI] [PubMed] [Google Scholar]
- 14.Farrag, N., I. Eltringham, and H. Liddy. 1996. Vancomycin-dependent Enterococcus faecalis. Lancet 348:1581–1582. [DOI] [PubMed] [Google Scholar]
- 15.Fines, M., B. Perichon, P. E. Reynolds, D. F. Sahm, and P. Courvalin. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 43:2161–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraimow, H. S., D. L. Jungkind, D. W. Lande, D. R. Delso, and J. L. Dean. 1994. Urinary tract infection with an Enterococcus faecalis isolate that requires vancomycin for growth. Ann. Intern. Med. 121:22–26. [DOI] [PubMed] [Google Scholar]
- 17.Galakatos, N. G., E. Daub, D. Botstein, and C. T. Walsh. 1986. Biosynthetic alr alanine racemase from Salmonella typhimurium: DNA and protein sequence determination. Biochemistry 25:3255–3260. [DOI] [PubMed] [Google Scholar]
- 18.Giles, N. H., C. W. Partridge, S. I. Ahmed, and M. E. Case. 1967. The occurrence of two dehydroquinases in Neurospora crassa, one constitutive and one inducible. Proc. Natl. Acad. Sci. USA 58:1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, M., J. Shlaes, K. Barnadore, and D. M. Shlaes. 1995. Bacteremia due to vancomycin-dependent Enterococcus faecium. Clin. Infect. Dis. 20:712–714. [DOI] [PubMed] [Google Scholar]
- 20.Grohs, P., L. Gutmann, R. Legrand, B. Schoot, and J. L. Mainardi. 2000. Vancomycin resistance is associated with serine-containing peptidoglycan in Enterococcus gallinarum. J. Bacteriol. 182:6228–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healy V. L., I. S. Park, and C. T Walsh. 1998. Active-site mutants of the VanC2 d-alanyl-d-serine ligase, characteristic of one vancomycin-resistant bacterial phenotype, revert towards wild-type d-alanyl-d-alanine ligases. Chem. Biol. 5:197–207. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 23.Lessard, I. A. D., V. L. Healy, I. Park, and C. T. Walsh. 1999. Determinants for differential effects on d-Ala-d-lactate vs d-Ala-d-Ala formation by the VanA ligase from vancomycin-resistant enterococci. Biochemistry 38:1406–14022. [DOI] [PubMed] [Google Scholar]
- 24.Lessard, I. A., and C. T. Walsh. 1999. Mutational analysis of active-site residues of the enterococcal d-Ala-d-Ala dipeptidase VanX and comparison with Escherichia coli d-Ala:d-Ala ligase and d-Ala-d-Ala carboxypeptidase VanY. Chem. Biol. 6:177–187. [DOI] [PubMed] [Google Scholar]
- 25.Lessard, I. A., S. D. Pratt, D. G. McCafferty, D. E. Bussiere, C. Hutchins, B. L. Wanner, L. Katz, and C. T. Walsh. 1998. Homologs of the vancomycin resistance d-Ala-d-Ala dipeptidase VanX in Streptomyces toyocaensis, Escherichia coli and Synechocystis: attributes of catalytic efficiency, stereoselectivity and regulation with implications for function. Chem. Biol. 5:489–504. [DOI] [PubMed] [Google Scholar]
- 26.McKessar, S. J., A. M. Berry, J. M. Bell, J. D. Turnidge, and J. C. Paton. 2000. Genetic characterization of vanG, a novel vancomycin resistance locus in Enterococcus faecalis. Antimicrob. Agents Chemother. 44:3224–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran, C. P., N. Lang, S. F. J. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339–346. [DOI] [PubMed] [Google Scholar]
- 28.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett. Appl. Microbiol. 8:151–156. [Google Scholar]
- 29.Reynolds P. E., C. A. Arias, and P. Courvalin. 1999. Gene vanXYc encodes both d,d-dipeptidase (VanX) and d,d-carboxypeptidase (VanY) activities in vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 34:341–349. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, P. E., H. A. Snaith, A. J. Maguire, S. Dutka-Malen, and P. Courvalin. 1994. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem. J. 301:5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds, P. E., F. Depardieu, S. Dutka-Malen, M. Arthur, and P. Courvalin. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol. Microbiol. 13:1065–1070. [DOI] [PubMed] [Google Scholar]
- 32.Roper, D., T. Huyton, A. Vagin, and G. Dodson. 2000. The molecular basis of vancomycin resistance in clinically relevant enterococci: crystal structure of d-alanyl-d-lactate ligase (VanA). Proc. Natl. Acad. Sci. USA 97:8921–8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, Y., and C. T. Walsh. 1995. Active site mapping of Escherichia coli d-Ala:d-Ala ligase by structure-based mutagenesis. Biochemistry 34:2768–2776. [DOI] [PubMed] [Google Scholar]
- 36.Sifaoui, F., and L. Gutmann. 1997. Vancomycin dependence in a VanA-producing Enterococcus avium strain with a nonsense mutation in the natural d-Ala:d-Ala ligase gene. Antimicrob. Agents Chemother. 41:1409.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umbarger, H. E. 1978. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 47:532–606. [DOI] [PubMed] [Google Scholar]
- 39.Van Bambeke, F., M. Chauvel, P. E. Reynolds, H. S. Fraimow, and P. Courvalin. 1999. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob. Agents Chemother. 43:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wasserman, S. A., C. T. Walsh, and D. Botstein. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zawadzke, L. E., T. D. Bugg, and C. T. Walsh. 1991. Existence of two d-alanine:d-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry 30:1673–1682. [DOI] [PubMed] [Google Scholar]