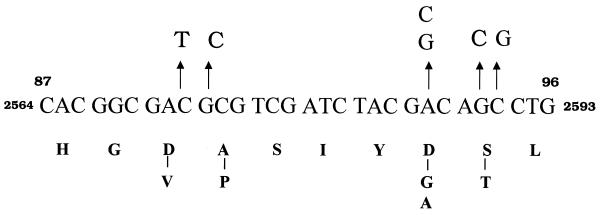Abstract
The World Health Organization has identified India as a major hot-spot region for Mycobacterium tuberculosis infection. We have characterized the sequences of the loci associated with multidrug resistance in 126 clinical isolates of M. tuberculosis from India to identify the respective mutations. The loci selected were rpoB (rifampin), katG and the ribosomal binding site of inhA (isoniazid), gyrA and gyrB (ofloxacin), and rpsL and rrs (streptomycin). We found known as well as novel mutations at these loci. Few of the mutations at the rpoB locus could be correlated with the drug resistance levels exhibited by the M. tuberculosis isolates and occurred with frequencies different from those reported earlier. Missense mutations at codons 526 to 531 seemed to be crucial in conferring a high degree of resistance to rifampin. We identified a common Arg463Leu substitution in the katG locus and certain novel insertions and deletions. Mutations were also mapped in the ribosomal binding site of the inhA gene. A Ser95Thr substitution in the gyrA locus was the most common mutation observed in ofloxacin-resistant isolates. A few isolates showed other mutations in this locus. Seven streptomycin-resistant isolates had a silent mutation at the lysine residue at position 121. While certain mutations are widely present, pointing to the magnitude of the polymorphisms at these loci, others are not common, suggesting diversity in the multidrug-resistant M. tuberculosis strains prevalent in this region. Our results additionally have implications for the development of methods for multidrug resistance detection and are also relevant in the shaping of future clinical treatment regimens and drug design strategies.
Recent years have witnessed a dramatic upsurge in cases of drug-resistant Mycobacterium tuberculosis infections. The acquisition of resistance by the bacterium is a random event, and in a given mycobacterial population, 1 in 106 bacteria mutates to develop isoniazid resistance, while 1 in 108 mutates to develop rifampin resistance (8). The chance that a bacterium will acquire multidrug resistance (defined as resistance to at least rifampin and isoniazid) is thus 10−14 (8). The drug-resistant phenotype may get selected due to single-drug therapy, poor patient adherence, and improper diagnosis. With the AIDS pandemic fuelling increasing numbers of multidrug-resistant (MDR) strains of M. tuberculosis, urgent measures need to be taken to contain this scourge (2). A recently published World Health Organization report reviewing the global status of tuberculosis has pointed to an increasing incidence of drug-resistant tuberculosis (5). The highest rates of MDR tuberculosis have been reported in Nepal (48.0%), Gujarat, India (33.8%), New York City (30.1%), Bolivia (15.3%), and Korea (14.5%). Furthermore, the report points to the alarming increase in the number of tuberculosis patients in the Indian subcontinent, with India being singled out as having the greatest burden of tuberculosis patients. Three different studies from North and Northwest India indicate an increasing incidence of acquired MDR tuberculosis (9, 12, 15). Furthermore, the incidence of primary MDR tuberculosis in North India was put at 3.3% in one of the studies (12).
While there is lot of literature on the molecular epidemiology and characterization of MDR isolates from the United States and Europe, the same is not true for the Indian strains. The prevalence of drug-resistant tuberculosis in North India is known, but no serious efforts have been made to identify the drug resistance genotypes or their prevalence in the community. The present study was undertaken to characterize mutations prevalent in patient isolates of M. tuberculosis from North India with respect to a few of these drug target loci. We have chosen to look at the drug target genes for the drugs rifampin, isoniazid, streptomycin, and fluoroquinolones, which are commonly prescribed for the treatment of tuberculosis in North India. The first three drugs are the frontline drugs in tuberculosis chemotherapy, while fluoroquinolones are prescribed for drug-resistant cases. The loci studied were rpoB (RNA polymerase B subunit), katG (catalase-peroxidase), inhA (enoyl coenzyme A reductase), rpsL (ribosomal protein S12), rrs (16S rRNA), and gyrAB (DNA gyrases A and B). The present study, in combination with the molecular epidemiology of the drug-resistant strains, will help track the routes of infection and the extent of drug-resistant tuberculosis in this region. The elucidation of common and novel mutations in these loci could form the basis for the creation of new diagnostic tools and the development of novel strategies that can be used to combat the menace of drug-resistant M. tuberculosis.
MATERIALS AND METHODS
Sources of Mycobacterium isolates.
Mycobacterium isolates were collected from patients reporting to the outpatient departments of hospitals in northern India, primarily New Delhi and its neighboring regions. Another source of samples was the National Mycobacterial Repository at the Central Jalma Institute for Leprosy, Agra, India. The samples collected over a 3-year period from 1995 to 1998 were included in the present study. A large number of the patients (75%) had histories of previous treatment and were on antitubercular treatment at the time of collection of their sputa. Most of these patients had been through various degrees of antitubercular drug therapy during the previous 20 months. Rifampin and isoniazid were the most common drugs used in these regimens. Sputum samples collected from patients reporting with pulmonary tuberculosis were processed by standard methods and were streaked onto Lowenstein-Jensen slants. Most of them were coded with ICC numbers (ICC01, ICC201, etc.). The samples were biochemically characterized as belonging to the M. tuberculosis complex by nitrate reduction, niacin production, and BACTEC NAP tests. Drug susceptibility profiles were evaluated by the proportion method. The drugs tested were rifampin (Lupin, India), isoniazid (Lupin), ofloxacin (Ranbaxy, India), and streptomycin (Lupin). The MICs at which the isolates were considered resistant were as follows: 10 μg/ml for rifampin, 1 μg/ml for isoniazid, 2 μg/ml for ofloxacin, and 2 μg/ml for streptomycin. The numbers of drug-resistant isolates included in the study were as follows: for rifampin, n = 94; for isoniazid, n = 74; for streptomycin, n = 14; and for ofloxacin, n = 68. A total of 126 isolates were tested. Thirty-six isolates were resistant to a single drug, 66 isolates were resistant to two drugs, 22 isolates were resistant to three drugs, and 4 isolates were resistant to four drugs.
DNA isolation and PCR.
The isolates were cultured on Lowenstein-Jensen slants. The colonies were scraped, resuspended in 500 μl of TE (10 mM Tris, 1 mM EDTA [pH 8]), and killed by freezing at −70°C followed by heating at 80°C. This cycle was repeated thrice to kill all the bacteria. The DNA was isolated (by tretament with cetyltrimethylammonium bromide in the presence of 0.7 M sodium chloride) and amplified by standardized protocols as reported previously (21).
Table 1 lists the sequences of the different primers used and their positions on the corresponding genes. It also lists the amplicon sizes generated and the annealing temperatures used for PCR cycling. The temperatures used for all cycles were identical for all PCRs except for that for annealing, the temperature of which varied for each primer pair. Briefly, 35 cycles of 94°C for 1 min, 45 to 60°C for 1 min, and 72°C for 2 min were used to amplify the loci. The samples were resolved in a 2% agarose gel, and the specific bands were excised. DNA was extracted from the gel slices with a QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.) according to the manufacturer’s instructions. The purified DNA was resuspended in sterile double-distilled water and was used for the sequencing studies.
TABLE 1.
Primers used in the study to amplify and sequence the different loci, amplicon sizes, annealing temperatures, and amplicon positions on the respective genes
| Gene (accession no.) | Primer | Sequence | Annealing temp (°C) | Position (nt) | Amplicon size (bp) |
|---|---|---|---|---|---|
| rpoB (L27989) | Forward | GGG AGC GGA TGA CCA CCC | 60 | 2266 | 350 |
| Reverse | GCG GTA CGG CGT TTC GAT GAA C | 2615 | |||
| katG (X68081) | Forward | GCC CGA GCA ACA CCC | 60 | 3 | 237 |
| Reverse | ATG TCC CGC GTC AGG | 239 | |||
| Forward | CGA GGA ATT GGC CGA CGA GTT | 55 | 1187 | 414 | |
| Reverse | CGG CGC CGC GGA GTT GAA TGA | 1600 | |||
| inhA regulator sequence | Forward | CCT CGC TGC CCA GAA AGG GA | 45 | Upstream of inhA gene | 248 |
| Reverse | ATC CCC CGG TTT CCT CCG GT | ||||
| gyrA (L27512) | Forward | CAG CTA CAT CGA CTA TGC GA | 45 | 2383 | 320 |
| Reverse | GGG CTT CGG TGT TAC CTC AT | 2702 | |||
| gyrB (L27512) | Forward | CCA CCG ACA TCG GTG GAT T | 55 | 1538 | 428 |
| Reverse | CTG CCA CTT GAG TTT GTA CA | 1965 | |||
| rpsl (X70995) | Forward | GGC CGA CAA ACA GAA CGT | 54 | 5′ noncoding region | 505 |
| Reverse | GTT CAC CAA CTG GGT GAC | S7 gene | |||
| rrs (Z83862) | Forward | TTG GCC ATG CTC TTG ATG CCC | 54 | 141 | 1140 |
| Reverse | TGC ACA CAG GCC ACA AGG GA | 1280 |
DNA sequencing.
Sequencing of the amplicons was carried out with an ABI Prism 377 automated DNA sequencer (ABI Prism). PCR sequencing was carried out with a BigDye terminator kit (ABI Prism) according to the manufacturer’s instructions. The Sequencing Analysis (version 3.3) software package was used to analyze the gel information. The sequences generated with the program were compared to their respective wild-type sequences by using MegAlign software (Lasergene; DNASTAR, Inc., Madison, Wis.).
RESULTS
Mutations in the hot-spot regions of various loci were characterized. The results are summarized in Table 2. On the basis of the drug susceptibility profile for an isolate, the corresponding loci (representing the drug target gene) were amplified and sequenced. The largest number of samples was obtained from New Delhi, followed by Chandigarh, Ahmedabad, Agra, Bangalore, and Shimla, with a few samples coming from Jaipur and Chennai. Except for Chennai and Bangalore, all the cities are located in North India. We could establish a previous treatment history for patients from whom 94 of the 126 isolates were recovered. These isolates probably represent those with acquired resistance, as the patients had at some time point been given antitubercular drug therapy.
TABLE 2.
Characteristics of M. tuberculosis isolates from patients
| Strain no. | Geographic location | Treatment historya | Drug susceptibilityb | Polymorphismc
|
|||
|---|---|---|---|---|---|---|---|
| rpo | katG or inhA | gyrA | rpsL | ||||
| ICC14 | New Delhi | + | Rr, Ir, Or | D516V | N35D, NA at second locus | S95T | |
| ICC19 | New Delhi | + | Rr, Ir, Or | L511L, S531L | R463L | S95T | |
| ICC23 | New Delhi | + | Rr, Ir | L511L, S531L | NA | ||
| ICC98 | New Delhi | − | Ir, Or | R463L, Inh (C/T) | S95T | ||
| ICC100 | New Delhi | + | Rr, Ir, Or, Sr | S531L | R463L | S95T | NM |
| ICC101 | New Delhi | + | Rr, Or | S531L | S95T | ||
| ICC102 | New Delhi | + | Rr, Ir, Or | S531L | NM | S95T | |
| ICC103 | New Delhi | + | Rr, Ir, Or | L511L, N518T | NM | A90A, S95T | |
| ICC104 | New Delhi | + | Rr, Ir, Or | D516V | Δ30C, R463L | S95T | |
| ICC105 | New Delhi | + | Rr, Ir | K527N | R463L | ||
| ICC107 | New Delhi | − | Rr, Or, Sr | N518T, R528P | S95T | NM | |
| ICC109 | New Delhi | + | Ir | NM | |||
| ICC111 | New Delhi | + | Rr, Ir, Sr | S531W | Insertion 185C | K121K | |
| ICC114 | New Delhi | + | Ir | R463L | |||
| ICC115 | New Delhi | + | Rr, Ir | S531W | Insertion 98A, R463L | ||
| ICC123 | New Delhi | + | Rr, Ir, Or | R528P | NA | S95T | |
| ICC124 | New Delhi | + | Rr, Ir | H526Y | R463L | ||
| ICC125 | New Delhi | + | Rr | L511L, S531L | |||
| ICC128 | New Delhi | + | Rr, Or, Sr | H526Y, R528H | S95T | K121K | |
| ICC129 | New Delhi | + | Rr, Ir, Sr | R528P | R463L | K121K | |
| ICC147 | New Delhi | − | Rr, Ir, Or | S531W | Δ109G, R463L | S95T | |
| ICC203 | New Delhi | + | Rr | D516V | |||
| ICC204 | New Delhi | + | Rr, Ir | L521L, K527N | R463L | ||
| ICC205 | New Delhi | + | Rr, Ir | D516V | NA | ||
| ICC206 | New Delhi | + | Rr, Ir | S531W | NM | ||
| ICC208 | New Delhi | − | Rr | D516V | R463L | ||
| ICC209 | New Delhi | − | Rr | D516V | |||
| ICC210 | New Delhi | + | Rr, Or | L511V, N518T | S95T | ||
| ICC211 | New Delhi | + | Ir, Or | R463L | S95T | ||
| ICC212 | New Delhi | − | Rr | S531L | |||
| ICC213 | New Delhi | + | Rr, Ir | R528H, S531W | R463L, Inh (C/T) | ||
| ICC214 | New Delhi | + | Ir | R463L | |||
| ICC215 | New Delhi | + | Rr | S531L | |||
| ICC216 | New Delhi | + | Rr, Sr | S531L | K121K | ||
| ICC217 | New Delhi | + | Rr, Ir, Or | S531L | R463L | S95T | |
| ICC218 | New Delhi | + | Rr, Ir, Or | S522Q | R463L | S95T | |
| ICC219 | New Delhi | + | Ir | T12P, R463L | |||
| ICC220 | New Delhi | + | Rr, Ir, Or, Sr | S531W | R463L, Inh (T/A) | D94G, S95T | NM |
| ICC221 | New Delhi | − | Rr, Or | L521L | D94A, S95T | ||
| ICC222 | New Delhi | − | Or | S95T | |||
| ICC223 | New Delhi | − | Rr, Or | H526Y | S95T | ||
| ICC225 | New Delhi | + | Rr, Or | S531L | S95T | ||
| ICC237 | NewDelhi | − | Rr, Or | D516G | S95T | ||
| ICC239 | NewDelhi | + | Rr, Ir | D516V | R463L | ||
| ICC240 | NewDelhi | + | Rr, Ir | D516V | R463L | ||
| ICC242 | NewDelhi | + | Rr, Or | L511V | S95T | ||
| ICC244 | NewDelhi | + | Rr, Ir, Or | S531L | A61T, R463L | S95T | |
| ICC246 | NewDelhi | + | Ir, Or | Insertion 185C,R463L | S95T | ||
| ICC275 | New Delhi | + | Rr,Or | H526Y | S95T | ||
| ICC277 | NewDelhi | + | Rr, Ir, Or, Sr | H526Y | Δ30C | D94A, S95T | K121K |
| ICC284 | New Delhi | − | Or | NM | |||
| ICC286 | NewDelhi | + | Rr,Ir | D516G | NM | ||
| ICC287 | NewDelhi | + | Rr,Ir | H526Y | NM | ||
| ICC325 | NewDelhi | + | Ir, Or,Sr | NM | NM | NM | |
| ICC326 | NewDelhi | + | Rr, Ir,Sr | H526L | Insertion 98A, R463L | NM | |
| ICC327 | New Delhi | + | Rr, Ir,Sr | S509R | R463L | K121K | |
| ICC328 | NewDelhi | − | Or | NM | |||
| ICC408 | NewDelhi | + | Rr, Ir, Or, Sr | Q510H, S531W | R463L | S95T | K121K |
| ICC425 | New Delhi | + | Rr,Ir | S531L | R463L | ||
| F4 | NewDelhi | + | Rr, Or | H526Y | D94G,S95T | ||
| F5 | New Delhi | + | Rr,Or | S531L | S95T | ||
| F7 | NewDelhi | + | Rr, Or | S531L | A90V,S95T | ||
| F8 | New Delhi | + | Rr, Or | S531L | A90V, S95T | ||
| F9 | New Delhi | + | Rr, Or | N518T | S91P, S95T | ||
| N31 | New Delhi | + | Ir | R463L | |||
| N33 | NewDelhi | + | Ir | R463L | |||
| N34 | NewDelhi | + | Ir | R463L | |||
| N35 | NewDelhi | + | Ir | D73N, R463L | |||
| N36 | NewDelhi | − | Ir | R463L | |||
| ICC32 | Ahmedabad | + | Rr,Ir | S531L | NA | ||
| ICC33 | Ahmedabad | + | Rr, Ir, Or | S509R | R463L | S95T | |
| ICC36 | Ahmedabad | + | Ir,Or | R463L | S95T | ||
| ICC37 | Ahmedabad | + | Rr,Ir | D516G | Insertion 98A,R463L | ||
| ICC131 | Ahmedabad | + | Rr,Ir | H526Y,R528P | R463L | ||
| ICC132 | Ahmedabad | − | Or | S95T | |||
| ICC133 | Ahmedabad | + | Rr,Ir | H526R | Δ30C,R463L | ||
| ICC134 | Ahmedabad | − | Rr, Or | S522Q | S95T | ||
| ICC136 | Ahmedabad | + | Rr, Ir | H526R | Insertion 98A,R463L | ||
| ICC137 | Ahmedabad | − | Or | S95T | |||
| ICC138 | Ahmedabad | − | Or | S95T | |||
| ICC226 | Ahmedabad | + | Rr,Ir | L511L, H526R | Δ109G,R463L | ||
| ICC233 | Ahmedabad | + | Rr, Ir | D516V, H526Y | Δ30C, R463L | ||
| ICC151 | Chandigarh | − | Or | S95T | |||
| ICC154 | Chandigarh | − | Rr,Ir, Or | S531L | Δ30C,R463L | S95T | |
| ICC155 | Chandigarh | + | Ir | Δ30C,R463L | |||
| ICC159 | Chandigarh | + | Rr,Or | H526Y | S95T | ||
| ICC161 | Chandigarh | + | Rr,Or | S522Q | S95T | ||
| ICC162 | Chandigarh | − | Rr,Or | S522Q | S95T | ||
| ICC164 | Chandigarh | − | Or | S95T | |||
| ICC165 | Chandigarh | − | Or | S95T | |||
| ICC166 | Chandigarh | − | Or | S95T | |||
| ICC167 | Chandigarh | + | Ir,Or | R463L | S95T | ||
| ICC168 | Chandigarh | + | Or | S95T | |||
| ICC169 | Chandigarh | + | Rr,Ir | N518T | R463L | ||
| ICC170 | Chandigarh | − | Or | S95T | |||
| ICC171 | Chandigarh | + | Rr,Ir | S531L | R463L | ||
| ICC172 | Chandigarh | + | Rr,Or | S522Q | S95T | ||
| ICC173 | Chandigarh | − | Rr,Sr | H526L | K121K | ||
| ICC174 | Chandigarh | + | Or | S95T | |||
| ICC175 | Chandigarh | − | Rr,Or | H526Y | S95T | ||
| ICC247 | Chandigarh | − | Rr,Or | D516V | S95T | ||
| ICC248 | Chandigarh | − | Rr,Or | D516V | S95T | ||
| ICC249 | Chandigarh | − | Or | S95T | |||
| ICC251 | Chandigarh | − | Or | NM | |||
| ICC254 | Chandigarh | + | Rr | S531L | |||
| ICC255 | Chandigarh | − | Rr,Or | N518T | NM | ||
| ICC256 | Chandigarh | + | Rr, Ir | H526Y | NM | ||
| ICC257 | Chandigarh | + | Rr,Or | Q510H,L511L | S95T | ||
| ICC262 | Chandigarh | + | Rr, Ir, Or | D516V | R463L | S95T | |
| ICC95 | Bangalore | − | Or | S95T | |||
| ICC96 | Bangalore | + | Rr,Or | S531L | S95T | ||
| ICC399 | Bangalore | + | Rr,Or | S531W | NM | ||
| ICC524 | Bangalore | + | Rr,Ir | S531L | R463L | ||
| ICC525 | Bangalore | + | Rr, Ir | S531L | R463L | ||
| ICC143 | Shimla | − | Or | NM | |||
| ICC144 | Shimla | + | Ir | R463L | |||
| ICC145 | Shimla | − | Or | NM | |||
| A3 | Agra | + | Rr,Ir, Sr | S531L | R463L | NM | |
| A4 | Agra | + | Rr,Ir | D516V | R463L | ||
| A9 | Agra | + | Rr,Ir | S531L | R463L | ||
| A11 | Agra | + | Rr,Ir | D516G | T11A,R463L | ||
| A12 | Agra | + | Rr,Ir | D516V | N35D,R463L | ||
| A13 | Agra | + | Rr,Ir | D516V,N518T | R463L | ||
| A14 | Agra | + | Rr,Ir | D516V | R463L | ||
| A15 | Agra | + | Rr,Ir | S531L | R463L | ||
| ICC332 | Jaipur | + | Rr,Ir | H526R | Insertion 185C, R463L | ||
| ICC337 | Jaipur | + | Rr,Ir | S531L | R463L | S95T | |
| ICC85 | Chennai | + | Rr,Ir, Or | H526R | NA | S95T | |
History of treatment in the previous 20 months.
Rr, rifampin resistant; Ir, isoniazid resistant; Or, ofloxacin resistant; Sr, streptomycin resistant.
NA, no amplification; NM, no mutation; Inh, mutation in the inhA ribosome binding site; Δ, deletion at the indicated nucleotide position.
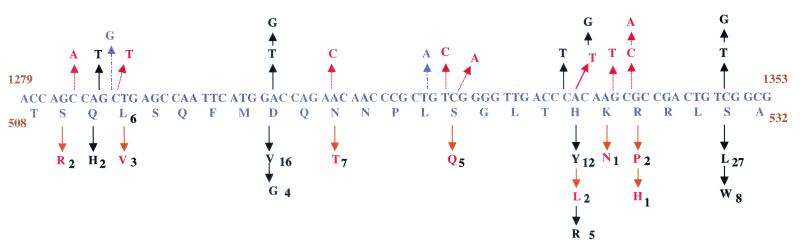
A stretch of 30 amino acids at the center of the amplicon for the rpoB locus was studied. Amino acids 432 to 458 comprised the hot-spot region for mutations. For the sake of comparison, we used the corresponding Escherichia coli numbering, which is amino acids 507 to 533. We identified previously reported mutations as well as certain novel mutations. Codon 531 seemed to be the most vulnerable to mutations, as most rifampin-resistant isolates had this mutation (Fig. 1). Of the 93 rifampin-resistant strains in our study, 28 had the missense mutation Ser531Leu and 8 had the substitution Ser531Trp. The next most common mutations were the amino acid substitutions Asp516Val or Asp516Gly (20 isolates) and His526Tyr, His526Leu, or His526Arg (19 isolates). We found two isolates with Gln510His changes. While all these mutations have been reported earlier, we also found mutations that have not been reported previously. These included Ser509Arg (isolate ICC33), Leu511Val (isolate ICC242), Asn518Thr (isolate ICC107), Ser522Gln (isolate ICC172), Lys527Asn (isolate ICC105), Arg528Pro (isolate ICC129), and Arg528His (isolate ICC213). Most of these mutations occurred less frequently, comprising about 24% of the total mutations in the 94 isolates studied. Other mutations identified in our study were silent mutations at amino acids Leu511 and Leu521. Interestingly, the mutation at position 511 never occurred alone and was present only in isolates with more than one mutation at the rpoB locus.
FIG. 1.
Summary of mutations at codons 508 to 532 in the rpoB gene. The wild-type sequence and amino acids are shown in the middle frame. Nucleotide changes are marked with arrows in the top frame, and the corresponding amino acid changes are denoted in the bottom frame. The amino acids are subscripted with numbers that indicate the number of isolates harboring the change. Changes marked with orange lines (dotted arrows) are novel mutations; silent mutations are marked with blue lines (dashed arrows). Codons 531, 526, and 516 exhibit high degrees of polymorphism. Codons 509, 511, 522, 527, and 528 show novel mutations.
An important outcome of these studies is the direct correlation of certain mutations to high MICs. Table 3 lists the isolates, their mutations, and the corresponding MICs at which they remained resistant. Mutations in codons 516 and 521 conferred low-level resistance (MIC, <40 μg/ml) to rifampin, whereas mutations in codons 510, 526, 527, 528, and 531 were seen to confer high levels of resistance (MICs, ≥64 μg/ml). Amino acids 526 to 531 appear to be very important in drug target interactions, and mutations in them result in MICs in the range of 64 μg/ml and above. In a few cases (e.g., for isolates ICC204, ICC257, and ICC128), double mutations were found to have an additive effect on the degree of resistance.
TABLE 3.
Correlation of specific mutations with rifampin MICsa
| Strain | Rifampin MIC (μg/ml) | Mutation | Mutation type | Amino acid change |
|---|---|---|---|---|
| ICC221 | 10 | G1317A | Novel | L521L |
| ICC208, ICC205 | 10 | A1304T | Reported | D516V |
| ICC37 | 10 | A1304G | Reported | D516G |
| ICC204 | 40 | G1317A | Novel | L521L |
| ICC204 | 40 | G1336T | Novel | K527N |
| ICC105 | 40 | G1336T | Novel | K527N |
| ICC129 | 40 | G1338C | Novel | R528P |
| ICC131 | 40 | C1331T | Reported | H526Y |
| ICC131 | 40 | G1338C | Novel | R528P |
| ICC123 | 64 | G1338C | Novel | R528P |
| ICC100 | 64 | C1349T | Reported | S531L |
| ICC213 | 64 | G1340A | Novel | R528H |
| ICC213 | 64 | C1349G | Reported | S531W |
| ICC218 | 64 | T1321C | Novel | S522Q |
| ICC218 | 64 | C1322A | Novel | S522Q |
| ICC257 | 64 | G1287T | Novel | Q510H |
| ICC257 | 64 | C1288T | Novel | L511L |
| ICC275 | 64 | C1333T | Reported | H526Y |
| ICC220 | 64 | C1349G | Reported | S531W |
| ICC128 | 128 | C1331T | Reported | H526Y |
| ICC128 | 128 | G1338A | Novel | R528H |
Missense mutations in the RpoB protein at amino acid positions 510, 511, 522, 526, 527, 528, and 531 confer higher levels of resistance (MICs, ≥40 μg/ml) than those at positions 509, 516, and 521 (MICs, ≤10 μg/ml).
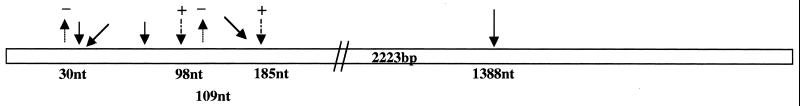
Insertion, deletion, and substitution mutations were mapped in the katG locus in 24 isoniazid-resistant isolates. In the present study we looked for mutations in the 5′ region (nucleotides [nt] 3 to 239) and the midregion (nt 1187 to 1600) of the katG gene, corresponding to amino acid positions 2 to 77 and 395 to 533, respectively. The results are summarized in Fig. 2. A C nucleotide at position 30 was deleted in six of the isolates. This deletion results in chain termination, thereby generating only a short polypeptide of 26 amino acids. Another deletion of a single nucleotide, a G residue at position 109, was observed in two isolates; this deletion would result in the production of a 45-amino-acid truncated polypeptide. Insertions were also observed at nt positions 98 (an A nucleotide) and 185 (a C nucleotide) in four and three isolates, respectively. Both of these insertions cause aberrant chain termination. Ala61Thr, Thr12Pro, Thr11Ala, Asp73Asn, and Asn35Asp missense mutations were observed in this locus in a few of the isolates. These are novel observations, as there are no reports of such mutations occurring in isoniazid-resistant strains from other parts of the world. We were unable to amplify this locus in six of the isolates (isolates ICC14, ICC23, ICC32, ICC85, ICC123, and ICC205), indicating a partial deletion of the gene. A common mutation in all these isolates was Arg463Leu. However, this mutation has been shown to have no direct consequence for drug resistance. To confirm this we sequenced this locus for all 126 isolates included in the study. It was found that the majority of the isolates carried this change. It has been argued previously that this polymorphism in the katG locus might be more important as a marker of evolution than as a marker of resistance (22). Three isoniazid-resistant isolates carried mutations in the ribosomal binding site upstream of the inhA gene. While two isolates showed a C-to-T transition, one had a T-to-A transversion. These mutations have previously been reported by other groups. The present understanding of these mutations is that they probably confer resistance by a drug titration effect.
FIG. 2.
Summary of mutations in the katG gene. Deletions are indicated by lines with a minus sign, while insertions are depicted by dashed lines with a plus sign. Solid lines show the substitutions. Codon 463 exhibited the highest degree of polymorphism, followed by the deletion at nucleotide 30.
Sixty-eight ofloxacin-resistant isolates were analyzed. The hot-spot region of the gyrA gene spanning codons 89 to 95 was sequenced to identify mutations. Most of the isolates showed a single mutation corresponding to the amino acid change Ser95Thr (Fig. 3). The second most common mutation, observed in four isolates, was Asp94Gly or Asp94Ala. Two isolates had an Ala90Val substitution, while one had a silent mutation at this codon. Seven isolates had double mutations, with the S95T change being common to all seven. These mutations were present in MDR isolates for which the MICs of the drugs were high, including the frontline drugs used in antituberculosis therapy. All strains were also checked for mutations in the gyrB locus, which is associated with low levels of resistance. However, we found no mutations in the gyrB loci of these isolates. It has been argued that the S95T mutation does not correlate with drug resistance (22). It therefore appears that the isolates have acquired resistance to ofloxacin via other mechanisms.
FIG. 3.
Summary of missense mutations in the gyrA locus. Nucleotide changes are indicated on top of the wild-type sequence, and the corresponding amino acid changes are shown at the bottom. The most common mutation in this locus is Ser95Thr.
We tested 14 isolates resistant to streptomycin for mutations in the rpsL and rrs loci. In eight strains we found a novel silent mutation at amino acid position 121 in the rpsL locus, where the codon AAA (Lys) was changed to AAG (Lys), but we found no mutations in the rrs genes. To our knowledge, there are no reports of this mutation. The reported mutations at the rpsL locus are generally Leu43Arg, Leu43Thr, or Lys88Arg. We are still not clear about how a mutation at this locus leads to the development of streptomycin resistance. The remaining isolates probably acquired resistance by other means, such as by the development of a permeability barrier or by the production of drug-altering enzymes.
A point to be kept in mind is that the majority of isolates included in the present study were from North India. Our data are therefore inherently biased toward drug-resistant strains from this region and should not be seen as representative for isolates from the whole of India.
DISCUSSION
The mycobacterium uses various mechanisms to evade killing by drugs, including mutations in genes that code for drug target proteins (20), a complex cell wall which blocks drug entry, and membrane proteins that act as drug efflux pumps (6, 14). The objective of the present study was to identify mutations in drug target loci in Indian strains of M. tuberculosis and to identify the different drug resistance genotypes. As in all such studies, the aim was to generate information about the markers associated with drug resistance, polymorphisms in the drug target genes, the association of the level of resistance with particular mutations, etc. Our findings of mutations in the rpoB, katG, and rpsL loci are similar to those reported from other parts of the world, especially the common mutations, which reflect a global pattern (20). Rifampin resistance is often regarded as an excellent surrogate marker for MDR tuberculosis (4, 10), and our study corroborates this hypothesis. The mutation frequency of codon 531 (rpoB) was similar to that reported earlier (13, 17, 19, 20, 21, 25). Significantly, the frequency of mutations (relative to those of other mutations) was higher at codon 516 and lower at codon 526 in Indian isolates compared to those reported elsewhere. We found novel mutations that broaden the range of known mutations at this locus. When taken together, these mutations were detected in a significant number of drug-resistant isolates, a fact that needs to be considered when designing tools for the detection of MDR M. tuberculosis. We found a definite correlation between MICs and the type of mutation in many isolates. As reported by previous investigators (24), mutations at positions 528 and 531 are important in the development of high MICs. Our findings further strengthen the belief that the degree of resistance to rifampin exhibited by an isolate is related to the type of mutation in the rpoB locus.
In isoniazid-resistant isolates, significantly more deletion and insertion mutations than substitution mutations were found, of which a few have been reported previously (11, 19). We observed that almost all isolates studied carried the Arg463Leu substitution, which is also present in isolates that were sensitive to isoniazid. This is in concordance with a report from Sreevatsan et al. (22), who argue that polymorphism at this residue does not contribute to resistance per se but is an important marker for evolutionary genetics. The insertions and deletions in the katG locus invariably resulted in chain truncation and termination, leading to the generation of dysfunctional polypeptides. We found changes in the putative ribosomal binding site of the inhA gene in three isolates. While the exact mechanism of how these mutations confer resistance to isoniazid is not clear, reports (1, 18, 20) indicate that they probably increase the levels of enoyl-acyl carrier protein reductase which in turn leads to resistance via a drug titration mechanism. In isolates with no mutations in the hot-spot region of the gene, the complete sequencing of the gene is being done. However, resistance to isoniazid can also be due to mutations in the ahpC-oxyR and kasA gene loci (7, 16).
Fluoroquinolones comprise the secondary drug regimen in the treatment of tuberculosis. A large number of isolates were resistant to ofloxacin, which could be due in part to the inaccurate diagnosis of tuberculosis as a bacterial infection and fluoroquinolone overuse in the population. Codons 89, 90, 91, 94, and 95 in the gyrA gene have been shown to be polymorphic (20, 23, 26). The most common mutation in ofloxacin-resistant isolates in the present study was Ser95Thr, which reportedly has no direct role in the development of drug resistance, as it also occurs in drug-sensitive strains (22). It seems likely that ofloxacin resistance possibly results due to mutations elsewhere in the gene or the presence of drug efflux pumps.
Mutations in codons 43 and 88 of the rpsL gene generally result in high levels of resistance to streptomycin, while mutations in the loop at codon 530 or the region at codon 915 of the rrs locus are associated with low levels of resistance (3). We did not find any of these mutations in the 14 streptomycin-resistant isolates included in our study. However, we did observe a silent mutation at codon 121 that has not been reported by any other group.
Our study provides valuable data on the different kinds of mutations occurring at various target loci in Indian clinical isolates of M. tuberculosis that enhance our understanding of the molecular mechanisms of drug resistance. The diversity of the polymorphisms exhibited at these loci by the drug-resistant strains indicates the prevalence of a large numbers of drug-resistant strains in this region. Additionally, our data will also assist in the process of designing new molecular biology-based techniques for the diagnosis of MDR tuberculosis. Such methods promise faster detection rates compared to those achieved by methods based solely on culture of the isolates.
Acknowledgments
This study was supported by research grants from the Department of Biotechnology, Government of India.
We thank Sunder S. Bisht and Mohammed Iliyas Ghazi, who helped with the generation of the sequencing data. We are also grateful to Ram Das, Kiran Srivastava, and V. Chauhan of the Central Jalma Institute of Leprosy, who provided us with the isolate DNAs.
REFERENCES
- 1.Basso, L. A., R. Zheng, J. M. Musser, W. R. Jacobs, Jr., and J. S. Blanchard. 1998. Mechanism of isoniazid resistance in Mycobacterium tuberculosis: enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J. Infect. Dis. 178:769–775. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, B. R., and J. L. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055–1064. [DOI] [PubMed] [Google Scholar]
- 3.Bottger, E. C. 1994. Resistance to drugs targeting protein synthesis in mycobacteria. Trends Microbiol. 2:416–421. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1993. Initial therapy for tuberculosis in the era of multidrug resistance. Recommendations of the Advisory Council for the Elimination of Tuberculosis. Morb. Mortal. Wkly. Rep. 42(RR-7). [PubMed]
- 5.Cohn, D. L., F. Bustreo, and M. C. Raviglione. 1997. Drug-resistant tuberculosis: review of the worldwide situation and the W.H.O./IULATD global surveillance project. Clin. Infect. Dis. 24:(Suppl. 1)S121–S130. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. (Erratum, 396: 190–198.) [DOI] [PubMed] [Google Scholar]
- 7.Collins, D. M., and T. M. Wilson. 1996. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol. Microbiol. 19:1025–1034. [DOI] [PubMed] [Google Scholar]
- 8.Harkin, T. J., and H. W. Harris. 1995. Treatment of multidrug resistant tuberculosis, p.843–850. In N. W. Rom and S. Garay (ed.), Tuberculosis. Little, Brown & Company, Boston, Mass.
- 9.Harris, K. A., Jr., U. Mukundan, J. M. Musser, B. N. Kreiswirth, and M. K. Lalitha. 2000. Genetic diversity and evidence for acquired antimicrobial resistance in Mycobacterium tuberculosis at a large hospital in South India. Int. J. Infect. Dis 4:140–147. [DOI] [PubMed] [Google Scholar]
- 10.Hasnain, S. E., A. Amin, N. Siddiqi, M. Shamim, N. K. Jain, A. Rattan, V. M. Katoch, and S. K. Sharma. 1998. Molecular genetics of multiple drug resistance (MDR) in Mycobacterium tuberculosis, p.35–40. In R. L. Singhal and O. P. Sood (ed.), Drug resistance: mechanism and management. Proceedings of the Fourth Annual Ranbaxy Science Foundation Symposium. Ranbaxy Science Foundation, New Delhi, India.
- 11.Heym, B., P. M. Alzari, N. Honore, and S. T. Cole. 1994. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15:235–245. [DOI] [PubMed] [Google Scholar]
- 12.Janmeja, A. K., and B. Raj. 1998. Acquired drug resistance in tuberculosis in Harayana, India. J. Assoc. Physicians India 46:194–198. [PubMed] [Google Scholar]
- 13.Kapur, V., L.-L. Li, S. Iordanescu, M. C. Hamrick, A. Wanger, B. N. Kreiswirth, and J. M. Musser. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis, K. 1994. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem. Sci. 19:119–123. [DOI] [PubMed] [Google Scholar]
- 15.Mathur, M. L., P. K. Khatri, and C. S. Base. 2000. Drug resistance in tuberculosis patients in Jodhpur District. Indian J. Med. Sci. 54:55–58. [PubMed] [Google Scholar]
- 16.Mdluli, K., R. A. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry III. 1998. Inhibition of Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science 280:1607–1610. [DOI] [PubMed] [Google Scholar]
- 17.Miller, L. P., J. T. Crawford, and T. M. Shinnick. 1994. The rpoB gene of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris, S., G. Han Bai, P. Suffys, L. P. Gomez, M. Fairchok, and D. Rouse. 1995. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J. Infect. Dis. 171:954–960. [DOI] [PubMed] [Google Scholar]
- 19.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic bases of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3–29. [DOI] [PubMed] [Google Scholar]
- 21.Siddiqi, N., M. Shamim, N. K. Jain, A. Rattan, A. Amin, V. M. Katoch, S. K. Sharma, and S. E. Hasnain. 1998. Molecular genetic analysis of multidrug resistance in Indian isolates of Mycobacterium tuberculosis. Mem. Inst. Oswaldo Cruz 93:589–594. [DOI] [PubMed] [Google Scholar]
- 22.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takiff, H. E., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of the Mycobacterium tuberculosis gyrA and gyrB genes and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi, H., H. Aramaki, Y. Nikaido, Y. Mizuguchi, M. Nakamura, T. Koga, and S. Yoshida. 1996. Rifampicin resistance and mutations of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 144:103–108. [DOI] [PubMed] [Google Scholar]
- 25.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. T. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650. [DOI] [PubMed] [Google Scholar]
- 26.Xu, C., B. N. Kreiswirth, S. Sreevatsan, J. M. Musser, and K. Drlica. 1996. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J. Infect. Dis. 174:1127–1130. [DOI] [PubMed] [Google Scholar]