Abstract
Biofilm formation mediated by polysaccharide intercellular adhesin (PIA) is the major virulence factor of Staphylococcus epidermidis and is often associated with methicillin resistance. Transposon Tn917 insertions leading to a biofilm-negative phenotype in the biofilm-producing S. epidermidis strain 1457 (mecA-negative) were transferred into the methicillin-resistant, biofilm-producing S. epidermidis 1057 (mecA-positive) by transduction. According to their phenotypes and genotypes, the mutants could be separated into genetic classes I to IV (D. Mack, H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K. M. Knobloch, H.-A. Elsner, and H. H. Feucht, Infect. Immun. 68:3799–3807, 2000). All transductants of S. epidermidis 1057 had phenotypes for biofilm formation similar to those of the corresponding mutants of S. epidermidis 1457. With a mecA-specific probe, identical hybridization patterns were observed for wild-type S. epidermidis 1057 and all the transductants. There were minor changes in oxacillin MICs for Class II and III transductants compared to those for wild-type S. epidermidis 1057. On population analysis, S. epidermidis 1057 displayed a heterogeneous expression type of resistance with an oxacillin MIC of ≥6 μg/ml for more than 90% of the cells. An almost identical profile was observed with biofilm-negative class I mutants, where the transposon insertions inactivate the icaADBC gene locus essential for PIA synthesis. In contrast, class III mutants were more sensitive to oxacillin with a MIC of ≤1 μg/ml for more than 90% of the cells. The class IV mutant displayed homogenous resistance with a MIC of ≥50 μg/ml for more than 90% of the cells. On oxacillin gradient plates, the class II mutant displayed decreased resistance. Apparently, different independent mutations leading to a biofilm-negative phenotype of S. epidermidis by influencing expression of icaADBC on the level of transcription significantly influence the expression of methicillin resistance. However, transcription of mecA was not significantly altered in the different transductants compared to the wild type, independent of mecA induction with oxacillin, indicating that other mechanisms influencing phenotypic expression of methicillin resistance are involved.
Today, coagulase-negative staphylococci, mostly Staphylococcus epidermidis, represent the most frequent causes of nosocomial sepsis and of infections of implanted medical devices (37). A major problem with these organisms is their pronounced antibiotic resistance, with up to 90% of nosocomial isolates being methicillin resistant (1, 4, 43).
Methicillin resistance is often associated with the ability of S. epidermidis to produce biofilms (6, 15). Biofilm formation is the preeminent virulence factor of S. epidermidis (23, 24, 38, 39), which is subject to phase variation leading to a biofilm-negative phenotype (5, 34, 46, 47). Pleiotropic phenotypic changes were observed with biofilm-negative phase variants of S. epidermidis RP62A, including decreased expression of methicillin resistance. However, the genetic mechanisms leading to these changes remain unknown (5, 34, 35).
Biofilm formation may be arbitrarily divided into two phases involving primary attachment of bacterial cells to a polymer surface, followed by accumulation of the attached bacteria in a multilayered biofilm (23, 24). The polysaccharide intercellular adhesin (PIA) is composed primarily of N-acetylglucosamine in β-1,6-glycosidic linkages containing deacetylated amino groups and succinate and phosphate substituents (26). PIA is functional in cell-to-cell adhesion and hemagglutination and is essential for biofilm accumulation of most clinical S. epidermidis strains (14, 27–29, 31, 32). The gene products of the icaADBC locus of S. epidermidis have enzymatic activity, which leads to synthesis of PIA in vivo and in vitro (17, 18, 29). Using transposon mutagenesis, we recently identified four unlinked genetic loci, whose inactivation led to a biofilm-negative phenotype (30). All class I mutants have insertions in the icaADBC locus, whereas apparently regulatory genetic loci in mutants of classes II, III, and IV are inactivated by the Tn917 insertions. These genetic loci control expression of PIA synthesis and biofilm formation by directly or indirectly influencing expression of icaADBC on the level of transcription (30).
In the present study, we investigated the role of these different genetic loci on expression of methicillin resistance by transfer of the respective transposon insertions into the biofilm-producing, methicillin-resistant, mecA-positive S. epidermidis 1057 by transduction.
(Part of this work was conducted by A. Sabottke in partial fulfillment of the requirements for a from Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.)
MATERIALS AND METHODS
Bacterial strains.
The biofilm-producing S. epidermidis 1457 and its isogenic biofilm-negative Tn917 insertion mutants 1457-M10 and -M13 (class I), -M12 (class II), -M15 (class III), and -M17 (class IV) have been described (28, 30). Biofilm-producing S. epidermidis 1057 is a clinical isolate from an infected central venous catheter (36), which was methicillin resistant and mecA positive as determined by PCR essentially as previously described (41). S. epidermidis 1457 was methicillin sensitive and mecA negative (data not shown).
Phage transduction.
Phage transduction with S. epidermidis phage 71, kindly provided by V. T. Rosdahl, Statens Seruminstitut, Copenhagen, Denmark, was performed as described previously (25, 36).
Adherence assay for measurement of biofilm production by S. epidermidis.
Biofilm production by S. epidermidis strains grown in Trypticase soy broth (Becton Dickinson, Cockeysville, Md.) was determined by a semiquantitative adherence assay with 96-well tissue culture plates (Nunc, Roskilde, Denmark) as described previously (7, 25).
Determination of oxacillin susceptibility.
The oxacillin (Sigma, Deisenhofen, Germany) MIC was determined by the broth microdilution method according to the procedure suggested by the NCCLS with Mueller-Hinton broth (Becton Dickinson) supplemented with 2% NaCl as the growth medium (19). MICs were read after 24 and 48 h of incubation at 35°C.
Phenotypic expression of methicillin resistance on oxacillin-gradient agar plates.
Oxacillin gradient agar plates (oxacillin, 0 to 100 μg/ml) were prepared using Mueller-Hinton agar (Becton Dickinson) supplemented with 2% NaCl. S. epidermidis strains were suspended in phosphate-buffered saline to a concentration of approximately 108 CFU/ml, and appropriate dilutions were streaked onto the agar plate parallel to the oxacillin gradient and incubated for 48 h at 37°C.
Population analysis of phenotypic expression of methicillin resistance.
S. epidermidis strains were subcultured overnight on blood agar and suspended in sterile phosphate-buffered saline to a concentration of 108 CFU/ml. Aliquots (100 μl) of these suspensions and of serial dilutions were plated onto Mueller-Hinton agar plates supplemented with 2% NaCl containing a range of concentrations of oxacillin (usually 1 to 800 μg/ml). Plates were incubated for 48 h at 37°C before the colonies were counted (42).
DNA isolation and Southern blot hybridization.
Isolation of chromosomal DNA, digestion with restriction enzymes, and Southern blot analysis with [32P]dCTP-labeled probes specific for Tn917 or mecA were performed as described previously (25, 28, 40).
RNA extraction and Northern blot analysis.
An overnight culture of the strains was diluted 1:100 in fresh, prewarmed Mueller-Hinton broth (Becton Dickinson) either unsupplemented or supplemented with oxacillin (1 μg/ml). Bacteria were cultivated under constant agitation at 160 rpm and 37°C and harvested after 6.5 h of incubation in mid-exponential growth phase (strain1457, optical density at 578 nm [OD578]of ∼1.0; strain1057 and 1057 mutants, OD578 of ∼2.5). Total bacterial RNA was extracted and Northern blot analysis was performed as described (13, 30). The blots were hybridized with a 32P-radiolabeled mecA-specific probe. This was prepared from a 1,071-bp XbaI/PstI fragment of plasmid pBBB85 composed of a 4.2-kb HindIII fragment containing mecA of Staphylococcus aureus BB270 cloned in shuttle vector pGC-2 (40), which was kindly provided by B. Berger-Bächi, Institute of Medical Microbiology, University of Zürich, Zürich, Switzerland. Hybridization was performed at 42°C with 50% formamide, 2% sodium dodecyl sulfate, 5× SSPE (1× SSPE is 0.15 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA) (pH 7.4), 10% dextran sulfate, and 1% nonfat dry milk as described (13).
RESULTS
Transfer of Tn917-insertions of biofilm-negative mutants into mecA-positive S. epidermidis strain.
Using S. epidermidis phage 71, the insertions leading to altered biofilm formation in isogenic mutants 1457-M10 and -M13 (class I), -M12 (class II), -M15 (class III), and -M17 (class IV) of S. epidermidis 1457 (30) were transduced into the biofilm-positive mecA-positive wild-type S. epidermidis 1057. The respective transductants 1057-M10, 1057-M13, 1057-M12, 1057-M15, and 1057-M17 had antibiotic resistance profiles identical to those of the wild type plus an additional macrolide resistance determinant introduced by Tn917. Similar hybridization patterns were detected with a Tn917-specific probe with all mutants and their respective transductants (data not shown). An identical EcoRI fragment of S. epidermidis 1057 and the respective transductants hybridized with a mecA-specific probe, indicating that replacement of the respective wild-type alleles by the different transposon insertions did not interfere with the mec locus in any transductant (data not shown). Compared to the wild-type strains S. epidermidis 1457 and 1057, the mutants and respective transductants displayed very similar phenotypic changes with regard to biofilm formation and colony morphology (Table 1).
TABLE 1.
Phenotypic properties of biofilm-producing S. epidermidis wild-type strains and their corresponding biofilm-negative transductants
| Mutant | Class |
S. epidermidis 1457
|
S. epidermidis 1057
|
||
|---|---|---|---|---|---|
| Biofilm (OD570) | Colony morphology | Biofilm (OD570) | Colony morphology | ||
| Wild type | 2.50 | White | 2.50 | White | |
| M10 | I | 0.06 | White | 0.06 | White |
| M13 | I | 0.02 | White | 0.02 | White |
| M12 | II | 0.03 | Grey | 0.03 | Grey |
| M15 | III | 0.10 | Grey | 0.10 | Grey |
| M17 | IV | 0.30 | White | 0.28 | White |
Oxacillin MICs.
MICs in broth microdilution were determined for wild-type S. epidermidis 1057 and the different transductants. After 24 h of incubation, a MIC of 256 μg/ml was determined for the wild type and transductants 1057-M10, 1057-M13, and 1057-M17, whereas the oxacillin MIC for transductants 1057-M12 and 1057-M15 was 128 μg/ml. After the strains underwent extended incubation for 48 h, the oxacillin MICs increased by 1 dilution, but the relative levels remained unchanged (Table 2).
TABLE 2.
Oxacillin MICs for methicillin-resistant, biofilm-producing S. epidermidis 1057 and its biofilm-negative transductants
| Strain | Class | MIC (μg/ml)
|
|
|---|---|---|---|
| After 24 h | After 48 h | ||
| 1057 | WTa | 256 | 512 |
| 1057-M10 | I | 256 | 512 |
| 1057-M13 | I | 256 | 512 |
| 1057-M12 | II | 128 | 256 |
| 1057-M15 | III | 128 | 256 |
| 1057-M17 | IV | 256 | 512 |
WT, wild type.
Population analysis of oxacillin resistance.
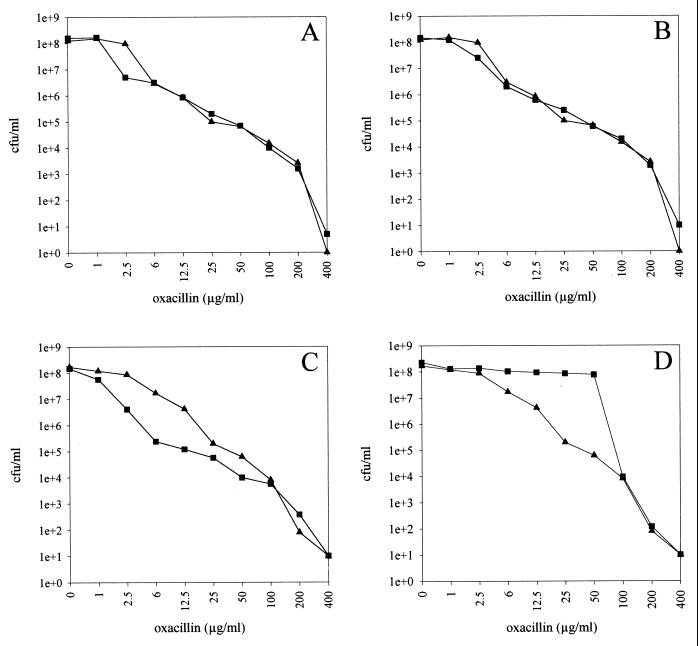
To investigate the resistance distribution within the bacterial cell populations of the different mutant classes, the efficiency of plating of bacterial cell populations on Mueller-Hinton agar plates supplemented with different concentrations of oxacillin was analyzed as described in Materials and Methods. The wild-type S. epidermidis 1057 displayed heterogeneous expression of oxacillin resistance with 90% of cells for which the MIC was ≥6 μg/ml (Fig. 1A and B). With the biofilm-negative transductants of class I 1057-M10 and 1057-M13, a very similar resistance distribution was observed (Fig. 1A and B). In contrast, the population of class III transductant 1057-M15 was shifted to a more sensitive distribution where the oxacillin MICs for more than 90% of the population were ≤1 μg/ml (Fig. 1C). Interestingly, the resistance curve of class IV transductant 1057-M17 was shifted to a more resistant population, with an oxacillin MIC of ≥50 μg/ml for more than 90% of the cell population (Fig. 1D).
FIG. 1.
Population analysis of oxacillin resistance. Suspensions of S. epidermidis 1057 (▴) (A to D) and biofilm-negative transductants (▪) 1057-M10 (class I) (A), 1057-M13 (class I) (B), 1057-M15 (class III) (C), and 1057-M17 (class IV) (D) were spread on agar plates containing different concentrations of oxacillin as described in Materials and Methods. The CFU per milliliter are plotted against oxacillin concentration.
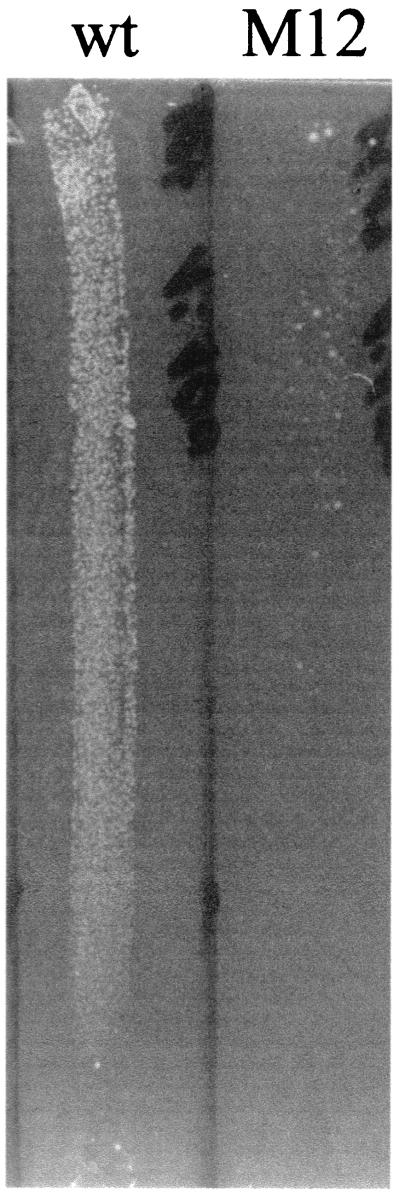
As S. epidermidis 1057-M12 (class II) tended to clump, the resistance distribution within the bacterial cell population of that mutant was analyzed with oxacillin gradient agar plates. A significant decrease in the size of the resistant cell population was observed for class II transductant 1057-M12 (Fig. 2). Changes in the resistance distribution of cell populations of mutants 1057-M15 (class III) and 1057-M17 (class IV) corresponding to those seen in population analysis were also observed when the oxacillin gradient plates were used (data not shown). The resistance distribution was unaltered when oxacillin gradient plates were used with mutant 1057-M10 (class I) compared to wild-type S. epidermidis 1057 (data not shown).
FIG. 2.
Resistance expression profile on oxacillin gradient plates. Similar aliquots of suspensions of S. epidermidis 1057 (wt) and biofilm-negative transductant 1057-M12 (class II) (M12) were spread on a oxacillin gradient plate (0- to 100-μg/ml oxacillin) as described in Materials and Methods.
Transcriptional analysis of mecA.
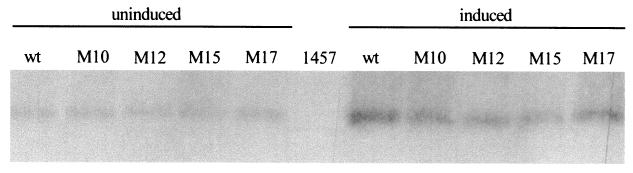
The Tn917 insertions of the biofilm-negative mutants of classes II to IV were shown to directly or indirectly influence transcription of icaADBC, as no icaADBC-specific transcript was observed in these mutants (30). We therefore investigated if there was a similar effect on transcription of mecA in the respective transductants. RNA was extracted in the mid-exponential growth phase in either the presence or absence of oxacillin (1 μg/ml) and probed with a mecA-specific probe. A significant induction in the presence of oxacillin of a mecA-specific transcript was detected in wild-type S. epidermidis 1057 and all transductants (Fig. 3). The amount of accumulating transcripts appeared to differ only marginally between the wild type and the mutants, indicating that the observed differences in the expression of methicillin resistance was not related to alterations of transcription of mecA.
FIG. 3.
Northern blot analysis of S. epidermidis 1057 (wt) and its isogenic biofilm-negative transposon mutants 1057-M10, 1057-M12, 1057-M15, and 1057-M17 with a mecA-specific probe. Bacterial cells were grown in Mueller-Hinton broth either uninduced or induced with oxacillin (1 μg/ml) to mid-exponential growth phase, and total cellular RNA was extracted. A total of 10 μg of RNA was separated on a 1% agarose-formaldehyde gel, blotted onto nylon membranes, and hybridized with a mecA-specific probe. The mecA-negative wild-type S. epidermidis 1457 grown in oxacillin-free medium was used as a negative control.
DISCUSSION
In the present study, we investigated the effect of four different classes of Tn917 insertions influencing expression of biofilm formation in S. epidermidis 1457 on the expression of methicillin resistance in the genetic background of a mecA-positive S. epidermidis wild-type strain. After transduction of the respective insertions into S. epidermidis 1057, changes in expression of biofilm formation were seen, similar to those observed in S. epidermidis 1457, indicating that the inactivated gene loci are also relevant for biofilm formation in the differing genetic backgrounds (Table 1).
In S. aureus, a variety of genetic loci influencing expression of methicillin resistance known as fem or aux factors have been described (2, 3, 8–11, 21, 33, 44). However, for S. epidermidis, the impact of these or other similar factors on expression of methicillin resistance has not been investigated.
S. epidermidis 1057 displayed a heterotypic expression profile of methicillin resistance as is observed with most clinical S. epidermidis isolates (12, 42). For the different biofilm-negative transductants, only minor changes in the oxacillin MIC were observed. However, with population profile analysis of oxacillin gradient agar plates and of plates with different oxacillin concentrations, significant differences in the resistance distribution of the cell populations were observed. Inactivation of the icaADBC locus, encoding the enzymes for PIA synthesis, did not lead to changes in the expression profile of methicillin resistance. In contrast, transductants 1057-M15 (class III) and 1057-M12 (class II) displayed a significant increase in the susceptible cell population compared to the wild type.
Recently, we identified the molecular basis of the phenotypic change of the class III biofilm-negative mutant M15 as an insertion of Tn917 19 bp downstream of the translation start codon of rsbU of the ςB operon of S. epidermidis (20). In the homotypic methicillin-resistant S. aureus strain COL, insertion of Tn551 into sigB and rsbU both led to a significant decrease in the methicillin MIC from 1600 to 50 μg/ml (45). The observed differences in MICs for S. aureus and S. epidermidis rsbU mutants may be related to the expression type of methicillin resistance of both strains—homotypic and heterotypic—or to differences in the regulatory pathways of the σB operon of S. aureus and S. epidermidis. However, inactivation of sigB in two other S. aureus strains resulted in less-pronounced alterations of expression of methicillin resistance (2; B. Berger-Bächi, personal communication). Our results demonstrate that with S. epidermidis, ςB activity significantly influences the expression of methicillin resistance.
The exact mechanism of the phenotypic change of class II Tn917 insertion of mutant 1057-M12 appears rather complex and is not yet fully elucidated. However, the respective gene locus and neighboring genes are different from known fem or aux factors of S. aureus described to date (2, 11; K. Bartscht, J. K.-M. Knobloch, and D. Mack, unpublished data), indicating that additional gene loci do influence expression of methicillin resistance in S. epidermidis.
In contrast to these other two mutant classes, the Tn917 insertion of 1057-M17 (class IV) led to a significant shift of the cell population to higher resistance, although no difference in the overall oxacillin MIC was observed. With S. aureus, a chromosomal gene locus referred to as chr* independent of mecA was identified as necessary for homotypic expression of methicillin resistance (40). The molecular basis of chr* may be related to two recently identified chromosomal genes, hmrA and hmrB (22). Overexpression of these cloned genes led to a homotypic expression profile of methicillin resistance in heterotype S. aureus LR5 (22). However, in the original strain N315 used to isolate these genes an additional gene locus was apparently altered, as both hmrA and hmrB were unchanged in this strain. hmrA has homology to amidohydrolase enzymes, whereas hmrB has homology to acyl carrier proteins (22). Additionally, deletion of the gene of lytic protein LytH led from heterotypic to homotypic expression of methicillin resistance in S. aureus SR17238 (16). The insertion site of mutant 1457-M17 is unknown at present; however, a regulatory function is conferred on expression of icaADBC (30). As the genes involved in transition of hetero- to homo-type expression of methicillin resistance of S. aureus apparently do not encode regulatory proteins, no obvious relation exists to the gene locus inactivated in S. epidermidis 1057-M17 (16, 20).
The three classes of transposon insertions of mutants 1057-M15, 1057-M12, and 1057-M17 were identified as mutations leading to a biofilm-negative phenotype of S. epidermidis 1457 (30). All three mutations affected biofilm formation by directly or indirectly influencing expression of icaADBC, as no icaADBC-specific transcript was detected in biofilm-negative mutants of classes II, III, and IV (30). Similar phenotypic changes regarding biofilm formation were induced in S. epidermidis 1057. It is therefore of interest that these transposon insertions did not significantly alter the level of transcription of mecA. Apparently, as induction of the mecA transcript occurred similarly in all mutants in the presence of oxacillin, neither of these transposon insertions acts via the regulatory mechanisms of mecA induction. Obviously, the mutations induced by the transposon insertions influenced methicillin resistance of S. epidermidis 1057 by mechanisms independent of expression of mecA. These observation are in contrast to results reported for biofilm-negative phase variants of S. epidermidis RP62A, where a reduced oxacillin MIC was related to a complete lack of mecA transcription (34, 35).
Our results show an interesting link between the regulation of the major virulence factor of S. epidermidis, biofilm formation mediated by PIA (23, 38, 39), and methicillin resistance, which contribute to increased survival and dissemination of S. epidermidis as a nosocomial pathogen (1). Further studies are warranted to explore this relation in more detail.
Acknowledgments
We thank Rainer Laufs for his continuous support. For the kind gift of plasmid pBBB85 containing mecA we thank Brigitte Berger-Bächi, University of Zürich, Zürich, Switzerland.
This work is supported by a grant of the Deutsche Forschungsgemeinschaft given to D.M.
REFERENCES
- 1.Archer, G. L., and M. W. Climo. 1994. Antimicrobial susceptibility of coagulase-negative staphylococci. Antimicrob. Agents Chemother. 38:2231–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger-Bächi, B. 1999. Genetic basis of methicillin resistance in Staphylococcus aureus. Cell. Mol. Life Sci. 56:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger-Bächi, B., A. Strassle, J. E. Gustafson, and F. H. Kayser. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, G. D., L. M. Baddour, B. M. Madison, J. T. Parisi, S. N. Abraham, D. L. Hasty, J. H. Lowrance, J. A. Josephs, and W. A. Simpson. 1990. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to beta-lactam antibiotics, and virulence. J. Infect. Dis. 161:1153–1169. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lencastre, H., B. L. de Jonge, P. R. Matthews, and A. Tomasz. 1994. Molecular aspects of methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 33:7–24. [DOI] [PubMed] [Google Scholar]
- 9.de Lencastre, H., A. M. Sa Figueiredo, C. Urban, J. Rahal, and A. Tomasz. 1991. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 35:632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lencastre, H., and A. Tomasz. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lencastre, H., S. W. Wu, M. G. Pinho, A. M. Ludovice, S. Filipe, S. Gardete, R. Sobral, S. Gill, M. Chung, and A. Tomasz. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5:163–175. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson, T. M., and G. L. Archer. 2000. Phenotypic expression of oxacillin resistance in Staphylococcus epidermidis: roles of mecA transcriptional regulation and resistant-subpopulation selection. Antimicrob. Agents Chemother. 44:1616–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobinsky, S., and D. Mack. 2001. Efficient RNA isolation method for analysis of transcription in sessile Staphylococcus epidermidis biofilm cultures. Methods Enzymol. 336:255–262. [DOI] [PubMed] [Google Scholar]
- 14.Fey, P. D., J. S. Ulphani, F. Götz, C. Heilmann, D. Mack, and M. E. Rupp. 1999. Characterization of the relationship between polysaccharide intercellular adhesin and hemagglutination in Staphylococcus epidermidis. J. Infect. Dis. 179:1561–1564. [DOI] [PubMed] [Google Scholar]
- 15.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimura, T., and K. Murakami. 1997. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J. Bacteriol. 179:6294–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerke, C., A. Kraft, R. Süssmuth, O. Schweitzer, and F. Götz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586–18593. [DOI] [PubMed] [Google Scholar]
- 18.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083–1091. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen, J. H., J. D. Turnidge, and J. A. Washington. 1999. Antibacterial susceptibility tests: dilution and disk diffusion methods, p.1526–1543. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 20.Knobloch, J. K. M., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsuzawa, H., M. Sugai, K. Ohta, T. Fujiwara, S. Nakashima, J. Suzuki, C. Y. Lee, and H. Suginaka. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2355–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, N., K. Kuwahara-Arai, H. Kuroda-Murakami, E. Tateda-Suzuki, and K. Hiramatsu. 2001. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob. Agents Chemother. 45:815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack, D. 1999. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J. Hosp. Infect. 43(Suppl.):S113–S125. [DOI] [PubMed] [Google Scholar]
- 24.Mack, D., K. Bartscht, S. Dobinsky, M. A. Horstkotte, K. Kiel, J. K. M. Knobloch, and P. Schäfer. 2000. Staphylococcal factors involved in adhesion and biofilm formation on biomaterials, p.307–330. In Y. H. An and R. J. Friedman (ed.), Handbook for studying bacterial adhesion: principles, methods, and applications. Humana Press, Totowa, N.J.
- 25.Mack, D., K. Bartscht, C. Fischer, H. Rohde, C. de Grahl, S. Dobinsky, M. A. Horstkotte, K. Kiel, and J. K. M. Knobloch. 2001. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzymol. 336:215–239. [DOI] [PubMed] [Google Scholar]
- 26.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear â-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack, D., M. Haeder, N. Siemssen, and R. Laufs. 1996. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J. Infect. Dis. 174:881–884. [DOI] [PubMed] [Google Scholar]
- 28.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mack, D., J. Riedewald, H. Rohde, T. Magnus, H. H. Feucht, H. A. Elsner, R. Laufs, and M. E. Rupp. 1999. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect. Immun. 67:1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mack, D., H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K. M. Knobloch, H.-A. Elsner, and H. H. Feucht. 2000. Identification of three essential regulatory gene loci governing expression of the Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68:3799–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mack, D., N. Siemssen, and R. Laufs. 1994. Identification of a cell cluster associated antigen specific for plastic-adherent Staphylococcus epidermidis which is functionally related to intercellular adhesion. Zentbl. Bakteriol. Suppl. 26:411–413. [Google Scholar]
- 33.Maki, H., and K. Murakami. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:6944–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mempel, M., H. Feucht, W. Ziebuhr, M. Endres, R. Laufs, and L. Grüter. 1994. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 38:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mempel, M., E. Müller, R. Hoffmann, H. Feucht, R. Laufs, and L. Grüter. 1995. Variable degree of slime production is linked to different levels of beta-lactam susceptibility in Staphylococcus epidermidis phase variants. Med. Microbiol. Immunol. (Berlin) 184:109–113. [DOI] [PubMed] [Google Scholar]
- 36.Nedelmann, M., A. Sabottke, R. Laufs, and D. Mack. 1998. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentbl. Bakteriol. 287:85–92. [DOI] [PubMed] [Google Scholar]
- 37.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231–243. [DOI] [PubMed] [Google Scholar]
- 38.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67:2656–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryffel, C., A. Strässle, F. H. Kayser, and B. Berger-Bächi. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz, F. J., C. R. Mackenzie, B. Hofmann, J. Verhoef, M. Finken-Eigen, H. P. Heinz, and K. Kohrer. 1997. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by a multiplex PCR. J. Med. Microbiol. 46:773–778. [DOI] [PubMed] [Google Scholar]
- 42.Tomasz, A., S. Nachman, and H. Leaf. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35:124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.U.S. Department of Health and Human Services. 1999. Intensive care antimicrobial resistance epidemiology (ICARE) surveillance report, data summary from January 1996 through December 1997. Am. J. Infect. Control 27:279–284. [DOI] [PubMed] [Google Scholar]
- 44.Vasilieva, E., H. Labischinski, and B. Berger-Bächi. 1997. Identification and mapping of new chromosomal sites affecting response to β-lactams in Staphylococcus aureus. Int. J. Antimicrob. Agents 8:13–21. [DOI] [PubMed] [Google Scholar]
- 45.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziebuhr, W., C. Heilmann, F. Götz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziebuhr, W., V. Krimmer, S. Rachid, I. Löβner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345–356. [DOI] [PubMed] [Google Scholar]