Abstract
Mycobacterium smegmatis is a fast-growing nonpathogenic species particularly useful in studying basic cellular processes of relevance to pathogenic mycobacteria. This study focused on the d-alanine racemase gene (alrA), which is involved in the synthesis of d-alanine, a basic component of peptidoglycan that forms the backbone of the cell wall. M. smegmatis alrA null mutants were generated by homologous recombination using a kanamycin resistance marker for insertional inactivation. Mutants were selected on Middlebrook medium supplemented with 50 mM d-alanine and 20 μg of kanamycin per ml. These mutants were also able to grow in standard and minimal media without d-alanine, giving rise to colonies with a drier appearance and more-raised borders than the wild-type strain. The viability of the mutants and independence of d-alanine for growth indicate that inactivation of alrA does not impose an auxotrophic requirement for d-alanine, suggesting the existence of a new pathway of d-alanine biosynthesis in M. smegmatis. Biochemical analysis demonstrated the absence of any detectable d-alanine racemase activity in the mutant strains. In addition, the alrA mutants displayed hypersusceptibility to the antimycobacterial agent d-cycloserine. The MIC of d-cycloserine for the mutant strain was 2.56 μg/ml, 30-fold less than that for the wild-type strain. Furthermore, this hypersusceptibility was confirmed by the bactericidal action of d-cycloserine on broth cultures. The kinetic of killing for the mutant strain followed the same pattern as that for the wild-type strain, but at a 30-fold-lower drug concentration. This effect does not involve a change in the permeability of the cell wall by this drug and is consistent with the identification of d-alanine racemase as a target of d-cycloserine. This outcome is of importance for the design of novel antituberculosis drugs targeting peptidoglycan biosynthesis in mycobacteria.
Mycobacteria include the facultative intracellular pathogens Mycobacterium tuberculosis and Mycobacterium avium. It has been estimated that up to a third of the world’s population is infected with M. tuberculosis (14). Microorganisms of the M. avium complex have achieved prominence as major opportunistic pathogens of AIDS patients. M. avium is naturally resistant to most first-line antituberculosis drugs (19). This threat to public health has been partially met by therapy with appropriate antimicrobial agents, but unfortunately drug-resistant M. avium and M. tuberculosis strains readily appear (6, 15), underscoring the need to develop new and more effective antimycobacterial agents.
Biosynthesis of the mycobacterial cell wall has received considerable attention in the search for inhibitors useful for drug therapy (7). These cell walls display a complex architecture of glycolipids and proteins linked to the mycolyl-arabinogalactan-peptidoglycan backbone (26). This structure is a barrier that contributes to drug resistance (43), and many of its components have been found to play a major role in pathogenesis (11). The analysis of the M. tuberculosis genome sequence suggests that peptidoglycan biosynthesis in mycobacteria follows the general pathway of other bacteria, including the formation of the basic building block d-alanyl-d-alanine (2, 9, 44). d-alanine racemase (Alr) catalyzes the conversion of l-alanine into d-alanine (22), and d-alanine-d-alanine ligase catalyzes the subsequent dimerization of d-alanine into the key dipeptide d-alanyl-d-alanine (27). The corresponding enzymes from both Escherichia coli (23, 29) and mycobacteria (5, 13) are inhibited by d-cycloserine (DCS), a d-alanine analog (28). The dipeptide is then added to the UDP-tripeptide precursor by the action of the d-alanine-d-alanine adding enzyme that completes the reactions of the d-alanine branch of peptidoglycan assembly (45).
DCS is particularly effective against mycobacteria albeit with marked side effects (10, 49). Moreover, overproduction of Alr in Mycobacterium smegmatis, Mycobacterium intracellulare, and Mycobacterium bovis BCG leads to a DCS-resistant phenotype. We have also shown that the M. smegmatis enzyme is inhibited by DCS in a concentration-dependent manner (5). Likewise, the M. avium and M. tuberculosis enzymes produced from E. coli recombinant clones are also inhibited by DCS (39). Nonetheless, the specific characteristics of the mycobacterial enzymes involved in peptidoglycan biosynthesis, including the essentiality of each of their functions, remain unknown. This knowledge is important to the design of specific inhibitors that would serve as novel bactericidal agents to treat M. tuberculosis and M. avium infections. Furthermore, the inactivation of the genes encoding for these enzymes may lead to the generation of attenuated strains of pathogenic mycobacteria that could serve as candidate vaccines against tuberculosis.
M. smegmatis has been extensively used as a model system for M. tuberculosis and other pathogenic mycobacteria. M. smegmatis is nonpathogenic, requiring less stringent containment facilities, and it grows at a relatively high rate in a variety of defined and nutrient-restricted media (20). M. smegmatis has been used to study drug resistance mechanisms (5, 34, 41) and basic physiological processes including the synthesis of peptidoglycan precursors (8, 32). Insights gained from these studies can then be applied to the pathogenic mycobacteria. Thus, we started the genetic analysis of the d-alanine branch of peptidoglycan biosynthesis in this model system and describe the generation, isolation, and characterization of M. smegmatis d-alanine racemase (alrA) mutants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani broth or agar. M. smegmatis strains were routinely grown at 37°C in Middlebrook 7H9 base broth or agar (BBL Microbiology Systems, Cockeysville, Md.) supplemented with 0.5% bovine serum albumin fraction V (EM Science, Gibbstown, N.J.), 0.01 M dextrose (Sigma Chemical Co., St. Louis, Mo.), 0.015 M sodium chloride, and 0.2% glycerol (MADC). Broth medium was also supplemented with 0.05% Tween 80 (Sigma), and solid medium was made with 1.5% Bacto Agar (Difco Laboratories, Detroit, Mich.). When required, d-alanine (Sigma) was used at 50 mM. Liquid cultures were incubated with shaking at 200 rpm in an Innova 4300 rotary incubator (New Brunswick Scientific Co. Inc., Edison, N.J.). As needed, the following antibiotics were used at the specified concentrations: ampicillin (Sigma), 50 μg/ml for E. coli; kanamycin A monosulfate (Sigma), 25 μg/ml for E. coli and 20 μg/ml for M. smegmatis; hygromycin B (Roche Molecular Biochemicals, Indianapolis, Ind.), 100 μg/ml for E. coli and M. smegmatis. For some experiments, M. smegmatis was grown in a broth minimal medium based on the formulation of Zygmunt (50), as modified by Cáceres (5a). Components and final concentrations were 5.0 mM ammonium chloride, 6.8 × 10−7 mM calcium chloride, 8.4 × 10−7 mM cobalt(II) chloride, 22 mM dibasic potassium phosphate, 2.5 × 10−10 mM ferric chloride, 21 mM glycerol, 2.4 mM magnesium sulfate, 1.0 × 10−5 mM manganese chloride, 16 mM monobasic potassium phosphate, 4.9 × 10−6 mM pyridoxal hydrochloride, 0.4 mM Tween 80, and 8.6 × 10−6 mM zinc sulfate. Individual chemicals were purchased from Sigma.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5a | F−lacZDM15 endA1 hsdR17 supE44 gyrA96 relA1 | Invitrogen Life Technologies |
| E. coli XL2-Blue MRF′ | D(mcrA)183 D(mcrCB-hsdSMR-mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB laclqZDM15 Tn10 Tetr Camr] | Stratagene |
| M. smegmatis mc2155 | Alr+; high-efficiency plasmid transformation mutant of M. smegmatis mc26 | 37 |
| M. smegmatis TAM20 | Alr− Kanr; M. smegmatis alr mutant derived from mc2155 | This study |
| M. smegmatis TAM23 | Alr− Kanr; M. smegmatis alr mutant derived from mc2155 | This study |
| M. smegmatis TAM23 (pTAMU3) | Alr+ Hygr Kanr; M. smegmatis alr mutant complemented with wild-type gene integrated at the mycobacteriophage L5 attB site | This study |
| Plasmids | ||
| pBUN82 | Kanr; recombinant plasmid carrying the alr gene from M. smegmatis mc2155 in a 1.9-kb BamHI/PvuII fragment | 5 |
| pBluescript II KS + | Ampr; standard E. coli cloning vector | Stratagene |
| pTAMU1 | Ampr; pBluescript II KS(+) with the 1.9-kb BamHI/PvuII fragment of pBUN82 in the BamHI/EcoRV site | This study |
| pTAMU2 | Ampr Kanr; pTAMU1 with the 1.24-kb PstI aph fragment of pUC4K (Pharmacia) in the PstI site | This study |
| pTAMU3 | Ampr Hygr; pYUB412 with the 1.9-kb BamHI/PvuII fragment of pBUN82 in the BclI/EcoRV site | This study |
| pYUB412 | Ampr Hygr; E. coli-Mycobacterium integration-proficient vector; integrates at the mycobacteriophage L5 attB site | 33 |
Bacterial transformation.
E. coli was transformed as previously described (1). For the generation of M. smegmatis alrA mutants, 50 ml of an early exponential phase culture of M. smegmatis mc2155 was washed twice and concentrated 100-fold in cold 10% ultrapure glycerol (Invitrogen Life Technologies, Carlsbad, Calif.). Concentrated cells were electroporated with approximately 5.0 μg of the BamHI-KpnI fragment of pTAMU2 carrying the inactivated alrA gene. Electroporation was carried out at 2,500 V, 100 μF, and 246 Ω in an electrocell manipulator (model 600; BTX Inc., San Diego, Calif.). Electroporated cells were allowed to recover at 37°C in MADC broth and plated on MADC agar supplemented with 50 mM d-alanine (Sigma) and kanamycin (20 μg/ml; Sigma). For the genetic complementation experiment, 50 ml of an early-exponential -phase culture of M. smegmatis TAM23 alrA mutant was electroporated with 1.5 mg of pTAMU3 DNA at 2,500 V, 25 μF, and 1,000 Ω in a Gene Pulser electrocell manipulator (Bio-Rad Laboratories, Richmond, Calif.), as previously described (16). Transformants were selected on MADC agar supplemented with hygromycin (100 μg/ml; Roche).
Nucleic acid manipulations.
Mycobacterial DNA was isolated by the standard method using cetyl trimethyl ammonium bromide (1). Plasmid DNA was isolated by an alkaline lysis method as previously described (36), using a large-scale isolation kit (Promega, Madison, Wis.) as recommended by the manufacturer. DNA fragments used for plasmid construction in E. coli and for recombination experiments in M. smegmatis were purified by gel electrophoresis and recovered by absorption to glass particles (GeneClean Bio 101, Vista, Calif.) as recommended by the manufacturer. Standard procedures were followed for restriction digestions, ligations, and agarose gel electrophoreses (36).
Amplification of the alrA gene was carried out with 50 ng of M. smegmatis genomic DNA template using primers NAN-1 (5′-TCTGCGGCCTCTGGGACAATGGG-3′) and NAN-2 (5′-GACACACCTGCCACGGTGCCGAC-3′) for 27 cycles in a thermal cycler (Perkin-Elmer Gene Amp PCR System 2400; Roche Molecular Systems, Branchburg, N.J.) as previously described (5). For Southern blotting analysis, approximately 3.0 μg of M. smegmatis genomic DNA was digested with SmaI, and DNA fragments were separated on a 0.8% agarose gel, subjected to an alkaline denaturing procedure, and transferred to Biotrans nylon membranes (ICN Biomedicals, Inc., Costa Mesa, Calif.). Membranes were hybridized with a probe corresponding to the 1.9-kb BamHI/PvuII fragment containing the wild-type M. smegmatis alr gene, which was radiolabeled with [α-32P]dCTP using the Rediprime DNA labeling II system (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) as recommended by the manufacturer. Prehybridization and hybridization were performed at 56°C. Washes were done under high-stringency conditions at 65°C as previously described (36).
Gram staining and acid-alcohol resistance testing.
M. smegmatis cells were stained by the crystal violet method using a Gram stain kit (Sigma) as recommended by the manufacturer. Acid-alcohol resistance was determined by the Ziehl-Neelsen acid-fast procedure using the TB Stain Kit ZN (Difco), as recommended by the manufacturer.
Electron microscopy.
Strains were grown to an optical density at 600 nm (OD600) of approximately 1.0 in MADC-Tween, with or without d-alanine. Cells were harvested, washed twice with phosphate-buffered saline (PBS)-0.05% Tween, and fixed for 1 h in 2.5% buffered glutaraldehyde, washed twice, and postfixed with 2.0% osmium tetroxide. After repeated washings in PBS, samples were dehydrated in a graded series of ethanol solutions, washed twice in propylene oxide, and embedded with Araldite resin. Thin sections were examined with a Philips 201 transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands) at an accelerating voltage of 60 kV.
d-Alanine racemase assays.
Approximately 200-ml cultures of M. smegmatis mc2155 or TAM23 were grown in MADC-Tween with or without d-alanine until they reached an OD600 of ca. 1.0. Cultures were washed twice and concentrated 50-fold in 50 mM Tris-HCl (pH 8.0). Cells were sonicated on a salt-ice-water bath with a Vibra-Cell model VC600 sonicator (Sonic and Materials, Inc., Danbury, Conn.). Sonication was carried out for 2 min at 80% power output and 50% duty cycle, and in the presence of 30% (vol/vol) type A-5 alumina (Sigma). The resulting active cell extracts were centrifuged at 4°C in a JA-17 rotor (Beckman Instruments, Inc., Fullerton, Calif.) for 30 min at 15,000 rpm, dialyzed against 50 mM Tris-HCl (pH 8.0), and sterilized by filtration through a 0.22-μm-pore-size filter (Advantec MFS Inc., Pleasanton, Calif.). Protein concentration was determined by the DC assay (Bio-Rad) as recommended by the manufacturer. Alr activity in the active cell extracts was assayed in the direction of the conversion of l-alanine into d-alanine by a modification of the coupled-spectrophotometric method previously described (5, 48). To start the reactions, active crude cell extracts were added to 1.0 ml of prewarmed mixtures containing 50 mM Tris-HCl (pH 8.0), 0.1 mM pyridoxal phosphate (Sigma), and 15 mM l-alanine (Sigma). After 15 min of incubation at 37°C, when the conversion of substrate into product remains linear, reactions were stopped by boiling for 10 min. Subsequently, 1 U of d-amino acid oxidase (Calzyme, San Luis Obispo, Calif.), 0.2 mM NADH (Roche Laboratories), and 10 U of rabbit muscle lactic dehydrogenase (Sigma) were added. The coupled reaction was measured by the change in absorbance at 340 nm after 5 h of incubation at 37°C. All samples were measured in triplicate. Specific activities (in micromoles of consumed substrate minute−1 milligram−1) were calculated as previously described (5).
LDH assays.
l-Lactate dehydrogenase (LDH) activity endogenous to crude cell extracts of M. smegmatis was measured in the direction of the conversion of pyruvate into lactate coupled to the oxidation of NADH as previously described in the Worthington enzyme manual (Worthington Biochemical Corp., Lakewood, N.J.). Crude cell extracts were added to 1.0 ml of prewarmed mixtures containing 50 mM Tris-HCl (pH 8.0), 1.0 mM sodium pyruvate (Sigma), and 0.2 mM NADH (Roche). The change in absorbance at 340 nm was measured after 1 h of incubation at 37°C. All samples were measured in triplicate. Specific activities (in micromoles of consumed substrate minute−1 milligram−1) were calculated as described in the enzyme manual mentioned above by subtracting the background change in absorbance (obtained from boiled inactivated extracts processed in identical manner) from the change in absorbance obtained with the active cell extracts.
Drug susceptibility assays.
MICs were determined by a microplate twofold dilution method, as described by Takiff et al. (40), with modifications. M. smegmatis mc2155 and TAM23 cells were grown in 25 ml of MADC-Tween with and without 50 mM d-alanine to an OD600 of approximately 1.0. Bacteria were washed with PBS-Tween, and 105 CFU was inoculated onto each well containing serial twofold dilutions of various antimicrobial agents. Plates were incubated at 37°C, and visual inspection to determine MICs was carried out at 48 h. The MIC is defined as the lowest concentration that prevents observable bacterial growth and is determined by the consistent result of three independent cultures, each assayed in triplicate.
Bactericidal action of DCS.
M. smegmatis strains mc2155 and TAM23 were grown in MADC-Tween broth at 37°C to an OD600 of ca. 0.5. These starter cultures were used to inoculate 200 ml of MADC broth, and cells were then grown to an OD600 of ca. 0.4. Two 50-ml aliquots of these exponentially growing cultures were transferred to separate flasks. DCS was added to one of the corresponding cultures, with the other serving as a growth control. Concentrations of DCS were 50 times the MIC for each strain (3.75 mg/ml for mc2155 and 128 μg/ml for TAM23). All cultures were incubated at 37°C in a shaking incubator for 72 h. The OD600 was measured for each culture at 3, 6, 9, 12, 24, 48, and 72 h. Concurrently, aliquots were taken, serially diluted, and plated onto MADC agar in triplicate to determine numbers of viable bacterial counts.
Uptake assays.
For d-alanine uptake assays, we used a modification of the method described by David (12). Cells exponentially growing in MADC-Tween (OD600, ca. 1.0) were collected by centrifugation at 4°C, washed once in ice-cold 50 mM Tris-HCl buffer (pH 8.0) containing 0.05% Tween 80, and concentrated 20 times in the same buffer. Samples were prewarmed for 10 min at 37°C, and d-alanine was added to a final specific activity of 3.0 μCi of [14C]-1-d-alanine (ICN) per μmol and 0.2 mM concentration. Samples were placed in a 37°C water bath. Aliquots were taken at various times and immediately placed on ice. The sample for each time point was divided in three subsample triplicates of 0.5 ml and filtered through 0.8-μm-pore-size membrane filters (Millipore Corp., Bedford, Mass.) in a manifold. Each filter was washed three times with 10 ml of cold buffer, dried under a heat lamp, and weighed. The cell-associated radioactivity was determined in a liquid scintillation counter (Wallac 1410; Pharmacia, Piscataway, N.J.) using EcoLite scintillation cocktail (ICN). Uptake data were expressed as micromoles of d-alanine per milligram (dry cell weight).
Since radiolabeled DCS is not commercially available, an uptake assay was developed based on the colorimetric determination of this compound (21). Cells exponentially growing in MADC-Tween (OD600 of ∼1.0) were harvested at 4°C by centrifugation, washed twice with water, and concentrated 40 times. Samples were prewarmed at 37°C for 10 min, and DCS was added to a final concentration of 250 μg/ml (∼2.5 mM). Cells were placed in a 37°C water bath. Aliquots were taken at various times and immediately placed on ice. After 20 min of incubation, clumping of TAM23 cells was evident and uptake determinations became unreliable. Each time point sample was washed twice with water, resuspended, and sonicated. Protein concentration was determined using the Bio-Rad DC assay, followed by protein removal through serial passages of both YM-10 and YM-3 Centricon concentrators (Millipore Corp.). The concentration of DCS was determined by measuring the OD620 after adding a specific color-developing reagent as previously described (21). A standard curve was generated by diluting a DCS standard in a cell extract that was prepared from cells not exposed to DCS. Uptake data were expressed as micromoles of DCS per milligram of protein. Since d-alanine and DCS uptake assays do not measure the same parameters, reported values for each assay cannot be directly compared.
RESULTS
Inactivation of the M. smegmatis alrA gene.
To inactivate the alrA gene in M. smegmatis, a DNA fragment carrying the wild-type gene was subcloned into the E. coli cloning vector pBluescript II KS(+) to yield the recombinant plasmid pTAMU1 (Table 1). Then, the 1.2-kb kanamycin resistance determinant from plasmid pUC4K containing the aminoglycoside 3′-phosphotransferase type I-coding gene from transposon Tn903 (30) was inserted at the unique PstI site internal to the alrA gene. The resulting construct, pTAMU2, carries an insertionally inactivated alrA gene, which can be excised as a linear 3.2-kb BamHI-KpnI fragment (Table 1). Based on the hypotheses that d-alanine is an essential component and that AlrA is the only enzyme responsible for d-alanine biosynthesis in M. smegmatis, it was predicted that an alrA mutant would be dependent on d-alanine for growth. Thus, the step to select for this mutant was carried out in MADC agar supplemented with 50 mM d-alanine in addition to 20 μg of kanamycin per ml. Transformation of M. smegmatis mc2155 with the 3.2-kb linear fragment carrying the inactivated alrA gene yielded 25 kanamycin-resistant transformants, and 2 of these, designated TAM20 and TAM23, were further analyzed.
To determine whether these transformants carry an inactivated alrA gene, genomic DNA was isolated and amplified by PCR. As expected for the inactivation of the alrA gene, genomic DNA from both TAM20 and TAM23 yielded the 2.4-kb product. Southern blotting analysis was used to verify the occurrence of these recombinational events in the appropriate M. smegmatis strains (data not shown). Genomic DNA was isolated; digested with SmaI, which cuts once within the alrA gene; transferred to a membrane; and hybridized with the wild-type alrA gene fragment as a probe. The wild-type strain mc2155 gave two homologous bands of approximately 15.0 and 1.8 kb, whereas TAM20 and TAM23 yielded a mutant-type pattern with three bands of approximately 15.0, 2.2, and 1.2 kb. These patterns were as expected for the predicted recombinational events, validating the construction of the strains described herein.
Phenotypic characterization of M. smegmatis alrA mutants: independence of d-alanine for growth.
The identification of only one d-alanine racemase gene in the mycobacterial genome sequencing projects suggests that M. smegmatis alrA mutants may be dependent on exogenous d-alanine for growth. Both mutant strains, TAM20 and TAM23, exhibited wild-type growth in MADC agar supplemented with d-alanine, giving rise to typical flat-border colonies after 3 days of incubation at 37°C. In the absence of d-alanine, TAM20 and TAM23 cells were also able to grow, but colonies displayed a drier appearance with more-raised borders. Complementation of TAM23 with the integrating construct pTAMU3 introduces a wild-type alrA gene at the mycobacteriophage L5 attachment site (24, 31) and fully restores wild-type colony morphology. Except for these differences in colony morphology, no other observable differences were detected by light or electron microscopy when cells were grown in the presence or absence of d-alanine. Bacilli from both wild-type and mutant strains were weakly gram positive, acid fast, and displayed the same aspect of elongated rods. Likewise, at the ultrastructural level, cells did not differ in either shape, size, or thickness of the cell walls. In summary, M. smegmatis alrA mutants are independent on exogenous d-alanine, a property that was further confirmed by their ability to grow in MADC (see below and Fig. 2) and minimal broth containing mineral salts, glycerol, pyridoxal phosphate, and Tween 80 (data not shown).
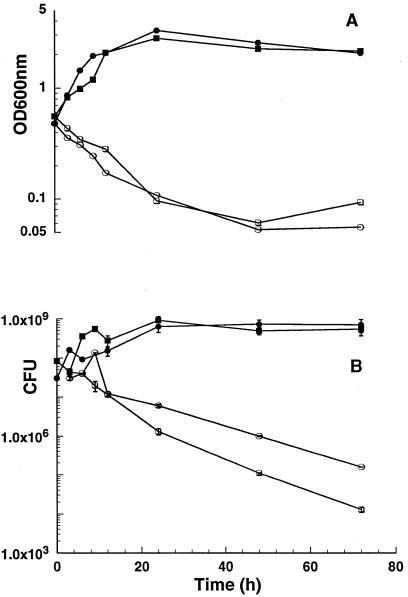
FIG. 2.
Bactericidal action of DCS on M. smegmatis wild-type and alr mutant strains. Cells were grown in MADC broth without d-alanine to an OD600 of ca. 0.4. At this time (time zero), cultures for each strain were split in two, and DCS was added to one of these subcultures at a concentration of 50 times the MIC for the corresponding strain (see Materials and Methods). ODs (A) and CFU per milliliter (means ± standard deviations [error bars] of triplicate measurements) (B) were determined for mc2155 in the subcultures with (open circles) or without (closed circles) DCS. Identical measurements were performed for the corresponding subcultures of TAM23 with (open squares) or without (closed squares) DCS.
M. smegmatis alrA mutants have no detectable d-alanine racemase activity.
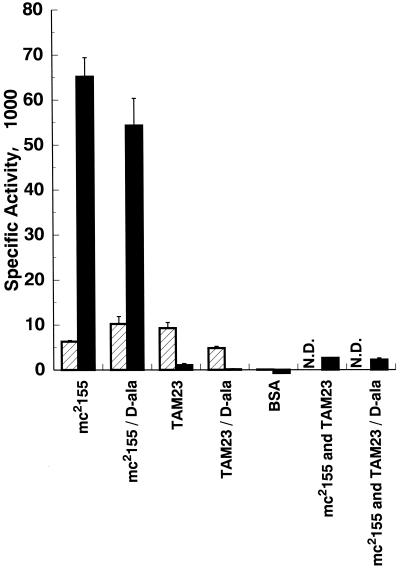
To determine whether M. smegmatis alrA mutants are or are not devoid of Alr activity, crude cell extracts from wild-type and mutant strains were prepared and assayed for enzyme activity (Fig. 1). Protein extracts prepared from wild type mc2155 cells grown in the presence or absence of d-alanine displayed approximately equal levels of Alr activity that matched the specific activities previously reported (5). In contrast, extracts from TAM23 prepared from cells grown in the presence or absence of d-alanine lacked any detectable Alr activity. These extracts yielded background levels of Alr activity not significantly different from the levels obtained by replacing bovine serum albumin for TAM23 extracts in the reaction mixture. This absence of Alr activity in TAM23 extracts was not due to the presence of an inhibitor since mixtures of TAM23 and mc2155 extracts gave the Alr activity proportional to the amount of enzyme present in the active extract from the wild-type strain. Furthermore, to rule out the possibility that sonic disruption may have damaged more readily protein extracts of potentially fragile TAM23 cells, LDH activity was also measured. The M. smegmatis LDH activity was found to be more sensitive to prolonged sonic disruption (data not shown) and provides a useful control to determine whether TAM23 extracts are enzymatically active. In contrast to the results obtained in the Alr assay, extracts from both mc2155 and TAM23 displayed similar levels of LDH activity above background levels. Furthermore, complementation of TAM23 with a wild-type alr gene restored Alr activity. Thus, it was concluded that inactivation of the alrA gene results in no detectable Alr activity in the M. smegmatis alrA mutant strain.
FIG. 1.
Specific activities of d-alanine racemase and LDH in M. smegmatis cell extracts. Mean specific activities (in micromoles) of substrate consumed minute−1 milligram−1, of d-alanine racemase (solid bars) and LDH (striped bars) from wild-type and mutant strains were determined in cell extracts prepared as described in Materials and Methods. Cells were grown to exponential phase in medium with or without d-alanine (50 mM) as indicated at the bottom of the figure. Extracts were prepared from three independent cultures for each strain and medium and assayed in triplicate. Combined extracts of mc2155 and TAM23 (ca. 1:24 [wt/wt] protein mixture ratio, with TAM23 extract as the predominant component) were also assayed in triplicate. A mock assay was also carried out with bovine serum albumin (BSA) in place of equivalent amounts of the corresponding cell extracts. N.D., not determined. Error bars, standard deviations.
M. smegmatis alrA mutants are hypersusceptible to d-cycloserine.
The inhibition of AlrA by DCS in a concentration-dependent manner indicates that this enzyme serves as one of the drug targets (5). However, the dispensability of the alrA gene for growth in vitro suggests that it is not the main target responsible for the bactericidal effect of DCS. Since overproduction of the enzyme leads to increased resistance to DCS, it appears that binding of DCS to AlrA protects another target within the cell. This hypothesis predicts that alrA null mutants would be hypersusceptible to DCS. Thus, the corresponding MICs for wild-type and TAM23 alrA mutant strains were determined.
For MIC tests, M. smegmatis cells grown with and without d-alanine were inoculated onto complete Middlebrook MADC broth without d-alanine. The MIC of DCS for the wild-type strain was 75 μg/ml, independent of the presence of d-alanine in the original inoculum (Table ). As expected, TAM23 was about 30-fold more sensitive (MIC, 2.56 μg/ml) when the inoculum was grown without d-alanine. Since TAM23 cells carry a mutation in a gene responsible for d-alanine biosynthesis, it is possible that growth in the presence of d-alanine partially restores wild-type MICs of DCS. To test this hypothesis, TAM23 was grown with d-alanine, harvested, washed extensively to prevent d-alanine carryover, and inoculated into the MIC test cultures. These conditions resulted in a fourfold increase in the MIC (MIC, 10.2), still about 7.5 times lower than the MIC for the wild-type strain. As a control, the MICs of the unrelated drugs amikacin, ethambutol, and rifabutin were also determined. The MICs for both wild-type and mutant strains were the same, independently of the presence or absence of d-alanine in the medium used to grow the inocula to determine the MICs. The effect of the addition of d-alanine directly into the MIC test cultures was also determined. d-Alanine would be expected to effectively compete with DCS and decrease the susceptibilities of both strains to DCS. As expected, under these conditions, the MIC of DCS for both wild-type and TAM23 strains increased dramatically to 1,200 μg/ml independently of prior growth conditions of the inocula. No significant differences were observed for the other unrelated drugs, demonstrating that the effect was specific for DCS. Complementation of strain TAM23 with the integrating vector pTAMU3, which carries the wild-type alr gene, resulted in a strain for which the MICs were identical to those for the wild type (data not shown).
TABLE 2.
Determination of MICs of selected antimycobacterial agents for M. smegmatis strains determined with inocula grown with or without d-alanine
| Drug | MICa (μg/ml) of drug for strain grown as indicated
|
|||
|---|---|---|---|---|
| Wild-type mc2155
|
TAM23 alr mutant
|
|||
| Without d-alanineb | With d-alaninec | Without d-alanineb | With d-alaninec | |
| Amikacin | 1.56 | NDd | 1.56 | 1.56 |
| DCS | 75.0 | 75.0 | 2.56 | 10.2 |
| Ethambutol | 3.13 | ND | 3.13 | 3.13 |
| Rifabutin | 2.00 | ND | 2.00 | 1.00 |
MICs were determined in complete Middlebrook 7H9 medium as described in Materials and Methods. For a given drug, MIC differences between the two strains are considered significant when values correspond to a separation of at least 2 doubling dilutions.
Inoculum was grown in complete Middlebrook 7H9 medium without d-alanine supplementation, as described in Materials and Methods.
Inoculum was grown in complete Middlebrook 7H9 medium supplemented with 50 mM d-alanine, as described in Materials and Methods.
ND, not determined.
This hypersusceptibility indicated by the MIC growth inhibition data was also confirmed by the analysis of the bactericidal action of DCS in broth cultures grown in absence of d-alanine. Pilot experiments were performed with each strain to determine the optimal DCS concentration that resulted in a strong bactericidal effect. In the final experiment, cultures were grown to an early exponential phase (OD600 of ∼0.4), split in two, and DCS was added to one of these subcultures at 50 times the MIC. In absence of DCS, both the wild-type strain mc2155 and the TAM23 mutant cells grew to an OD600 of >2.0 (Fig. 2A) and reached saturation at a cell density approximately above 5.0 × 108 (Fig. 2B). These data also confirmed the independence of M. smegmatis alr mutants on d-alanine for growth and further demonstrate that alr mutants can grow in absence of d-alanine at approximately the same growth rate as wild-type cells. In contrast, in the presence of DCS both cells underwent rapid death by lysis as revealed by both the drastic decrease observed in optical density and viable counts. Furthermore, the kinetic of killing was similar for both the wild-type and mutant strains. However, it must be emphasized that considering absolute drug concentrations, the effect on the mutant strain is observed at a 30-fold-lower concentration than for the wild-type strain. This pattern suggests that the bactericidal action of DCS is due to the inhibition of a more fundamental target different from AlrA.
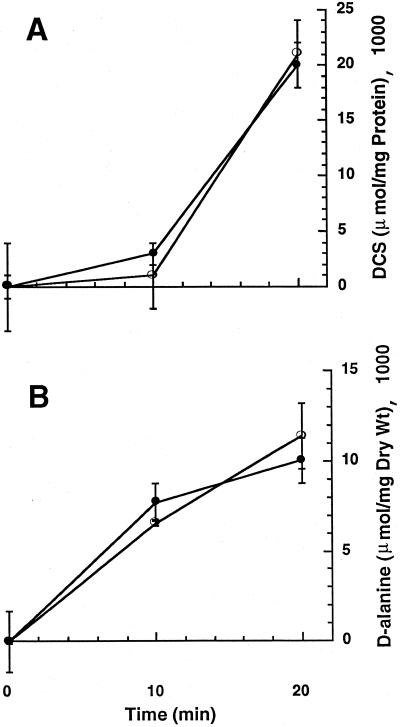
An alternative explanation for the increased susceptibility of the null mutants to DCS is that the inactivation of the alrA gene changes the permeability of the cell wall to DCS. This alteration may not be detectable by morphological studies. To test this hypothesis, we developed a methodology to perform DCS uptake assays. The results obtained were similar for both wild-type and mutant strains (Fig. 3A). We also carried out d-alanine uptake assays using a standardized procedure obtaining similar results for both strains (Fig. 3B). Thus, it is not likely that a permeability change is responsible for the observed phenotype of the null mutants.
FIG. 3.
Uptake of d-alanine and DCS by M. smegmatis wild-type and alr mutant strains. Uptake assays for DCS (A) or d-alanine (B) for strains mc2155 (closed circles) and TAM23 (open circles) were carried out in as described in Materials and Methods. Values are means ± standard deviations (error bars) of triplicate measurements.
DISCUSSION
In this study, it was shown that the M. smegmatis alrA gene can be insertionally inactivated to generate null mutants. PCR and Southern blotting analyses of wild-type and mutant strains confirmed the inactivation of the alrA gene in TAM20 and TAM23 by gene replacement via a double-crossover event between the M. smegmatis chromosome and the linear fragment carrying the inactivated gene. Thus, alrA mutants are viable and can grow in medium without d-alanine. Complementation of the alrA mutant strain with a wild-type alrA copy restores the wild-type phenotype, indicating that properties of the mutant strain are due to the inactivation of the alrA gene rather than a polar effect on the expression of a downstream gene. The independence of M. smegmatis alrA mutants of d-alanine for growth indicates that this mutation does not impose an auxotrophic requirement for d-alanine. This phenotype has significant implications for the synthesis of d-alanine and peptidoglycan in M. smegmatis and possibly other mycobacterial species.
d-Alanine is an essential component for bacteria with a peptidoglycan layer structure. The essentiality of d-alanine stems from the key role of the dipeptide d-alanyl-d-alanine in the cross-linking of peptidoglycan strands (38). The repeating unit of the peptidoglycan from M. smegmatis has a d-alanine moiety (35), and d-alanyl-d-alanine is the only product detected in a biochemical assay using partially purified extracts of M. smegmatis d-alanine ligase (34). These properties support the hypothesis that d-alanine is also an essential component for M. smegmatis. In the context of this hypothesis, the independence of alrA mutants of d-alanine for growth suggests that M. smegmatis may have alternative pathways for the biosynthesis of d-alanine. Listeria monocytogenes, for example, has been shown to possess such metabolic routes (42).
Our studies are consistent with the existence of one d-alanine racemase in M. smegmatis, in contrast to the two alanine racemases identified in E. coli (4, 25) and Salmonella enterica serovar Typhimurium (17, 46, 47). Our Alr assay, as shown by the mixing of active and inactive extracts, was capable of detecting specific activities in the range of 0.002 μmol mg−1 min−1, corresponding to about 4% of the maximal activity obtained with extracts of the wild-type strain. Thus, although our data cannot completely rule out the presence of a second low-level Alr activity, this possibility is not likely. The independence of M. smegmatis alrA mutants from d-alanine for growth differs from the absolute dependency of Lactobacillus plantarum alr mutants (18). This is consistent with the existence of only one pathway for d-alanine biosynthesis in L. plantarum catalyzed by its sole d-alanine racemase, encoded by a single copy of the gene. The likely existence of a second pathway of d-alanine biosynthesis in M. smegmatis would leave unresolved whether endogenous d-alanine biosynthesis is or is not an essential function. In contrast, endogenous biosynthesis of diaminopimelate was shown to be an essential function of M. smegmatis since ask single mutants, auxotrophic for diaminopimelate, could not be obtained even in medium supplemented with this nutrient (32). It remains to be tested if the inactivation of the gene(s) responsible for a putative second pathway of d-alanine biosynthesis in an alrA− background would render viable mutants.
The uptakes of d-alanine and DCS in both wild-type and mutant strains were not significantly different, indicating that changes in cell wall permeability are not a likely explanation for the DCS hypersusceptible phenotype of the alr mutant strain (Fig. 3). Thus, the hypersusceptibility of TAM23 cells to DCS is consistent with the existence of multiple targets for DCS. Our previous studies identified d-alanine racemase as one of these targets (5). The lack of the racemase protein in the alrA mutant strain may lead to a hypersusceptible phenotype since more DCS would be required to inhibit both the racemase and an additional target(s) in the wild-type strain. In contrast, the bactericidal effect of DCS suggests the existence of another lethal target. In this context, d-alanine ligase is an attractive candidate since this enzyme activity is also inhibited by DCS (13). More importantly, the construction of a conditionally lethal mutant bank led to the isolation of a thermosensitive mutant impaired in this gene function (3). Furthermore, DCS hypersusceptibility may reflect an alteration of the peptidoglycan structure of TAM23 cells, as a direct consequence of the inactivation of the alr gene. In this context, TAM23 cells were also more susceptible to DCS than were wild-type cells when grown in medium with d-alanine, but mutant cells grown without d-alanine became hypersusceptible to both growth inhibition and the bactericidal action of DCS (Table 3; Fig. 2).
In summary, the d-alanine independent phenotype of M. smegmatis alrA mutants suggests that M. smegmatis has another pathway of d-alanine biosynthesis. Given the conservation of basic physiological processes, this finding could also be of significance for pathogenic mycobacteria and the design of attenuated strains and antimycobacterial agents.
Acknowledgments
Research was supported by funds from the University of Nebraska Department of Veterinary and Biomedical Sciences, the Nebraska Agriculture Experiment Station Interdisciplinary Research award, the Texas Agricultural Experimental Station-Texas Cattle and Deer Tuberculosis Management Plan, and USDA Cooperative State Research Service Project NEB 14-108. O.C. was also partially supported by loan fellowships from Colciencias and Colfuturo (Bogotá, Colombia). Z.F. is a recipient of the Maude Hammond Fling-Bukey Memorial Fund fellowship from the University of Nebraska-Lincoln Graduate Studies Program.
We thank T. A. Ficht, D. N. McMurray, and A. R. Rice-Ficht for useful discussions. We thank J. Zabaleta for technical assistance in preliminary experiments.
Footnotes
Journal Series no. 13366, Agricultural Research Division, University of Nebraska—Lincoln.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 2.Belanger, A. E., and J. M. Inamine. 2000. Genetics of cell wall biosynthesis, p.191–202. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 3.Belanger, A. E., J. C. Porter, and G. F. Hatfull. 2000. Genetic analysis of peptidoglycan biosynthesis in mycobacteria: characterization of a ddlA mutant of Mycobacterium smegmatis. J. Bacteriol. 182:6854–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., V. Burland, G. Plunkett III, H. J. Sofia, and D. L. Daniels. 1993. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 21:5408–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cáceres, N. E., N. B. Harris, J. F. Wellehan, Z. Feng, V. Kapur, and R. G. Barletta. 1997. Overexpression of the d-alanine racemase gene confers resistance to d-cycloserine in Mycobacterium smegmatis. J. Bacteriol. 179:5046–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cáceres, N. E. 1999. Ph.D. thesis. University of Nebraska, Lincoln.
- 6.Chaisson, R. E., C. A. Benson, M. P. Dube, L. B. Heifets, J. A. Korvick, S. Elkin, T. Smith, J. C. Craft, and F. R. Sattler. 1994. Clarithromycin therapy for bacteremic Mycobacterium avium complex disease. A randomized, double-blind, dose-ranging study in patients with AIDS. AIDS Clinical Trials Group Protocol 157 Study Team. Ann. Intern. Med. 121:905–911. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, D. 1997. The mycobacterial cell wall: structure, biosynthesis and sites of drug action. Curr. Opin. Chem. Biol. 1:579–588. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo J. D., T. R. Weisbrod, L. Pascopella, B. R. Bloom, and W. R. Jacobs, Jr. 1994. Isolation and characterization of the aspartokinase and aspartate semialdehyde dehydrogenase operon from mycobacteria. Mol. Microbiol. 11:629–639. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, D. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, R. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, M. M., R. A. Patnode, and P. C. Hudgins. 1955. Effects of cycloserine on Mycobacterium tuberculosis in vitro. Anibiot. Chemother. 5:198–203. [PubMed] [Google Scholar]
- 11.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131–203. [DOI] [PubMed] [Google Scholar]
- 12.David, H. L. 1971. Resistance to D-cycloserine in the tubercle bacilli: mutation rate and transport of alanine in parental cells and drug-resistant mutants. Appl. Microbiol. 21:888–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David, H. L., T. Takayama, and D. S. Goldman. 1969. Susceptibility of mycobacterial D-alanyl-D-alanine synthetase to D-cycloserine. Am. Rev. Respir. Dis. 100:579–581. [DOI] [PubMed] [Google Scholar]
- 14.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677–686. [DOI] [PubMed] [Google Scholar]
- 15.Espinal, M. A., S. J. Kim, P. G. Suarez, K. M. Kam, A. G. Khomenko, G. B. Migliori, J. Baez, A. Kochi, C. Dye, and M. C. Raviglione. 2000. Standard short-course chemotherapy for drug-resistant tuberculosis: treatment outcomes in 6 countries. JAMA 283:2537–2545. [DOI] [PubMed] [Google Scholar]
- 16.Foley-Thomas, E. M., D. L. Whipple, L. E. Bermudez, and R. G. Barletta. 1995. Phage infection, transfection, and transformation of Mycobacterium avium complex and M. paratuberculosis. Microbiology 141:1173–1181. [DOI] [PubMed] [Google Scholar]
- 17.Galakatos, N. G., E. Daub, D. Botstein, and W. T. Walsh. 1986. Biosynthetic alr alanine racemase from Salmonella typhimurium: DNA and protein sequence determination. Biochemistry 25:3255–3260. [DOI] [PubMed] [Google Scholar]
- 18.Hols, P., C. Defrenne, T. Ferain, S. Derzelle, B. Delplace, and J. Delcour. 1997. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J. Bacteriol. 179:3804–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 3:266–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs, W. R., Jr. 2000. Mycobacterium tuberculosis: a once genetically intractable organism, p.1–36. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 21.Jones, L. R. 1956. Colorimetric determination of cycloserine, a new antibiotic. Anal. Chem. 28:39–41. [Google Scholar]
- 22.Julius, M., C. A. Free, and G. T. Barry. 1970. Alanine racemase (Pseudomonas). Methods Enzymol. 17:171–176. [Google Scholar]
- 23.Lambert, M. P., and F. C. Neuhaus. 1972. Mechanism of d-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. H., and G. F. Hatfull. 1993. Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J. Bacteriol. 175:6836–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobocka, M., J. Hennig, J. Wild, and T. Klopotowski. 1994. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J. Bacteriol. 176:1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNeil, M. R., and P. J. Brennan. 1991. Structure, function, and biogenesis of the cell envelope of mycobacteria in relation to bacterial physiology, pathogenesis and drug resistance; some thoughts and possibilities arising from recent structural information. Res. Microbiol. 142:451–463. [DOI] [PubMed] [Google Scholar]
- 27.Neuhaus, F. C. 1962. The enzymatic synthesis of D-alanyl-D-alanine. I. Purification and properties of D-alanyl-D-alanine synthetase. J. Biol. Chem. 237:778–786. [PubMed] [Google Scholar]
- 28.Neuhaus, F. C. 1967. D-cycloserine and O-carbamyl-D-serine, p.40–83. In D. Gottlieb and P. L. Shaw (ed.), Antibiotics: mechanisms of action, vol. 1. Springer-Verlag, Heidelberg, Germany.
- 29.Neuhaus, F. C., and J. L. Lynch. 1964. The enzymatic synthesis of D-alanyl-D-alanine. III. On the inhibition of D-alanyl-D-alanine synthetase by the antibiotic D-cycloserine. Biochemistry 3:471–480. [DOI] [PubMed] [Google Scholar]
- 30.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217–226. [DOI] [PubMed] [Google Scholar]
- 31.Pascopella, L., F. M. Collins, J. M. Martin, M. H. Lee, G. F. Hatfull, C. K. Stover, B. R. Bloom, and W. R. Jacobs, Jr. 1994. Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect. Immun. 62:1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavelka, M. S., and W. R. Jacobs, Jr. 1996. Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J. Bacteriol. 178:6496–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavelka, M. S., and W. R. Jacobs, Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guérin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peteroy, M., A. Severin, F. Zhao, D. Rosner, U. Lopatin, H. Scherman, A. Belanger, B. Harvey, G. F. Hatfull, P. J. Brennan, and N. D. Connell. 2000. Characterization of a Mycobacterium smegmatis mutant that is simultaneously resistant to D-cycloserine and vancomycin. Antimicrob. Agents Chemother. 44:1701–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petit, J. F., A. Adam, J. Wietzerbin-Falszpan, E. Lederer, and J. M. Ghuysen. 1969. Chemical structure of the cell wall of Mycobacterium smegmatis. I. Isolation and partial characterization of the peptidoglycan. Biochem. Biophys. Res. Commun. 35:478–485. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911–1919. [DOI] [PubMed] [Google Scholar]
- 38.Strominger, J. L. 1962. Biosynthesis of bacterial cell walls, p.413–470. In I. C. Gunzalus and R. Y. Stanier (ed.), The bacteria, vol. III. Academic Press, Inc., New York, N.Y.
- 39.Strych, U., R. L. Penland, M. Jimenez, K. L. Krause, and M. J. Benedik. 2001. Characterization of the alanine racemases from two mycobacteria. FEMS Microbiol. Lett. 196:93–98. [DOI] [PubMed] [Google Scholar]
- 40.Takiff, H. E., M. Cimino, M. C. Musso, T. Weisbrod, R. Martinez, M. B. Delgado, L. Salazar, B. R. Bloom, and W. R. Jacobs, Jr. 1996. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 93:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Telenti, A., W. J. Philipp, S. Sreevatsan, C. Bernasconi, K. E. Stockbauer, B. Wieles, J. M. Musser, and W. R. Jacobs, Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3:567–570. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, R. J., H. G. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocitogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trias, J., and R. Benz. 1994. Permeability of the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 14:283–290. [DOI] [PubMed] [Google Scholar]
- 44.Van Heijenoort, J. 1996. Murein synthesis, p.1025–1034. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 45.Walsh, C. T. 1989. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem. 264:2393–2396. [PubMed] [Google Scholar]
- 46.Wasserman, S. A., E. Daub, P. Grishif, D. Botstein, and C. T. Walsh. 1984. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry 23:5182–5187. [DOI] [PubMed] [Google Scholar]
- 47.Wasserman, S. A., C. T. Walsh, and D. Botstein. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wijsman, H. J. W. 1972. The characterization of an alanine racemase mutant of Escherichia coli. Genet. Res. 20:269–277. [DOI] [PubMed] [Google Scholar]
- 49.Yew, W. W., C. F. Wong, P. C. Wong, J. Lee, and C. H. Chau. 1993. Adverse neurological reactions in patients with multidrug-resistant pulmonary tuberculosis after coadministration of cycloserine and ofloxacin. Clin. Infect. Dis. 17:288–289. [DOI] [PubMed] [Google Scholar]
- 50.Zygmunt, W. A. 1963. Antagonism of d-cycloserine inhibition of mycobacterial growth by d-alanine. J. Bacteriol. 85:1217–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]