Abstract
Mechanisms of resistance were studied in 22 macrolide-resistant mutants selected in vitro from 5 parental strains of macrolide-susceptible Streptococcus pneumoniae by serial passage in various macrolides (T. A. Davies, B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum, Antimicrob. Agents Chemother., 44:414–417, 2000). Portions of genes encoding ribosomal proteins L22 and L4 and 23S rRNA (domains II and V) were amplified by PCR and analyzed by single-strand conformational polymorphism analysis to screen for mutations. The DNA sequences of amplicons from mutants that differed from those of parental strains by their electrophoretic migration profiles were determined. In six mutants, point mutations were detected in the L22 gene (G95D, P99Q, A93E, P91S, and G83E). The only mutant selected by telithromycin (for which the MIC increased from 0.008 to 0.25 μg/ml) contained a combination of three mutations in the L22 gene (A93E, P91S, and G83E). L22 mutations were combined with an L4 mutation (G71R) in one strain and with a 23S rRNA mutation (C2611A) in another strain. Nine other strains selected by various macrolides had A2058G (n = 1), A2058U (n = 2), A2059G (n = 1), C2610U (n = 1), and C2611U (n = 4) mutations (Escherichia coli numbering) in domain V of 23S rRNA. One mutant selected by clarithromycin and resistant to all macrolides tested (MIC, >32 μg/ml) and telithromycin (MIC, 4 μg/ml) had a single base deletion (A752) in domain II. In six remaining mutants, no mutations in L22, L4, or 23S rRNA could be detected.
Resistance to macrolides is increasingly reported in clinical isolates of Streptococcus pneumoniae worldwide (10, 14). Resistance to macrolides was primarily related to modification of the ribosomal targets of these antibiotics. This mechanism relies on N-6 dimethylation of a specific adenine residue in 23S rRNA which confers cross-resistance to macrolides, lincosamides, and streptogramins B, the so-called MLSB phenotype, and is encoded in pneumococci by genes belonging to the erm(B) or erm(A) class (11, 18, 26). Subsequently, target modification was reported in the majority if not all macrolide-resistant pneumococci (11, 26). More recently, a mechanism of resistance by active efflux of erythromycin due to the mefE gene, renamed mef(A) (18), was reported in S. pneumoniae and appeared to be predominant in the United States and Canada, with prevalences ranging from 41 to 85% (9, 19, 20). The efflux phenotype, which is also called the M phenotype, is characterized by resistance to 14-membered-ring (erythromycin, clarithromycin, and roxithromycin) and 15-membered-ring (azithromycin) macrolides only. Ribosomal mutation has been reported only recently in a few clinical isolates of S. pneumoniae (5, 21, 22). The changes were clustered in a highly conserved sequence of L4 and in nucleotide residues of domain V of 23S rRNA which have a key role in macrolide binding. A recent study by Davies et al. (4) showed that mutants can readily be selected in pneumococci in the presence of any of several MLSB antibiotics in vitro. Those investigators isolated 26 mutants from 5 parent strains susceptible to macrolides. The aim of the present study was to characterize the mechanisms of macrolide resistance in these strains.
MATERIALS AND METHODS
Bacterial strains.
In the present study we examined 22 erythromycin-resistant mutants derived from 5 susceptible S. pneumoniae parent strains (strains 1, 2, 3, 4, and 5) obtained from Hershey Medical Center (4). Briefly, the mutants were obtained by serial passages in the presence of subinhibitory concentrations of macrolides (azithromycin, clarithromycin, erythromycin, and roxithromycin), a ketolide (telithromycin), or a lincosamide (clindamycin). Resistance remained stable in mutants after 10 consecutive subcultures on antibiotic-free medium. Of the 27 mutants selected in the original study, 4 could not be subcultured from frozen collection tubes. The MICs for the mutants were determined following the recommendations of the National Committee for Clinical Laboratory Standards (13). The authenticities of the mutants were checked by determination of the MICs before analysis of mutations. The passage-derived mutants did not contain the erm(B) or the mef(A) gene. These genes usually confer macrolide resistance in pneumococci (4). S. pneumoniae laboratory strain CP1000, which is susceptible to erythromycin (MIC, 0.05 μg/ml), was used as a recipient strain in transformation experiments (12).
MIC determinations.
In transformation experiments, the MICs of the antibiotics for the pneumococci were determined by the agar dilution method with an inoculum of 104 cells per spot on Mueller-Hinton medium (Bio-Rad, Marnes la Coquette, France) supplemented with 5% sheep blood (3). The plates were incubated for 24 h at 37°C in 5% CO2. The following antibiotics were provided by their manufacturers: azithromycin (Pfizer, Orsay, France), clarithromycin (Abbott, St. Rémy sur Avre, France), clindamycin (Pharmacia Upjohn, Val de Reuil, France), erythromycin (Sigma Chemical Co., St. Louis, Mo.), and pristinamycin and telithromycin (Aventis, Paris, France).
PCR conditions.
The genomic DNA was extracted with an Instagene Matrix kit (Bio-Rad Laboratories, Hercules, Calif.). Nucleotide sequences for 23S rRNA and L4 and L22 ribosomal proteins in Escherichia coli and S. pneumoniae were obtained from The Institute for Genomic Research website (http://www.tigr.org.). Specific oligonucleotide primers were designed from these sequences. Primer sequences and conditions for PCR amplifications are shown in Table 1.
TABLE 1.
Primer sequences and PCR conditions used in this study
| Gene (ribosomal protein or rRNA) | Primer position (nucleotide no.) | Primer sequence | Product size (bp) | PCR conditions
|
|
|---|---|---|---|---|---|
| MgCl2 concn (mM) | Amplification conditions | ||||
| rplV (L22) | −41a | 5′-GCAGACGACAAGAAAACACG-3′ | 437 | 1 | 1 cycle of 3 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 61°C, and 30 s at 72°C; 1 cycle of 10 min at 72°C |
| +396a | 5′-GCCGACACGCATACCAATTG-3′ | ||||
| rplD (L4) | −93a | 5′-AAAGGTAACGTACCAGGTGC-3′ | 478 | ||
| +385a | 5′-GCGTGGTGGTGGTGTTG-3′ | ||||
| 5′-CACGAGTGTCAACTTCAAATAC-3′ | 472 | Same as for L22 | |||
| +248a | 5′-GAGCGTCTACAGCTACG-3′ | ||||
| +720a | |||||
| rrl (23S rRNA domain II) | 578–850b | 5′-CGGCGAGTTACGATTATGATGC-3′ | 273 | 2 | 1 cycle of 3 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 59°C, and 30 s at 72°C; 1 cycle of 10 min at 72°C |
| 5′-CTCTAATGTCGACGCTAGCC-3′ | |||||
| rrl (23S rRNA domain V) | 1990–2134b | 5′-CTGTCTCAACGAGAGACTC-3′ | 144 | 1.5 | 1 cycle of 3 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 57°C, and 30 s at 72°C; 1 cycle of 10 min at 72°C |
| 5′-CTTAGACTCCTACCTATCC-3′ | |||||
| 2331–2769b | 5′-GTATAAGGGAGCTTGACTG-3′ | 439 | 1.5 | 1 cycle of 3 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 51°C, and 30 s at 72°C; 1 cycle of 10 min at 72°C | |
| 5′-GGGTTTCACACTTA GATG-3′ | |||||
Base relative to ATG.
E. coli numbering.
DNA sequencing.
PCR products were purified through a spin column and were sequenced by the Rhodamine dye terminator method with an ABI Prism 377 sequencer (Perkin-Elmer Corp., Norwalk, Conn.) The oligonucleotides used for PCR were also used as primers for DNA sequencing.
PCR-SSCP analysis.
The amplimers were analyzed by single-strand conformational polymorphism (SSCP) analysis. Aliquots of 20 μl of H2O containing 20 ng of PCR product were mixed with 20 μl of denaturant solution (95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol, 20 mM EDTA). The mixture was heated for 10 min at 100°C and cooled on ice, and the single-strand PCR product was then separated by nondenaturant polyacrylamide gel electrophoresis (10% acrylamide 29-bisacrylamide 1 in Tris-borate-EDTA buffer) by using a vertical slab gel unit (model SE 400; Hoefer Scientifics Instruments, San Francisco, Calif.). The gel was run for 12 to 15 h at 200 V and 4°C. The bands were then visualized by ethidium bromide staining.
Transformation experiments.
To assess the role of L22 mutations in antibiotic resistance, three resistant alleles of the L22 gene (rplV) were amplified and introduced into CP1000 by transformation, as described previously (12, 17). Competent cells were prepared as follows. A single colony of S. pneumoniae CP1000 was grown in 10 ml of caseine hydrolysate yeast extract tryptone medium (CAT) supplemented with 0.2% glucose and 15 mM K2HPO4 at 37°C (CATI). A 100-μl aliquot of an overnight culture was inoculated in 10 ml of CATI supplemented with 0.2% neutral bovine serum albumin and 1 mM CaCl2 (CATII). The broth culture was incubated to an optical density at 550 nm of 0.15. The cells were centrifuged at 3,000 × g for 10 min; the pellet was then resuspended in 1 ml of CATII with 15% glycerol. The competent cells were split into 100-μl aliquots, and the aliquots were stored at −80°C. For transformation, competent cells were thawed on ice and 10 μl was added to 10 ml of CATII that had been adjusted to pH 7.8; the mixture was then incubated at 37°C for 15 min. Purified PCR products (10 μl) were added to 1 ml of the culture, and the reaction mixture was incubated at 37°C to an optical density at 550 nm of 0.2. One hundred microliters was then plated onto 20 ml of Trypticase soy agar containing 5% sheep blood, and the plate was incubated for 2 h at 37°C to allow expression of resistance before 10 ml of selective medium containing erythromycin (1.5 μg/ml) was overlaid. The transformants were extracted from the agar and isolated on Trypticase soy agar plates supplemented with erythromycin (0.5 μg/ml) and 5% sheep blood; the plates were then incubated for 48 h at 37°C with CO2. The MICs of the macrolides for the transformants were determined. The rplV genes of the transformants were amplified and sequenced.
RESULTS
Screening of mutations by PCR-SSCP analysis.
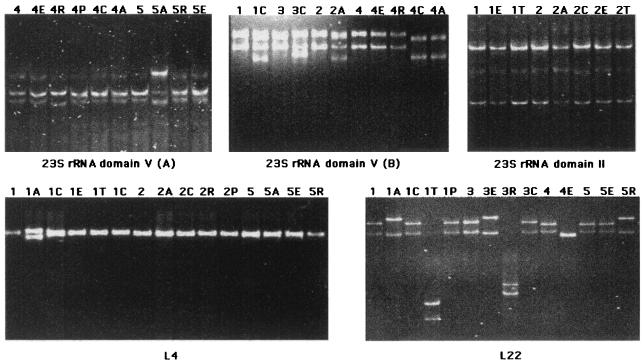
PCR-SSCP analysis is a rapid and convenient technique for detection of mutations and allelic variants (16). We used this technique to screen for mutations in 23S rRNA and ribosomal proteins L4 and L22. Since better discrimination between mutated alleles is obtained for denatured DNA fragments with sizes between 150 and 500 bp, portions of the rrl gene (domains II and V of 23S rRNA), the entire rplV gene, and two overlapping fragments of the L4 gene (rplD) were amplified (Table 1). The three fragments amplified from rrl, two for domain V and one for domain II, included bases critical for erythromycin resistance: G2057, A2058, A2062, G2505, C2611, and A752. The PCR-SSCP analysis was carried out for each parent strain and the corresponding mutants (Fig. 1). The migration patterns of the rplD and the rrl (domains V and II) genes from all parent strains were identical, confirming that the amplified regions are highly conserved within the species S. pneumoniae. In contrast, differences in L22 migration patterns were observed which were related to a silent mutation at codon N72 (C216T) in strains 2, 4, and 5, while the sequences of strains 1 and 3 were identical to that of the reference strain S. pneumoniae type 4 (http://www.tigr.org.). The migration patterns of the passage mutants were compared to those of the corresponding parent strains. Sequencing showed that the different mobilities in the mutants were associated with point mutations or deletions. The sequences of some DNA fragments from parents and mutants with identical profiles did not show any differences, confirming the specificity of the PCR-SSCP technique. Overall, analysis of migration profiles suggested that nine mutants had mutations in domain V, one mutant had a mutation in domain II, four mutants had mutations in rplV, and two strains displayed combinations of mutations, one in the rplV gene and the rplD gene and the other in the rplV gene and in domain V. The sequences of the amplified fragments were determined and showed that differences in migration profiles were related to mutations. Detailed results are presented in the following paragraphs and in Table 2. No difference in the migration profiles was observed for six mutants.
FIG. 1.
Analysis of DNAs from parental strains and mutants by PCR-SSCP. Two portions of the rrl gene corresponding to 23S rRNA domain V (from nucleotides 1990 to 2134 [upper left] and from nucleotides 2331 to 2769 [upper middle]), a portion of the rrl gene for domain II of 23S rRNA (upper right), a part of the L4 gene, rplD (lower left), and the entire L22 gene, rplV (lower right), were amplified by PCR. Examples of the migration patterns obtained for these genes after separation of the denaturated PCR product by nondenaturant polyacrylamide gel electrophoresis are shown. The DNAs from the parent strains are designated by a number; the DNAs from the corresponding mutants are designated by the same number followed by the initial of the selector antibiotic: A, azithromycin; C, clarithromycin; E, erythromycin; R, roxithromycin; P, pristinamycin; T, telithromycin. Migration patterns for mutants 5A in the upper left gel, mutants 1C, 3C, 2A, 4C, and 4A in the upper middle gel, mutant 2C in the upper right gel, mutant 1A in the lower left gel, and mutants 1A, 1T, 3E, 3R, 4E, and 5R in the lower right gel (where C, A, T, E, and R correspond to clarithromycin, azithromycin, telithromycin, erythromycin, and roxithromycin, respectively) differ from the patterns of the parent DNAs.
TABLE 2.
Ribosomal target mutations and MICs of macrolides and related antibiotics for parent and mutant strains
| Straina | Relevant mutation (heterozygosity) | MIC (μg/ml) (increase in MIC)b
|
||||||
|---|---|---|---|---|---|---|---|---|
| AZM | CLR | ERY | ROX | TEL | PRI | CLX | ||
| Parent | ||||||||
| 1 | None | 0.03 | 0.015 | 0.03 | 0.06 | 0.008 | 0.5 | 0.03 |
| 2 | None | 0.06 | 0.03 | 0.06 | 0.125 | 0.008 | 0.25 | 0.125 |
| 3 | None | 0.03 | 0.015 | 0.03 | 0.06 | 0.008 | 0.5 | 0.03 |
| 4 | None | 0.015 | 0.015 | 0.015 | 0.03 | 0.004 | 0.25 | 0.03 |
| 5 | None | 0.015 | 0.015 | 0.015 | 0.06 | 0.004 | 0.5 | 0.03 |
| Mutants | ||||||||
| 23SrRNA | ||||||||
| 1 CLR | A2058U (1A:3U) | >32 (>1,000) | 16 (>1,000) | >32 (>1,000) | >32 (>500) | 1 (128) | 0.5 (1) | 1 (32) |
| 3 CLR | A2058U (1A:3U) | >32 (>1,000) | >32 (>2,000) | >32 (>1,000) | >32 (>500) | 0.25 (32) | 0.5 (1) | 1 (32) |
| 5 AZM | A2058G (1A:3G) | >32 (>2,000) | >32 (>2,000) | >32 (>2,000) | >32 (>500) | 0.06 (16) | 0.5 (1) | 4 (133) |
| 4 ROX | A2059G (0A:4G) | >32 (>2,000) | 2 (133) | 8 (500) | 16 (500) | 0.015 (4) | 0.25 (1) | 2 (66) |
| 3 CLX | C2610U (2C:2U) | 0.125 (4) | 0.03 (2) | 0.06 (2) | 0.125 (2) | 0.008 (1) | 0.5 (1) | 0.5 (16) |
| 1 CLX | C2611U (2C:2U) | 0.5 (16) | 0.03 (2) | 0.06 (2) | 0.125 (2) | 0.08 (10) | 0.5 (1) | 0.5 (16) |
| 4 CLX | C2611U (0C:4U) | 0.5 (32) | 0.06 (4) | 0.06 (4) | 0.5 (16) | 0.015 (4) | 0.5 (2) | 2 (66) |
| 4 AZM | C2611U (0C:4U) | 0.5 (32) | 0.06 (4) | 0.125 (8) | 0.06 (2) | 0.015 (4) | 0.5 (2) | 2 (66) |
| 2 AZM | C2611U (2C:2U) | 0.5 (8) | 0.06 (2) | 0.125 (2) | 0.125 (1) | 0.08 (10) | 0.5 (2) | 2 (16) |
| 2 CLR | A 752 Deletion | >32 (500) | >32 (1,000) | >32 (500) | >32 (256) | 4 (500) | 1 (4) | 1 (8) |
| L22 ribosomal protein | ||||||||
| 3 ERY | G95D | 1 (32) | 1 (64) | 1 (32) | — | 0.12 (16) | 1 (2) | 0.125 (4) |
| 5 ROX | G95D | 0.06 (4) | 0.125 (8) | 0.25 (16) | 0.5 (8) | 0.125 (32) | 2 (4) | 0.03 (1) |
| 4 ERY | P99Q | 0.125 (8) | 0.125 (8) | 0.25 (16) | 0.5 (16) | 0.06 (16) | 1 (4) | 0.125 (4) |
| 1 TEL | A93E, P91S, G83E | 0.25 (8) | 0.25 (16) | 0.5 (16) | 2 (32) | 0.25 (32) | 2 (4) | 0.03 (1) |
| Mutants with combined mutations | ||||||||
| 1 AZM | L22, G95D | 0.5 (16) | 0.125 (8) | 0.25 (8) | 0.5 (4) | 0.06 (8) | 1 (2) | 0.06 (2) |
| L4, G71R | ||||||||
| 3 ROX | L22, A93E | 0.5 (16) | 0.5 (32) | 1 (32) | 2 (32) | 0.125 (16) | 1 (2) | 0.06 (2) |
| 23S, C2611A (1C:3A) | ||||||||
Strains 1, 2, 3, 4, and 5 are macrolide-susceptible parent strains; mutants are designated by the parent strain number followed by the selector antibiotic.
AZM, azithromycin; CLR, clarithromycin; ERY, erythromycin; ROX, roxithromycin; TEL, telithromycin; PRI, pristinamycin; CLX, clindamycin; —, not determined. The increase in the MIC was expressed for each macrolide as a ratio of the MIC for the mutant to the MIC for the parent strain.
23S rRNA mutations.
Among the nine mutants with point mutations in domain V, one selected from passage in azithromycin had an A2058G mutation, two selected from passage in clarithromycin had an A2058U substitution, one selected from passage in roxithromycin had an A2059G mutation, four selected from passage in azithromycin and clindamycin had a C2611U mutation, and one selected from passage in clindamycin had a C2610U mutation. Conversion of A2058 to guanine and transversion of A2058 to uridine yielded significant increases in the MICs of 14- and 15-membered macrolides (MICs, >32 μg/ml). The activities of clindamycin and telithromycin were moderately affected, whereas pristinamycin remained active.
The A2059G mutant was resistant to erythromycin, azithromycin, and roxithromycin, with MICs for the mutant equal to or greater than 8 μg/ml. Although smaller increases in the MICs of clarithromycin and clindamycin (MICs, 2 μg/ml) were noted, the strain was categorized as resistant to these antimicrobials. Pristinamycin and telithromycin remained active, with MICs equal to 0.25 and 0.015 μg/ml, respectively.
The C2611U and C2610U substitutions had small impacts on the activities of macrolides and clindamycin and did not lead to categorization of the strains as resistant to these antimicrobials.
One mutant selected by clarithromycin had a deletion of one adenine in the series of four located at positions 749 to 752 in the hairpin 35 of domain II. The MICs of macrolides (erythromycin, clarithromycin, and roxithromycin) increased about 250- to 1,000-fold (MICs, >32 μg/ml). The telithromycin MIC for this strain also increased significantly (500-fold), and the strain was resistant to this antimicrobial (MIC, 4 μg/ml). The MIC of clindamycin was increased only 10 times, which was sufficient to categorize the strain as resistant (MIC, 1 μg/ml). The activity of pristinamycin was only partially altered, with a fourfold higher MIC (1 μg/ml).
Mutations in ribosomal protein L22.
Six mutants selected from passage in azithromycin, erythromycin, roxithromycin, and telithromycin contained mutations in L22 protein. Four patterns were detected by SSCP analysis, and these reflected the presence of four different mutations (Fig. 1). The locations of the mutations are shown in Table 3. Three mutants had a point mutation (G284A) that led to a single amino acid change from glycine to aspartic acid at position 95 (G95D) (S. pneumoniae numbering); two others presented a P99Q substitution and an A93E substitution, corresponding to point mutations C296A and C278A in the rrlV gene, respectively. The last mutant with an L22 mutation was the only one selected in the presence of telithromycin and had three point mutations in combination, resulting in three substitutions (A93E, P91S, and G83E) in the amino acid sequence. The MICs of azithromycin, clarithromycin, erythromycin, and roxithromycin for the four mutants with only an rplV mutation were from 4- to 64-fold higher than those for the wild-type strains. The clindamycin MIC was increased one to four times but remained in the susceptible range. Significant increases in the MICs of pristinamycin, which were equal to 1 or 2 μg/ml, were seen for all L22 mutants. Although the MIC of telithromycin was multiplied by 16 or 32, all mutants, including that selected from passage in telithromycin, were still susceptible to the ketolide.
TABLE 3.
Mutations in L22 ribosomal proteins
| Straina | C-terminal conserved sequence of L22 proteinb |
|---|---|
| Wild type | 69DKANLVVSEAFANEGPTMKRFRPRAKGSASPINKRTAHITVAVAEK114 |
| 1 AZM | 69DKANLVVSEAFANEGPTMKRFRPRAKDSASPINKRTAHITVAVAEK114 |
| 3 ERY | 69DKANLVVSEAFANEGPTMKRFRPRAKDSASPINKRTAHITVAVAEK114 |
| 5 ROX | 69DKANLVVSEAFANEGPTMKRFRPRAKDSASPINKRTAHITVAVAEK114 |
| 4 ERY | 69DKANLVVSEAFANEGPTMKRFRPRAKGSASQINKRTAHITVAVAEK114 |
| 3 ROX | 69DKANLVVSEAFANEGPTMKRFRPREKGSASPINKRTAHITVAVAEK114 |
| 1 TEL | 69DKANLVVSEAFANEEPTMKRFRSREKGSASPINKRTAHITVAVAEK114 |
The mutant designations are explained in footnote a of Table 2. AZM, azithromycin; ERY, erythromycin; ROX, roxithromycin; TEL, telithromycin.
The mutations in the L22 proteins are indicated in boldface and underlined.
Role of L22 protein in MLS resistance.
To assess the effects of mutations of the L22 protein on the activities of macrolides, lincosamides, and streptogramins (MLS), we amplified by PCR the entire L22 gene from mutants harboring three different types of mutations (G95D, P99Q, and A93E) and the triple mutation (A93E-P91S-G83E). The amplified fragments were then introduced by transformation into S. pneumoniae CP1000. Transformants with all amplified fragments except that containing the P99Q mutation could be selected on agar plates containing low concentrations of erythromycin. The L22 genes of the transformants were reamplified by PCR and sequenced to confirm that the expected amino acid substitutions were present. The MICs of the MLS antibiotics for the transformants were similar to those for the donor mutants (Table 4). Regardless of the type of mutation, including the triple mutation, the activities of the macrolides and clindamycin were only slightly altered. By contrast, the increases in the MICs of pristinamycin for the transformants were approximately 20 to 30 times, even greater than those for the passage mutants, confirming the role of L22 mutations in streptogramin resistance.
TABLE 4.
MICs of antibiotics for S. pneumoniae CP1000 and transformants
| Donor straina | Recipient strain or transformant (L22 mutation) | MIC (μg/ml)b
|
|||||
|---|---|---|---|---|---|---|---|
| ERY | CLR | AZM | TEL | CLX | PRI | ||
| NA | CP1000 (none) | <0.015 | <0.015 | <0.015 | 0.03 | <0.015 | 0.06 |
| 1 AZM | CP1000 (G95D) | 0.5 | 0.125 | 0.5 | 0.25 | 0.03 | 2 |
| 3 ERY | CP1000 (G95D) | 0.5 | 0.125 | 0.25 | 0.125 | 0.06 | 1 |
| 3 ROX | CP1000 (A93E) | 0.5 | 0.125 | 0.5 | 0.25 | 0.03 | 1 |
| 1 TEL | CP1000 (A93E, P91S, G83E) | 0.5 | 0.125 | 0.5 | 0.25 | 0.03 | 2 |
Combined mutations in different ribosomal targets.
The results of SSCP analysis led to the suspicion that combinations of mutations are present in ribosomal proteins and/or rRNA in two strains, i.e., L4 and L22 mutations and L22 and domain V mutations. The sequences of the PCR fragments confirmed this suspicion. In one mutant, selected by passage in azithromycin, a G211A mutation that led to a single amino acid change from glycine to arginine at position 71 (S. pneumoniae numbering) of L4 was combined with a G95D mutation in the L22 protein. The second strain, selected by passage in roxithromycin, had an A93E mutation in the L22 protein combined with a C2611A mutation in domain V of 23S rRNA. As shown in Table 2, a combination of mutations did not contribute in a more than additive fashion to macrolide resistance. Strain 3 ERY had the same G95D mutation in the L22 protein as strain 5 ROX, but the MICs for strain 3 ERY were higher, suggesting a combination of that mutation with another mutation. Introduction of the rplV allele from mutant 3 ERY into S. pneumoniae CP1000 yielded macrolide MICs similar to those for the other transformants, confirming that this mutation alone does not explain the macrolide resistance level of strain 3 ERY (Table 4). However, the nucleotide sequences of domains V and II of 23S rRNA and of the rplD gene were identical to the sequences of those regions of parent strain 3.
DISCUSSION
Study of a large number of mutants selected in the presence of various macrolides revealed that mutation of a variety of structures including domains V and II of 23S rRNA and proteins L22 and L4, which are part of the binding sites of macrolides, could be responsible for resistance to MLS antibiotics. Mutations in domain V of 23S rRNA were the most frequent, in particular, substitutions of A2058, A2059, and C2611. Similar mutations have been identified in erythromycin-resistant strains belonging to a wide variety of species including clinical isolates (A2059G, A2062C) and in vitro mutants (A2058G, C2611A, C2611G) of S. pneumoniae (5, 21, 22, 25). Here we have reported on a C2610U mutation which conferred a small increase in the MICs of macrolides and clindamycin. To our knowledge, this mutation has never been reported in S. pneumoniae or any other microorganism. The role of the mutation in macrolide resistance remains to be confirmed.
The phenotype conferred by modification of the 23S rRNA target varies according to the mutated base. Change of the adenine at position 2058 for a G or U conferred the MLSB phenotype, defined as high-level resistance to all drugs in this group. This phenotype is similar to that conferred by dimethylation of A2058 encoded by erm genes, confirming the key role of this adenine residue in binding of MLSB antibiotics. By contrast, the A2059G mutation conferred a lower level of resistance to macrolides, in particular, clarithromycin. The phenotype conferred by this mutation has previously been called ML since streptogramins B remain active (21). Substitution at position 2611 results in low-level resistance to 14-membered-ring macrolides and to clindamycin. C2611 is a residue that pairs with G2057 in the secondary structure of 23S rRNA, and the C2611U mutation results in a disruption in the rRNA structure at the end of the stem preceding the single-strand portion of the peptidyl transferase region containing A2058 and A2059 (6). So far, the C2611U mutation has been characterized only in laboratory mutants (22, 24, 25). Possibly, the mild macrolide resistance conferred by this mutation might explain why such mutants have not emerged during macrolide therapy. None of the mutations found in the 23S rRNA conferred resistance to pristinamycin, which is a mixture of streptogramins A and B.
Interestingly, the telithromycin MICs for all except one of the mutants with mutations in domain V ranged from 0.008 to 0.25 μg/ml; for one strain the telithromycin MIC was equal to 1 μg/ml and was therefore far lower than those of the other macrolides. The presence of an 11,12-carbamate extension in the telithromycin molecule likely explains this property. Binding of drugs to resistant ribosomes is improved by an alternative and effective contact with A752 in hairpin 35 of domain II via the 11,12-carbamate chain (6, 8). This finding explains why, in our study, the mutant which had an adenine deletion in hairpin 35 was the only one to be resistant to telithromycin (MIC, 4 μg/ml). These data are consistent with the report that a single point mutation (U754A) in a laboratory strain of E. coli was sufficient to render the cells resistant to the ketolide telithromycin (28). To the best of our knowledge, mutations have never been reported in domain II of macrolide-resistant pneumococci.
In addition to the nature of the mutated base and to its location in the 23S rRNA, differences in the number of mutated copies of 23S rRNA could lead to differences in MICs. Initially, domain V mutations have been reported in species harboring one or two copies of the rRNA genes operon, including Brachyspira, Helicobacter pylori, various species of atypical Mycobacterium, Propionibacterium, and Treponema (25). Analysis of mutants of S. pneumoniae which harbor four rrn copies has shown that a minimum of two mutated copies was sufficient to confer macrolide resistance (22). In our study, careful analysis of the SSCP profiles revealed that some domain V fragments containing mutations yielded heterogeneous profiles composed in part of the wild-type profile, suggesting that not all 23S rRNAs were mutated [Fig. 1, lanes 1C, 3C, and 2A in analysis of domain V (B)]. This heterogeneity was confirmed by analysis of the sequence data, which showed a mixture of bases at the altered residue. To determine the number of mutated copies, we have amplified each of the four rrn copies by using specific primers reported previously by Tait-Kamradt et al. (22). Analysis of the PCR products by SSCP analysis showed that a minimum of two rrn copies were mutated for each of the 10 strains studied (Table 2). Mutants 1 CLR, 3 CLR, and 5 AZM contained three alleles of A2058G or A2058U, while mutant 4 ROX was homozygous. A gene dosage effect could be seen for the mutants with the C2611U mutation. The MICs of azithromycin, clarithromycin, and erythromycin for homozygotous mutants were twice those for derivatives with two mutated copies.
Interestingly, several mutants selected by passage in various macrolides contained mutations in the L22 protein alone or, in two mutants, in combination with an L4 or a domain V mutation. L22 is a small 114-amino-acid protein of the 50S ribosomal subunit. Alignment of amino acid sequences deduced from the sequences of the L22 genes showed that mutations were clustered in the C terminus of the protein. These mutations are located in a highly conserved portion of the L22 protein (23). In E. coli, a deletion of 3 amino acids in this conserved region conferred resistance to erythromycin (2). Recently, laboratory mutants of S. pneumoniae derived by serial passage with telithromycin were also characterized by L22 mutations of G95D or A97D (J. Sutcliffe, A. Tait-Kamradt, A. Walker, and J. Petitpas, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1925, 2000). To assess the role of the L22 protein in macrolide resistance, we transformed S. pneumoniae CP1000 with the wild-type L22 gene and four mutated alleles. These experiments confirmed that mutations in the C terminus of the L22 protein conferred resistance to streptogramins and to low levels of macrolides and clindamycin. The recent elucidation of the structure of the Thermus thermophilus L22 protein and of the ribosome by X-ray crystallography allows clarification of the interactions of the L22 protein with the 23S rRNA and its role in macrolide resistance (1, 23). The L22 protein consists of a small α plus β domain and of a protruding β hairpin. All mutated amino acids characterized in our study were located in the β hairpin and are conserved in the various bacterial species. The glycine 91 in the L22 protein of T. thermophilus, which corresponds to glycine 95 in S. pneumoniae, has been postulated to have an important role for the turn of the β hairpin (23). Interestingly, this amino acid is preceded by the highly conserved β-bulge residue Pro 87 (Pro 91 in S. pneumoniae), which was mutated in one of our isolates. In addition, glycine 79 of T. thermophilus (glycine 83 in S. pneumoniae), which was mutated in one derivative, seems to be important in the high local twist of the β hairpin in combination with proline 80 (proline 84 in S. pneumoniae). L22 is the only protein to interact with RNA sequences belonging to all six domains of 23S rRNA and is important for the folding of the 23S rRNA (1). The β hairpin of the protein contributes to the formation of the polypeptide tunnel exit at the surface of the ribosome. The mutation might change the surface properties or perturb the three-dimensional structure of 23S rRNA at multiple sites, as proposed in E. coli, and therefore prevent antibiotic binding (7). The only mutant selected by telithromycin contained a combination of three L22 mutations, possibly accounting for the large number of serial passages (n = 44) required to select for resistance to this antibiotic. Two of these mutations occurred at positions 83 and 91, which are critical for the structure of L22, as discussed above. Telithromycin MICs were increased 8 to 64 times but remained equal to or less than 0.25 μg/ml. This triple mutation did not confer any advantage to the mutant in terms of resistance to telithromycin over single L22 mutations or other mutations. In particular, the mutation of L22 was less efficient than mutation of hairpin 35 in domain II at producing telithromycin resistance (Table 2). However, mutants selected by telithromycin in our study and in another independent experiment displayed L22 mutations (Sutcliffe et al., 40th ICAAC). The reason for this preferential selection of L22 mutations by telithromycin remains unknown. It should be stressed that mutants were less often selected by passage in telithromycin than by passage in other macrolides (4).
Although several in vitro and in vivo S. pneumoniae mutants with L4 mutations resistant to macrolides have been reported, only one of our mutants displayed the L4 mutation combined with an L22 mutation. The G71R mutation occurred in a highly conserved region and was previously reported in pneumococcal mutants (21, 22). L4 is an important player in the maintenance of the ribosome structure and is a partner of L22 in the probable formation of a gated opening for the tunnel exit (7, 15, 27).
The observation that no L22, L4, or 23S rRNA mutations could be detected in the remaining six mutants and that a relationship between the type of mutation and the level of macrolide resistance could not always be found suggested that additional unidentified mutations are involved.
In conclusion, the present study showed that several various ribosomal mutations can lead to macrolide resistance. Many of those found in the present study have never been reported either in laboratory mutants or in clinical strains of S. pneumoniae. Macrolides demonstrated different capacities to select for resistance, and each type of mutation resulted in a specific MLS resistance profile. The availability of primers and the availability of methods such as SSCP analysis to easily screen for mutations in ribosomal structures involved in macrolide binding allow assessment of the contributions of mutations to macrolide resistance in clinical isolates of S. pneumoniae.
Acknowledgments
This work was supported in part by grants from Aventis Pharma and the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–920. [DOI] [PubMed] [Google Scholar]
- 2.Chittum, H. S., and W. S. Champney. 1994. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 176:6192–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comité de l’Antibiogramme de la Société Française de Microbiologie. 1996. 1996 report of the Comité de l’Antibiogramme de la Société française de Microbiologie. Technical recommendations for in vitro susceptibility testing. Clin. Microbiol. Infect. 2(Suppl. 1):11–25. [Google Scholar]
- 4.Davies, T. A., B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2000. In vitro development of resistance to telithromycin (HMR 3647), four macrolides, clindamycin, and pristinamycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183–192. [DOI] [PubMed] [Google Scholar]
- 7.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J. Mol. Biol. 289:827–834. [DOI] [PubMed] [Google Scholar]
- 8.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 3:623–631. [DOI] [PubMed] [Google Scholar]
- 9.Johnston, N. J., J. C. De Azavedo, J. D. Kellner, and D. E. Low. 1998. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2425–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klugman, K. P. 1990. Pneumococcal resistance to antibiotics. Clin. Microbiol. Rev. 3:171–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison, D. A., M. C. Trombe, M. K. Hayden, G. A. Waszak, and J. D. Chen. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM β1. J. Bacteriol. 159:870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1998. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. Document M7–A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Nishijima, T., Y. Saito, A. Aoki, M. Toriya, Y. Toyonaga, and R. Fujii. 1999. Distribution of mefE and ermB genes in macrolide-resistant strains of Streptococcus pneumoniae and their variable susceptibility to various antibiotics. J. Antimicrob. Chemother. 43:637–643. [DOI] [PubMed] [Google Scholar]
- 15.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–930. [DOI] [PubMed] [Google Scholar]
- 16.Ouabdesselam, S., D. C. Hooper, J. Tankovic, and C. J. Soussy. 1995. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob. Agents Chemother. 39:1667–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestova, E., R. Beyer, N. P. Cianciotto, G. A. Noskin, and L. R. Peterson. 1999. Contribution of topoisomerase IV and DNA gyrase mutations in Streptococcus pneumoniae to resistance to novel fluoroquinolones. Antimicrob. Agents Chemother. 43:2000–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts, M., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Haij, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin- resistant M phenotype in Streptococcus pneumoniae Antimicrob. Agents Chemother. 41:2251–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unge, J., A. Aberg, S. Al-Kharadaghi, A. Nikulin, S. Nikonov, N. Davydova, N. Nevskaya, M. Garber, and A. Liljas. 1998. The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance. Structure 6:1577–1586. [DOI] [PubMed] [Google Scholar]
- 24.Vannuffel, P., M. Di Giambattista, E. A. Morgan, and C. Cocito. 1992. Identification of a single base change in ribosomal RNA leading to erythromycin resistance. J. Biol. Chem. 267:8377–8382. [PubMed] [Google Scholar]
- 25.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worbs, M., R. Huber, and M. C. Wahl. 2000. Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon. EMBO J. 19:807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633–639. [DOI] [PubMed] [Google Scholar]