Abstract
Most Aeromonas strains isolated from two European rivers were previously found to be resistant to nalidixic acid. In order to elucidate the mechanism of this resistance, 20 strains of Aeromonas caviae (n = 10), A. hydrophila (n = 5), and A. sobria (n = 5) complexes, including 3 reference strains and 17 environmental isolates, were investigated. Fragments of the gyrA, gyrB, parC, and parE genes encompassing the quinolone resistance-determining regions (QRDRs) were amplified by PCR and sequenced. Results obtained for the six sensitive strains showed that the GyrA, GyrB, ParC, and ParE QRDR fragments of Aeromonas spp. were highly conserved (≥96.1% identity), despite some genetic polymorphism; they were most closely related to those of Vibrio spp., Pseudomonas spp., and members of the family Enterobacteriaceae (72.4 to 97.1% homology). All 14 environmental resistant strains carried a point mutation in the GyrA QRDR at codon 83, leading to the substitution Ser-83→Ile (10 strains) or Ser-83→Arg. In addition, seven strains harbored a mutation in the ParC QRDR either at position 80 (five strains), generating a Ser-80→Ile (three strains) or Ser-80→Arg change, or at position 84, yielding a Glu-84→Lys modification. No amino acid alterations were discovered in the GyrB and ParE QRDRs. Double gyrA-parC missense mutations were associated with higher levels of quinolone resistance compared with the levels associated with single gyrA mutations. The most resistant strains probably had an additional mechanism(s) of resistance, such as decreased accumulation of the drugs. Our data suggest that, in mesophilic Aeromonas spp., as in other gram-negative bacteria, gyrase and topoisomerase IV are the primary and secondary targets for quinolones, respectively.
Mesophilic motile Aeromonas is a normal inhabitant of freshwaters and may be considered representative of the riverine autochthonous flora (19). These organisms are also human pathogens and are mainly responsible for gastroenteritis, skin and soft tissue infections, and a variety of clinical syndromes in compromised patients (21); quinolones are the drugs of choice for the treatment of Aeromonas-induced infections (22). In a previous study (14), as many as 59% of Aeromonas spp. isolated from two European rivers (the Arga River in Spain and the Garonne River in France) were found to be resistant to nalidixic acid; most of them were also resistant to other narrow-spectrum quinolones but remained susceptible to fluoroquinolones. Quinolones are synthetic antibiotics which are normally absent from freshwaters, and quinolone resistance is essentially due to chromosomal mutations (26, 32). Therefore, discharge of quinolones in rivers, probably from an agricultural source, must have selected resistant mutants among indigenous bacterial populations (13). Indeed, quinolones are widely used in veterinary medicine in Europe (30) and more recently in the United States (4); these antibiotics are mostly excreted as unchanged substances and are among the most persistent drugs in the environment (17).
Mutations that confer quinolone resistance principally alter the target enzymes, the type II bacterial topoisomerases. Both of these enzymes, DNA gyrase and topoisomerase IV, catalyze topological changes in DNA via an ATP-dependent double-strand cleavage and rejoining mechanism. DNA gyrase is primarily engaged in the control of negative supercoiling of DNA, and topoisomerase IV is essentially involved with the decatenation of the interlinked daughter chromosomes (5, 10, 20). Both enzymes are heterotetramers consisting of two types of subunits, GyrA and GyrB in DNA gyrase and the respective homologous proteins ParC and ParE in topoisomerase IV. Gyrase appears to be the primary target for quinolones in gram-negative bacteria since missense mutations in gyrA genes are sufficient to render these organisms quinolone resistant (1, 10, 24, 27, 29, 36). In contrast, mutations in the parC quinolone resistance-determining region (QRDR) are expressed only in the presence of gyrA mutations (10). Virtually all mutations responsible for quinolone resistance have been mapped in a small N-terminal region of the gyrA gene, close to the catalytic site Tyr-122, called the QRDR (12, 42). Alterations in the domains of the GyrB and ParE subunits which interact with GyrA and ParC, respectively, can also contribute to quinolone resistance (1, 7, 10, 27, 28, 41, 43).
The aim of the present study was to identify the mutations in the type II topoisomerase genes that confer quinolone resistance in our riverine Aeromonas strains. The sequence of the gyrA gene of the psychrophilic fish pathogen A. salmonicida has been previously established (31), and partial sequences of A. hydrophila gyrB have become available in GenBank since the beginning of the study; but so far, nothing has been reported on the topoisomerase IV-encoding genes in Aeromonas spp. Thus, DNA fragments encompassing the gyrA, gyrB, parC, and parE QRDRs of A. caviae, A. hydrophila, and A. sobria complexes were first determined for reference and environmental susceptible strains and were then compared with those of environmental isolates exhibiting various levels of quinolone resistance.
MATERIALS AND METHODS
Bacterial strains.
Of the 20 Aeromonas strains examined in this study, 17 (9 A. caviae, 4 A. hydrophila, and 4 A. sobria strains) were selected because they reflected the various antibiotic and quinolone resistance patterns detected among 138 environmental isolates (104 A. caviae, 12 A. hydrophila, and 22 A. sobria strains) previously collected from either the Arga River (Spain) or the Garonne River (France) in 1996 (13, 14). Strains were identified by the criteria of Popoff and Véron (34), as recommended by Holmes et al. (19) for environmental isolates. Briefly, aeroanaerobic gram-negative bacteria with positive oxidase reactions (bioMérieux) were tested for tolerance to 6% NaCl and compound O/129 (Diagnostic Pasteur). The enzymatic activities of ornithine decarboxylase, lysine decarboxylase, arginine dihydrolase, and urease, the Voges-Proskauer test, utilization of citrate, indole formation, H2S production, and carbohydrate fermentation were investigated with the API 20E and API 20NE systems (bioMérieux). The motility was observed on mannitol-motility-nitrate agar (bioMérieux). Hydrolysis of esculin (Merck) and gas formation from glucose metabolism (purple broth base was from BBL; carbohydrate was from Sigma) were also studied. Three type strains of Aeromonas were purchased from the Collection de l'Institut Pasteur (CIP) and were included as reference strains: A. caviae CIP 7616, A. hydrophila CIP 7614, and A. sobria CIP 7433. Escherichia coli XL1-Blue was used as the recipient in transformation experiments for cloning of the gyrB and parC gene fragments. All strains were grown on Mueller-Hinton agar.
Antibiotic susceptibility testing.
Antibiotic resistance patterns were determined by the disk diffusion method, and MICs were determined by an agar dilution method, according to official guidelines (http://www.sfm.asso.fr). The 10 quinolones tested and their respective manufacturers were as follows: nalidixic acid, Chirex; oxolinic acid, Parke-Davis; pipemidic acid, pefloxacin, and sparfloxacin, Rhône Poulenc Rorer; flumequine, Sigma; norfloxacin, MSD-Chibret; ofloxacin, Roussel Uclaf; and ciprofloxacin and enrofloxacin, Bayer Pharma.
Total DNA extraction.
Total cellular DNA was isolated from 3-ml cultures of Aeromonas strains grown overnight. Cells were harvested, suspended in TEG buffer (Tris-HCl, 10 mM; EDTA, 1 mM; glucose, 50 mM [pH 8.0]), and lysed with lysozyme (1 mg/ml; 30 min at 37°C) and proteinase K (25 μg/ml; 1 h at 37°C). The DNA was purified by phenol-chloroform extraction, followed by ethanol precipitation.
PCR amplification of QRDRs.
PCR amplification of QRDRs was performed with a series of primers, listed in Table 1. Primers AsalgyrAF and AsalgyrAR were designed from the nucleotide sequence of the A. salmonicida gyrA gene (31) to amplify the gyrA QRDRs of all Aeromonas strains. Amplification of the gyrB QRDR was carried out first for A. caviae 4 with degenerate primers DgyrBF and DgyrBR. After cloning of the QRDR fragment in the PGEM-T vector (see the following section), the PCR product was amplified by using the universal primers (M13 forward and reverse primers) and sequenced. Then, specific primers AcgyrBF and AcgyrBR were designed from this sequence to amplify the gyrB QRDRs of all other strains. Similarly, PCR amplification of the parC QRDR was performed at first for A. caviae 4 by using primers EcparCF and EcparCR, based on the parC QRDR of E. coli (23). After cloning and sequencing of the PCR product as indicated above, another specific primer, AcparCF, was designed to amplify with primer EcparCR the parC QRDRs of all Aeromonas isolates. Degenerate primer DparEF and specific primer EcparER, designed from the 3′ region of the E. coli parE QRDR (23), allowed amplification of the parE QRDRs of all Aeromonas strains. PCR amplifications were done under standard conditions. After a denaturation step of 5 min at 94°C, amplification was achieved, depending on the primers, over 35 cycles, with each cycle consisting of 1 min at 94°C, 1 min at 50 to 58°C, and 1 min at 72°C, with a final extension step of 10 min at 72°C. The PCR products were analyzed by electrophoresis on 2% (wt/vol) agarose gels.
TABLE 1.
Primers used to amplify by PCR and to sequence the topoisomerase II fragments of the mesophilic Aeromonas spp.
| Gene | Primer | Sequence (5′ to 3′) | Positionsc |
|---|---|---|---|
| gyrA | AsalgyrAFa | TCCTATCTTGATTACGCCATG | 58-78 |
| AsalgyrARa | CATGCCATACCTACCGCGAT | 520-539 | |
| gyrB | DgyrBFb | CCKGGMAARCTKGCRGAVTG | 1207-1226 |
| DgyrBRb | RTCYACRTCYGCRTCRGTCAT | 1483-1503 | |
| AcgyrBFa | CGGAATGCCAGGAGAAAGA | 1220-1238 | |
| AcgyrBRa | GGTCATGATGATGATGTTG | 1470-1488 | |
| parC | EcparCF | GAAACCTGTTCAGCGCCGCAT | 139-159 |
| EcparCRa | TTCGGTGTAACGCATTGCCGC | 371-391 | |
| AcparCFa | GTTCAGCGCCGCATCATCTAC | 146-166 | |
| parE | DparEFa,b | GAYGCCTTYATCCTGTGGCTG | 1118-1138 |
| EcparERa | GTCCGCATCCGCGAGGATACA | 1520-1540 |
Cloning of PCR products.
The products obtained by PCRs with primer pairs DgyrBF-DgyrBR and EcparCF-EcparCR were ligated in the PGEM-T vector with the pGEM-T Vector System cloning kit (Promega), according to the manufacturers' instructions. The ligation mixture was used to transform E. coli XL1-Blue by electroporation. Transformed cells were selected on Luria-Bertani agar plates supplemented with ampicillin (100 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (0.2 mg/liter), and isopropyl-β-d-thiogalactopyranoside (1 mM). The white colonies were replicated, and the recombinant plasmid DNA was extracted and purified with a Midi kit (Qiagen).
DNA sequencing and sequence analysis.
PCR amplification products for DNA sequencing were purified with Microspin S-400 HR columns (Amersham Pharmacia Biotech). Automated sequencing of both strands was carried out with an AmpliTaq DNA polymerase FS Dye Terminator Cycle Sequencing Ready Reaction kit and an ABI Prism 377 sequencer (Applied Biosystems Division, Perkin-Elmer), according to the manufacturer's recommendations. To avoid possible misreadings by Taq polymerase, three recombinant plasmids originating from different PCR and cloning experiments were selected for sequencing of the products that were obtained by PCR with DgyrBF-DgyrBR and EcparCF-EcparCR and cloned in E. coli XL1-Blue. Nucleotide and deduced amino acid sequences were compared by use of Sequence Navigator software (Perkin-Elmer).
Nucleotide sequence accession numbers.
The gyrA, gyrB, parC, and parE nucleotide sequences of the three reference strains are available in the GenBank nucleotide sequence databases with the following accession numbers: AY027899, AY027902, AF435418, and AF435421, respectively, for A. caviae CIP 7616; AY027901, AY027904, AF435419, and AF435422, respectively, for A. hydrophila CIP 7614; and AY027900, AY027903, AF435420, and AF435423, respectively, for A. sobria CIP 7433.
RESULTS
Antibiotic and quinolone resistance patterns of Aeromonas strains.
The antibiotic resistance patterns and the MICs of quinolones for the 20 strains of the A. caviae (n = 10), A. hydrophila (n = 5), and A. sobria (n = 5) complexes tested are indicated in Tables 2, 3, and 4, respectively. These isolates included six sensitive strains (one type and one environmental strain for each species), seven strains resistant to a single quinolone (three A. caviae, two A. hydrophila, and two A. sobria strains), and seven strains resistant to multiple drugs (including quinolones) (five A. caviae strains, one A. hydrophila strain, and one A. sobria strain). Nalidixic acid MICs for quinolone-resistant strains of Aeromonas were ≥128 mg/liter; most of them remained clinically susceptible to fluoroquinolones, despite a 10- to 4,000-fold increase in the MICs compared with those for susceptible strains.
TABLE 2.
Antibiotic resistance patterns, quinolone susceptibilities, and substitutions in the GyrA and ParC QRDRs for strains of the A. caviae complex
| A. caviae strain | Antibiotic resistance patterna | Quinolone MICb (mg/liter)
|
Amino acid in QRDRs
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GyrA
|
ParC
|
|||||||||||||||||
| NAL | OA | PI | UB | NOR | PEF | OFX | SPX | CIP | ENR | Position 83 | Position 92 | Position 80 | Position 84 | |||||
| CIP 7616 | 0.2 | ≤0.1 | ≤0.1 | 0.02 | ≤0.001 | ≤0.001 | ≤0.001 | ≤0.001 | ≤0.001 | 0.005 | Ser | Met | Ser | Glu | ||||
| 4 | ≤0.1 | ≤0.1 | ≤0.1 | ≤0.01 | 0.002 | ≤0.001 | ≤0.001 | ≤0.001 | 0.002 | 0.002 | ||||||||
| 242 | Q | 128 | 1 | 8 | 2 | 0.2 | 0.5 | 0.2 | 0.2 | 0.05 | 0.1 | Arg | ||||||
| 94 | Q | 128 | 8 | 128 | 16 | 2 | 2 | 1 | 0.5 | 0.5 | 1 | Ile | Lys | |||||
| 100 | Q | >512 | 32 | 256 | 512 | 4 | 16 | 4 | 4 | 1 | 4 | Ile | Leu | Ile | ||||
| 542 | Q, Ctx, (Tc), (C) | 128 | 4 | 16 | 4 | 4 | 4 | 1 | 1 | 1 | 0.5 | Arg | ||||||
| 33 | Q, Ctx, Sxt, KGT | 128 | 8 | 128 | 16 | 4 | 4 | 2 | 2 | 1 | 2 | Ile | Arg | |||||
| 2 | Q, Tc, (C), (Sxt), T | 512 | 32 | 64 | 64 | 4 | 2 | 1 | 4 | 0.5 | 2 | Ile | Ile | |||||
| 495 | Q, (Tc), Fos | >512 | 8 | 32 | 8 | 4 | 1 | 0.5 | 1 | 0.1 | 1 | Ile | ||||||
| 198 | Q, (Tc) | >512 | 32 | 128 | 512 | 16 | 16 | 2 | 2 | 4 | 4 | Ile | Leu | Lys | ||||
Q, quinolones; Ctx, cefotaxime; Tc, tetracycline; C, chloramphenicol; Sxt, co-trimoxazole; K, kanamycin; G, gentamicin; T, tobramycin; Fos, fosfomycin; parentheses indicate low-level resistance.
NAL, nalidixic acid; OA, oxolinic acid; PI, pipemidic acid; UB, flumequine; NOR, norfloxacin; PEF, pefloxacin; OFX, ofloxacin; SPX, sparfloxacin; CIP, ciprofloxacin; ENR, enrofloxacin.
QRDRs of quinolone-susceptible Aeromonas strains.
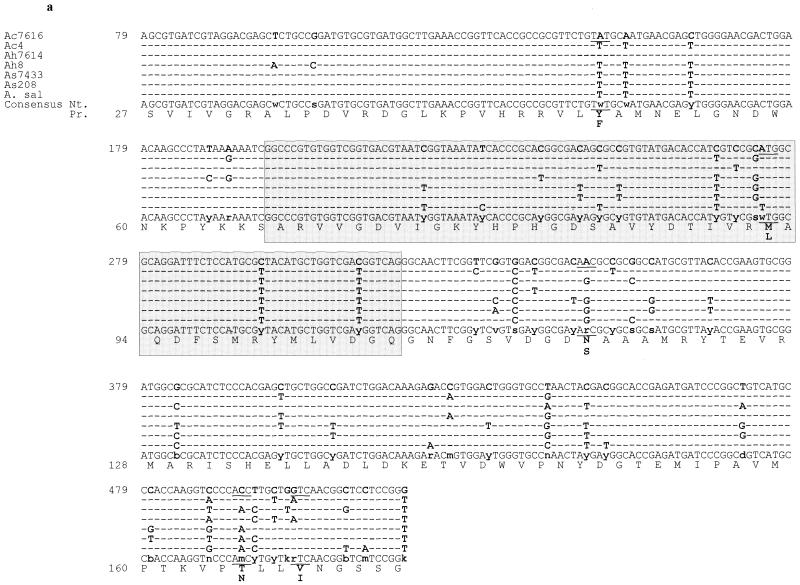
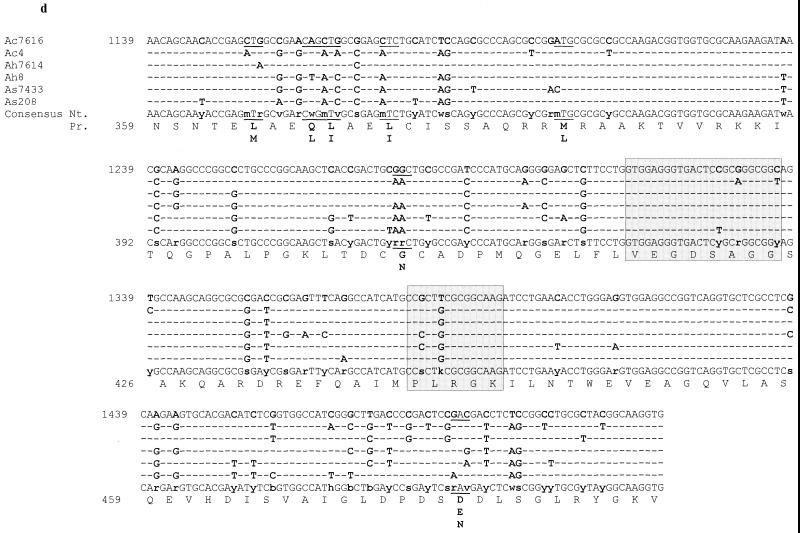
The 441-bp gyrA nucleotide sequences (the sizes of the amplified fragments excluding the primers) differed by 49 mismatches, including 12 in the QRDRs (Fig. 1a).The 231-bp gyrB sequences varied at 26 positions, 4 of which were located in the QRDRs (Fig. 1b). The 204-bp parC sequences of the six mesophilic Aeromonas strains exhibited 25 nucleotide changes, including 12 in the QRDRs (Fig. 1c). The 381-bp parE sequences showed as many as 69 base differences, although only 5 were situated in the QRDRs (Fig. 1d). These sequences shared 85.4 to 98.7% identity. The levels of similarity between the homologous QRDR fragments of each strain were calculated (data not shown), and the nucleotide variations did not appear to be correlated with the species complex. Analysis of the PCR product generated from A. caviae 4 DNA with primer pair DparEF-EcparCR revealed that the parE gene was present at less than 80 bp from the 5" end of the parC gene.
FIG.1.
Nucleotide sequences of gyrA (a), gyrB (b), parC (c), and parE (d) fragments containing the QRDRs of Aeromonas spp. The sequences of the six sensitive strains of A. caviae (Ac), A. hydrophila (Ah), and A. sobria (As) are as determined in the present study; Ac7616, Ah7614, and As7433 are the reference CIP strains. The A. salmonicida (A. sal) gyrA sequence is as reported by Oppegaard and Sørum (30). The E. coli numbering system is used (GenBank accession numbers are given in footnote c of Table 1). QRDRs are framed and shaded. Nucleotide differences are indicated with boldface characters and are underlined when the difference led to amino acid changes. The corresponding nucleotide (Nt.) and proteic (Pr.) consensus sequences are shown.
The 147 predicted peptide GyrA fragments (amino acids 27 to 173 [E. coli numbering]) differed at only four positions outside the QRDR, at positions 50 (Phe or Tyr), 116 (Asn or Ser), 165 (Thr or Asn), and 168 (Val or Ile); compared with A. salmonicida, a single change was noted within the QRDR, at position 92 (Met-92→Leu). The 76 peptide GyrB fragments (amino acids 415 to 491) were strictly identical to each other and to seven of the eight homologous sequences of A. hydrophila GyrB available in GenBank, with the sequence of the remaining one (GenBank accession no. AF208258) differing at three positions outside the QRDR. The 68 peptide ParC fragments (amino acids 48 to 115) exhibited a single substitution within the QRDR, at position 80 for A. hydrophila CIP 7614 (Ser to Ile). The 127 peptide ParE fragments (amino acids 359 to 485) varied at seven positions outside the QRDR: positions 364 (Leu or Met), 367 (Gln or Leu), 368 (Leu or Ile), 371 (Met or Leu), 380 (Met or Leu), 405 (Gly or Asn), and 475 (Asp, Glu, or Asn). The overall homology of the QRDR fragments of mesophilic Aeromonas was at least 96.1%. When the most closely related sequences were searched, the QRDR fragments of Aeromonas spp. were found to be 72.4 to 97.1% identical to those of Vibrio spp., Pseudomonas aeruginosa, and E. coli but only 37.3 to 74.1% identical to those of Bacillus subtilis and Streptococcus pneumoniae, which were used as examples of gram-positive organisms.
Mutations in type II topoisomerase QRDRs of quinolone-resistant Aeromonas strains.
Comparison of the deduced amino acid sequences of the GyrA, GyrB, ParC, and ParE sequences with those of the six sensitive strains described above showed that all quinolone-resistant strains of the A. caviae (Table 2), A. hydrophila (Table 3), and A. sobria (Table 4) complexes carried at least one amino acid substitution in the GyrA QRDR, at position 83: for 10 strains (six A. caviae, one A. hydrophila, and three A. sobria strains) a GC-to-TT mutation at nucleotide positions 248 and 249 resulted in a Ser-to-Ile substitution; in 4 other mutants (two A. caviae and and two A. hydrophila strains), a A-to-C mutation at nucleotide position 247 led to a Ser-to-Arg substitution. In two A. caviae strains, Met-92 was replaced by Leu, as in the gyrA QRDR of A. salmonicida. In addition, seven strains (five A. caviae strains, one A. hydrophila strain, and one A. sobria strain) exhibited amino acid modifications in the QRDR of ParC, either at position 80 or at position 84: in five strains, a A-to-C transversion at position 263 or a G-to-T transversion at nucleotide position 264 gave rise to a substitution of Ser-80 to Arg or Ile, respectively; in two strains, a G-to-A transition at nucleotide position 275 yielded a replacement of Glu-84 by Lys. Multiple amino acid variations were found outside the QRDRs of GyrA and ParE at the same positions and with the same residues as in quinolone-susceptible strains of Aeromonas, except that Leu was always present at position 364 and Ile rather than Met was found at position 371 of ParE (data not shown). No amino acid changes were found in the GyrB sequences of the 14 quinolone-resistant strains of mesophilic Aeromonas spp.
TABLE 3.
Antibiotic resistance patterns, quinolone susceptibilities, and mutations in the GyrA and ParC QRDRs for strains of the A. hydrophila complex
| A. hydrophila strain | Antibiotic resistance patterna | Quinolone MICb (mg/liter)
|
Amino acid in QRDRs
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | OA | PI | UB | NOR | PEF | OFX | SPX | CIP | ENR | GyrA position 83 | ParC position 80 | ||
| CIP 7614 | 0.2 | ≤0.1 | 0.5 | 0.05 | 0.002 | 0.002 | 0.002 | 0.005 | ≤0.001 | 0.005 | Ser | Ile | |
| 8 | ≤0.1 | ≤0.1 | 0.2 | ≤0.01 | ≤0.001 | ≤0.001 | ≤0.001 | 0.002 | ≤0.001 | ≤0.001 | Ser | ||
| 256 | Q | >512 | 4 | 32 | 4 | 2 | 2 | 2 | 4 | 1 | 2 | Arg | Ser |
| 209 | Q | 256 | 64 | 256 | 512 | 4 | 4 | 2 | 4 | 1 | 2 | Ile | |
| 34 | Q, Ctx, Tc, (C), Sxt | >512 | 8 | 64 | 32 | 8 | 16 | 4 | 2 | 2 | 2 | Arg | Ser |
TABLE 4.
Antibiotic resistance patterns, quinolone susceptibilities, and mutations in the GyrA and ParC QRDRs for strains of the A. sobria complex
| A. sobria strain | Antibiotic resistance patterna | Quinolone MICb (mg/liter)
|
Amino acid in QRDRs
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | OA | PI | UB | NOR | PEF | OFX | SPX | CIP | ENR | GyrA position 83 | ParC position 80 | ||
| CIP 7433 | ≤0.1 | ≤0.1 | 0.2 | 0.02 | 0.002 | ≤0.001 | ≤0.001 | 0.005 | ≤0.001 | ≤0.001 | Ser | Ser | |
| 208 | ≤0.1 | ≤0.1 | 0.5 | ≤0.01 | 0.002 | ≤0.001 | ≤0.001 | 0.002 | ≤0.001 | ≤0.001 | |||
| 536 | Q | 256 | 1 | 8 | 4 | 0.5 | 0.5 | 0.2 | 0.2 | 0.1 | 0.2 | Ile | |
| 367 | Q | 128 | 8 | 128 | 512 | 2 | 2 | 0.5 | 0.5 | 0.5 | 1 | Ile | Arg |
| 384 | Q, Tc, Sxt | 128 | 4 | 32 | 32 | 4 | 2 | 1 | 1 | 1 | 1 | Ile | |
Among the A. caviae isolates tested (Table 2), strain 100, which carried a mutation in both the GyrA (Ser-83→Ile) and the ParC (Ser-80→Ile) QRDRs, was more resistant to all quinolones tested than strain 94, which differed by a mutation in the ParC QRDR (Glu-84→Lys). The MICs of all drugs were higher for strain 94 than for strain 242, which carried a single mutation in the GyrA QRDR (Ser-83→Arg). Strain 542, which presented with an associated low level of resistance to tetracycline and chloramphenicol, was 5 to 20 times more resistant to fluoroquinolones than strain 242, despite an identical substitution in the GyrA QRDR (Ser-83→Arg). Similarly, among the A. hydrophila strains tested (Table 3), strain 209, which had a double mutation (Ser-83→Ile in GyrA, Ser-80→Ile in ParC), was 8 to 128 times more resistant to oxolinic acid, pipemidic acid, and flumequine than strain 256, which had a single mutation, Ah256 (Ser-83→Ile in GyrA). The MICs of most quinolones were higher for strain 34 than strain 256, although both strains carried the same mutation in GyrA (Ser-83→Arg), but the former strain was multidrug resistant, with resistance to chloramphenicol at low levels. Likewise, among the A. sobria isolates tested (Table 4), strain 536 (which had Ser-83→Ile in GyrA) was less resistant than strain 367 (which had Ser-83→Ile in GyrA and Ser-80→Arg in ParC) and strain 384 (which had Ser-83→Ile in GyrA and which was multidrug resistant).
DISCUSSION
Aeromonas spp. are exquisitely susceptible to quinolones (39), and until now they have been found to be consistently susceptible to these antibiotics in most parts of the world (22). However, the first nalidixic acid-resistant strain of A. hydrophila was reported in 1987 among clinical isolates from Asia (8), and in 1996, 4% of the clinical mesophilic strains of Aeromonas isolated in Taiwan were resistant to more than 2 mg of pefloxacin per liter (25). The mechanism of this resistance has not been investigated. We have found a very elevated numbers of quinolone-resistant strains among mesophilic Aeromonas strains isolated from two European rivers (13, 14). Such resistance is of clinical concern, since freshwaters are the main source of Aeromonas-induced infections in humans. Because mutations in the type II topoisomerase genes are the principal mechanisms of quinolone resistance, the sequences of the gyrA, gyrB, parC, and parE QRDRs of three mesophilic Aeromonas species were determined, and mutations associated with quinolone resistance were investigated.
The nucleotide sequences of the gyrA, gyrB, parC, and parE QRDR fragments of susceptible A. caviae, A. hydrophila, and A. sobria strains exhibited some genetic heterogeneity. In contrast, Oppegaard and Sørum (30) have reported a remarkable degree of nucleotide sequence identity among gyrA fragments from different strains of A. salmonicida. These strain-to-strain variations might be related to taxonomic uncertainties or gene polymorphism, or both. Indeed, the taxonomy of mesophilic Aeromonas strains has undergone deep revisions in the last two decades. While the psychrophilic aeromonads represent a homogeneous collection of strains, mesophiles are a heterogeneous cluster and have been allocated to 13 genomic species by DNA-DNA hybridizations (21). Biochemical identification is considered >85% accurate to the phenospecies level, e.g., to the A. hydrophila, A. caviae, and A. veronii (“A. sobria”) complex levels (21). Analysis of the restriction fragment length polymorphisms of PCR-amplified fragments of the 16S rRNA gene (6, 15) did not allow further specification of this identification (data not shown), and thus, strains were finally referred to as “complexes” rather than “species.” On the other hand, substantial polymorphism of the topoisomerases genes, including the QRDR sequences, has previously been described in a number of organisms (1, 9, 27, 29, 38). Surprisingly, the parC and parE genes were found to be contiguous on the chromosome of the mesophilic Aeromonas strains, whereas this feature is mainly observed in gram-positive organisms (10, 20).
The deduced amino acid sequences of the GyrA, GyrB, ParC, and ParE fragments of the A. caviae, A. hydrophila, and A. sobria complexes were highly homologous to each other; mostly similar to those of Vibrio spp., Pseudomonas spp., and members of the family Enterobacteriaceae; and more distant from those of gram-positive organisms, in agreement with the phylogenetic relationships of these bacteria. Within the GyrA QRDRs of Aeromonas spp. (30, 31), a serine residue was present at position 83, as in Vibrio parahaemolyticus (29) and most members of the family Enterobacteriaceae (38), whereas a threonine is present at position 83 in P. aeruginosa and some other organisms (10, 20). The GyrA QRDRs of mesophilic Aeromonas strains differed from that of A. salmonicida by a single amino acid change, Met-92→Leu, as, among members of the family Enterobacteriaceae, is found in Providencia stuartii (38). Multiple variations were found outside the QRDRs of GyrA (amino acids 50, 116, 165, and 168) and ParE (amino acids 364, 368, 371, 380, 405, and 475) and involved like residues that are also present in the most closely related species.
All highly quinolone-resistant strains of mesophilic Aeromonas examined in the present study carried an amino acid change in the GyrA subunit, at position 83. This observation strongly suggests that in mesophilic Aeromonas strains, as in other gram-negative bacteria, DNA gyrase is the primary target of quinolones (32). All mutations in gyrA responsible for high-level quinolone resistance are clustered within the QRDR (amino acids 67 to 106) (42), and those that alter residue 83 are both the most frequently encountered and those that confer the most significant increase in the level of quinolone resistance, followed by substitutions of amino acid 87 (1, 2, 10, 27, 42). For 10 strains of mesophilic Aeromonas the substitution was Ser-83→Ile, and for 4 strains the substitution was Ser-83→Arg. Quinolone resistance-determining mutations at positions 83 and 87 are always to hydrophobic amino acids, but the type of allele varies widely (1, 10, 11, 27, 29, 38, 42); in A. salmonicida, all quinolone-resistant strains investigated so far carried a Ser-83→Ile substitution (30). Double missense mutations in the gyrA QRDR, particularly at both position 83 and position 87, have been associated with an increase in the level of resistance to fluoroquinolones (1, 2, 11, 27, 32, 37). However, other amino acid modifications found in the GyrA fragments of quinolone-resistant Aeromonas strains have previously been identified in wild-type strains of Aeromonas and/or in other quinolone-susceptible species and, therefore, are unlikely to result in decreased quinolone susceptibility. Rarely, quinolone resistance-determining mutations have been mapped in GyrB, at Asp-426 and Lys-447 in E. coli (10, 37, 41) or at Ser-464 in P. aeruginosa (1, 27). Nevertheless, no substitutions were discovered in the GyrB sequences of our quinolone-resistant strains of Aeromonas.
In fact, among highly fluoroquinolone-resistant strains, mutants with double gyrA-parC mutations occur more frequently than those with double gyrA-gyrB mutations (1, 10, 24, 27, 29, 36). Actually, alterations in the ParC QRDR (amino acids 64 to 103) were detected in 7 of the 14 quinolone-resistant strains investigated. For five strains, the substitution was at position 80, either Ser-to-Ile (three strains) or Ser-to-Arg; for two strains, the substitution was Glu-84 to Lys. Amino acids 80 and 84 in ParC are homologous to residues 83 and 87 in GyrA (10). Substitution to hydrophobic and positively charged amino acids at these codons, respectively, are both the most common and those that convey the highest levels of quinolone resistance (1, 18, 24, 27, 29, 36). Accordingly, a mutation at position 80 conferred more resistance than a mutation at position 84. For Aeromonas strains carrying a double gyrA-parC mutation, quinolone MICs were higher than those for strains with a single gyrA mutation (1, 2, 10, 24, 27). No resistant mutants with a parC mutation alone were observed. Moreover, wild-type strain A. hydrophila CIP 7614 carried a substitution of Ser-80 to Ile in the ParC QRDR. These data support the view that topoisomerase IV is a secondary target for quinolones in Aeromonas spp. (10, 24). Exceptionally, quinolone resistance-determining mutations in parE have been characterized in gram-negative organisms: at Leu-445 in E. coli (7, 11, 35) or at Asp-420 in P. aeruginosa (1). However, no substitutions were detected within the ParE QRDRs of our quinolone-resistant strains of Aeromonas, and the amino acid variations found outside the QRDRs have already been recognized in quinolone-susceptible strains or species.
Finally, reduced levels of uptake or an active efflux system(s) (33) might explain why some multidrug-resistant strains of Aeromonas were more resistant to fluoroquinolones than other strains were, despite identical target modifications. Indeed, many of our riverine isolates exhibited low levels of resistance to tetracycline and/or chloramphenicol, and the resistance was not transferable (13). Low levels of resistance to multiple antibiotics, including quinolones, tetracyclines, and chloramphenicol, have been found to be associated with changes in outer membrane protein profiles and have been ascribed to decreased permeability in A. hydrophila (3, 16, 40).
In conclusion, the GyrA, GyrB, ParC, and ParE QRDRs of the A. caviae, A. hydrophila, and A. sobria complexes were highly similar or identical, despite some degree of genetic heterogeneity. Quinolone resistance was primarily related to mutations in the gyrA gene since all quinolone-resistant strains carried a substitution of the Ser at position 83, with a mutation to Ile being more frequent than that to Arg. The presence of an additional mutation in ParC, either a Ser-80→Ile or Arg change or a Glu-83→Lys change, was demonstrated in seven strains for which quinolone MICs were higher. Additional mechanisms such as decreased levels of drug accumulation probably account for the highest levels of quinolone resistance.
Acknowledgments
We thank Sophie Dessus-Babus and Florence Jude for helpful discussions and Catherine André for technical assistance.
This work was supported by a Ph.D. grant to M.G.-U. from the Navarra Regional Council.
REFERENCES
- 1.Akasaka, T., M. Tanaka, A. Yamaguchi, and K. Sato. 2001. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagel, S., V. Hüllen, B. Wiedemann, and P. Heisig. 1999. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, A. C., C. S. Lewin, T. S. Hastings, and S. G. B. Amyes. 1990. Cross resistance between oxytetracycline and oxolinic acid in Aeromonas salmonicida associated with alterations in outer membrane proteins. FEMS Microbiol. Lett. 72:337-340. [DOI] [PubMed] [Google Scholar]
- 4.Beam, T. R. 1994. Fluoroquinolones in animal feeds. ASM News. 60:348-349. [Google Scholar]
- 5.Berger, J. M. 1998. Structure of DNA topoisomerases. Biochim. Biophys. Acta 1400:3-18. [DOI] [PubMed] [Google Scholar]
- 6.Borrell, N., S. G. Acinas, M.-J. Figueras, and A. J. Martinez-Murcia. 1997. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J. Clin. Microbiol. 35:1671-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breines, D. M., S. Ouabdesselam, E. Y. Ng, J. Tankovic, S. Shah, C. J. Soussy, and D. C. Hooper. 1997. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 41:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, B. J., and S. M. Bolton. 1987. Plasmids and resistance to antimicrobial agents in Aeromonas sobria and Aeromonas hydrophila clinical isolates. Antimicrob. Agents Chemother. 31:1281-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, M. E., A. W. Wyke, R. Kuroda, and L. M. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica, K. and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, M. J., Y. Fang Jin, V. Ricci, and L. J. V. Piddock. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, S. M., T. Lu, and K. Drlica. 2001. Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother. 45:2378-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goñi-Urriza, M., M. Capdepuy, C. Arpin, N. Raymond, P. Caumette, and C. Quentin. 2000. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl. Environ. Microbiol. 66:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goñi-Urriza, M., L. Pineau, M. Capdepuy, C. Roques, P. Caumette, and C. Quentin. 2000. Antimicrobial resistance of mesophilic Aeromonas spp. isolated from two European rivers. J. Antimicrob. Chemother. 46:297-301. [DOI] [PubMed] [Google Scholar]
- 15.Graf, J. 1999. Diverse restriction fragment length polymorphism patterns of the PCR-amplified 16S rRNA genes in Aeromonas veronii strains and possible misidentification of Aeromonas species. J. Clin. Microbiol. 37:3194-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths, S. G., and W. H. Lynch. 1989. Characterization of Aeromonas salmonicida mutants with low-level resistance to multiple antibiotics. Antimicrob. Agents Chemother. 33:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halling-Sørensen, B., S. Nors Nielsen, P. F. Lanzky, F. Ingerslev, H. C. Holten Lützhøft, and S. E. Jørgensen. 1998. Occurrence, fate and effects of pharmaceutical substances in the environment—a review. Chemosphere 36:357-393. [DOI] [PubMed] [Google Scholar]
- 18.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40:879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, P., L. M. Niccolls, and D. P. Sartory. 1996. The ecology of mesophilic Aeromonas in the aquatic environment, p. 127-150. In B. Austin et al. (ed.) The genus Aeromonas. John Wiley & Sons Ltd., New York, N.Y.
- 20.Huang, W. M. 1996. Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 30:79-107. [DOI] [PubMed] [Google Scholar]
- 21.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 22.Jones, B. L., and M. H. Wilcox. 1995. Aeromonas infections and their treatment. J. Antimicrob. Chemother. 35:453-461. [DOI] [PubMed] [Google Scholar]
- 23.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Higara, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in Escherichia coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 24.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko, W. C., K. W. Yu, C. Y. Liu, C. T. Huang, H. S. Leu, and Y. C. Chuang. 1996. Increasing antibiotic resistance in clinical isolates of Aeromonas strains in Taiwan. Antimicrob. Agents Chemother. 40:1260-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 27.Mouneimné, H., J. Robert, V. Jarlier, and E. Cambau. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura, S., M. Nakamura, T. Kojima, and H. Yoshida. 1989. GyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob. Agents Chemother. 33:254-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuda, J., E. Hayakawa, M. Nishibuchi, and T. Nishino. 1999. Sequence analysis of the gyrA and parC homologues of a wild-type strain of Vibrio parahaemolyticus and its fluoroquinolone-resistant mutants. Antimicrob. Agents Chemother. 43:1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oppegaard, H., and H. Sørum. 1994. gyrA mutations in quinolone-resistant isolates of the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 38:2460-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oppegaard, H., and H. Sørum. 1996. Cloning and nucleotide sequence of the DNA gyrase gyrA gene from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 40:1126-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piddock, L. J. V. 1999. Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs. 58(Suppl. 2):11-18. [DOI] [PubMed] [Google Scholar]
- 33.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popoff, M., and M. Véron. 1976. A taxonomic study of the Aeromonas hydrophila-Aeromonas punctata group. J. Gen. Microbiol. 94:11-22. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz, J., S. Casellas, M. T. Jimenez de Anta, and J. Vila. 1997. The region of the parE gene, homologous to the quinolone-resistant determining region of the gyrB gene, is not linked with the acquisition of quinolone resistance in Escherichia coli clinical isolates. J. Antimicrob. Chemother. 39:839-840. [DOI] [PubMed] [Google Scholar]
- 36.Vila, J., J. Ruiz, P. Goñi, and M. T. Jimenez de Anta. 1996. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vila, J., J. Ruiz, F. Marco, A. Barcelo, P. Goñi, E. Giralt, and M. T. Jimenez de Anta. 1994. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob. Agents Chemother. 38:2477-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigel, L. M., C. D. Steward, and F. C. Tenover. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfson, J. S., and D. C. Hooper. 1989. Fluoroquinolone antimicrobial agents. Clin. Microbiol. Rev. 2:378-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood, S. C., R. N. McCashion, and W. H. Lynch. 1986. Multiple low-level antibiotic resistance in Aeromonas salmonicida. Antimicrob. Agents Chemother. 29:992-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagishi, J., H. Yoshida, M. Yamayoshi, and S. Nakamura. 1986. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol. Gen. Genet. 204:367-373. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida, H., M. Nakamura, M. Bogaki, and S. Nakamura. 1990. Proportion of DNA gyrase mutants among quinolone-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:1273-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]