Abstract
Over a 12-year period (1989 to 2000), 159 Klebsiella pneumoniae isolates harboring extended-spectrum β-lactamases (ESBLs) (4.8% of the total number of K. pneumoniae isolates obtained) were recovered from 58 patients, who were mainly hospitalized in intensive care and surgery units. For 62 representative isolates from 58 patients, 31 clonal types harboring TEM-4 (n = 5), SHV-2 (n = 7), SHV2a (n = 4), SHV-5 (n = 1), CTX-M-10 (n = 13), or CTX-M-9 (n = 1) β-lactamases were identified by pulsed-field gel electrophoresis. This is the first report to document the presence of the CTX-M-10 or the CTX-M-9 β-lactamase in K. pneumoniae. These β-lactamases were previously identified in Escherichia coli isolates from Spain. Only two of five K. pneumoniae TEM-4 clones caused more than a single case of infection, with one of them spreading for 9 months. A single plasmid was detected among these TEM-4 clones. Only two of seven K. pneumoniae clones containing SHV-2 and three of four strains harboring SHV-2a were detected in more than one case of infection. Plasmids encoding SHV-2 or SHV-2a were unrelated. Four of 13 K. pneumoniae CTX-M-10 clones were found in more than one patient, with two of them recovered 2 and 5 years apart. As in the case of the SHV-2 isolates, we were unable to document a common transmissible genetic element that could explain the polyclonal structure of our isolates. Nevertheless, the spread of a single gene may be suggested by the presence of a conserved set of noncoding polymorphisms in the sequences. Most ESBL-producing K. pneumoniae clones were ephemeral, being poorly selected and maintained in the hospital setting, but the genes encoding ESBL persisted successfully over the years that the strains were recovered, probably as a minority gene population in the hospital metagenome.
Since extended-spectrum β-lactamases (ESBLs) were initially reported in Klebsiella pneumoniae in Germany in 1983 (36), they have been increasingly described worldwide. Most ESBLs from K. pneumoniae are plasmid-encoded enzymes derived from the classical TEM- and SHV-type β-lactamases, which confer different levels of resistance to cefotaxime, ceftazidime, and other broad-spectrum cephalosporins and monobactams (36). In 1989, a novel type of ESBL that conferred a high level of resistance to cefotaxime but that had a low level of activity against ceftazidime was nearly simultaneously identified in an Escherichia coli strain isolated in Germany and in a Salmonella enterica serovar Typhimurium isolate recovered in Argentina (46). This new family of plasmid-mediated ESBLs of Ambler class A has been named the cefotaximase (CTX-M) family. The ESBL distribution varies from country to country. In general, TEM-type ESBLs predominate in the United States, while SHV-type ESBLs are the ESBLs most frequently isolated in Western Europe (36). Cefotaximases (CTX-M) have mainly been detected in South America, Eastern Europe, Japan, and more recently, Spain and Kenya (6, 21, 33, 35, 38, 46).
Genes encoding ESBLs are usually located on transferable plasmids that may also carry other resistance determinants such as those for resistance to aminoglycosides, tetracyclines, chloramphenicol, trimethoprim, and sulfonamides (13, 19, 30, 36, 49). The locations of these genes on transferable elements (1, 7, 19, 26, 35, 36, 45) may enhance the spread of ESBLs among different enterobacterial strains (35, 47). The epidemiology of ESBL-producing K. pneumoniae is complex and varies among institutions. Although identical genes encoding drug resistance have been found in different isolates and plasmid dissemination has been documented (1, 7, 11, 16, 35, 47), the spread of an epidemic strain remains the most commonly reported mechanism of ESBL dissemination (49). Most outbreaks are limited to areas where high-risk patients are cared for (36), but several epidemics have been detected among neonates, elderly patients, and even outpatients (2, 36, 47). In addition, the spread of ESBL-producing K. pneumoniae strains between hospitals (1, 7, 15–17, 36) and, more rarely, between countries (36, 40) has been demonstrated. Despite these facts, the possibility that the concurrent detection of different ESBL variants in multiple geographic locations may also be the result of the convergent evolution of proteins from widely spread broad-spectrum non-ESBL enzymes cannot be excluded.
The possibility of exploring the population biology of ESBL-harboring K. pneumoniae in a single institution from the time when these enzymes were first detected to the present may be essential to obtain an understanding of the evolution of the highly complex interplay among genes, plasmids, and clones involved in ESBL-mediated resistance. That was the goal of the present research.
MATERIALS AND METHODS
Setting.
The Hospital Ramón y Cajal is a 1,200-bed university teaching hospital (Alcalá de Henares University) that provides medical and surgical care for the pediatric and adult populations in the northeast region of Madrid, Spain. The institution provides specialized health care for a population of about 500,000.
Clinical isolates.
All K. pneumoniae isolates that harbored ESBL isolates and that were identified in the Microbiology Department from January 1989 to December 2000 were included in the study. One isolate per patient (or more than one isolate per patient when different pI patterns were observed) was selected for characterization of its ESBL and genetic studies in order to avoid overrepresentation of a particular strain. Preliminary identification and susceptibility tests were performed by using the automated microdilution PASCO system (Difco, Detroit, Mich.) or the WIDER system (Fco. Soria Melguizo, Madrid, Spain). The putative production of an ESBL was ordinarily detected by the double-disk synergy test (20).
Antimicrobial susceptibility.
The MICs of different β-lactam antibiotics were determined by the NCCLS agar dilution method (27). The amoxicillin-clavulanate and piperacillin-tazobactam combinations were used at a 2:1 ratio and a fixed concentration of 4 mg/liter, respectively. Susceptibilities to non-β-lactam antibiotics were tested by the standard disk diffusion method (28). The corresponding manufacturers provided the β-lactams and the β-lactam inhibitors as powders with stated potencies, whereas disks were purchased from Oxoid (Basingstoke, England). E. coli ATCC 25922, Enterococcus faecalis ATCC 29212, and Staphylococcus aureus ATCC 29213 were used as control strains.
Conjugation experiments.
Conjugation experiments were carried out by a broth or filter mating method with ESBL-producing K. pneumoniae isolates corresponding to different clonal types or to the same clonal type with different susceptibilities or different pI patterns. E. coli BM21 (nalidixic acid resistant, lactose fermentation positive, and plasmid free) (3) or a rifampin-resistant mutant of that strain (BM21R) was used as the recipient for mating experiments with ESBL-producing K. pneumoniae isolates susceptible and resistant to nalidixic acid, respectively. Overnight cultures of donor and recipient strains grown on brain heart infusion (BHI) broth (Difco) at 37°C were added to 3 ml of fresh BHI broth at a donor-recipient ratio of 1:2 (25 μl of the donor strain culture and 50 μl of the recipient strain culture), and then the mixture was incubated for 18 h at 37°C. One hundred microliters of this mixed culture was plated onto MacConkey agar (Difco) containing 64 mg of nalidixic acid per liter and 2 mg of cefotaxime or 1 mg of ceftazidime per liter. Colonies growing on the selection plates and again on subculture on MacConkey agar supplemented with cefotaxime or ceftazidime were subjected to the double-disk synergy test to confirm the presence of ESBL transconjugants. Resistance was considered nontransferable when the isolates failed to transfer it by both the broth and filter mating methods in more than two experiments each. The frequency of transfer was expressed relative to the number of donor cells.
Plasmid DNA analysis.
Plasmid DNA from the K. pneumoniae isolates and their corresponding E. coli transconjugants was obtained by the method described by Kado and Liu (44). Plasmid sizes were estimated from standard curves of the logarithm of the molecular sizes of the plasmids from E. coli V517 (eight plasmids ranging from 2.1 to 54.2 kb) and E. coli NCTC 50192 (four plasmids of 7, 36.2, 63.8, and 148.5 kb, respectively) against the relative mobilities of the tested isolates (44). For fingerprinting analysis, plasmid DNA was obtained by using a QIAgen Plasmid Midi kit (QIAGEN, Hilden, Germany), digested with EcoRI and PstI (from Roche Diagnostics, Barcelona, Spain), and electrophoresed in 1% agarose at 120 V for 2.5 h.
IEF.
β-Lactamases were characterized by isoelectric focusing (IEF) by applying the supernatant of a crude sonic extract to Phast gels (pH gradient, 3 to 9) in a Phast system (Pharmacia AB, Uppsala, Sweden). β-Lactamases with known pIs were included in each assay as controls. Gels were stained with 500 μg of nitrocefin (Oxoid) per ml to identify the bands corresponding to β-lactamases.
PCR detection of genes coding for ESBLs.
Detection of gene sequences coding for the TEM, SHV, and CTX-M enzymes was performed by PCR with genomic DNA from K. pneumoniae isolates (Instagene; Bio-Rad, La Jolla, Calif.) and plasmid DNA from the corresponding transconjugants (High Pure plasmid isolation kit; Roche Diagnostics). The oligonucleotide primer sets specific for the TEM and SHV genes and the cycling conditions used in the PCR assays have been described previously: 5′-ATA AAA TTC TTG AAG AC-3′ and 5′-TTA CCA ATG CTT AAT CA-3′ for amplification of a 1,076-bp sequence of blaTEM (25) and 5′-GGG TTA TTC TTA TTT GTC GC-3′ and 5′-TTA GCG TTG CCA GTG CTC-3 for amplification of a 930-bp sequence of blaSHV (37). For the PCR for the CTX-M-10 β-lactamase, the following primers were used: primer CTX-M-F8 (5′-CCG CGC TAC ACT TTG TGG C-3′) and primer CTX-M-R3 (5′-TTA CAA ACC GTT GGT GAC G-3′). The cycling conditions used for the PCR with these primers were 35 cycles of 94°C for 45 s, 56°C for 45 s, and 72 for 45 s, with a final extension of 72°C for 10 min (33; this study). For CTX-M-9, we used primers 5′-GTG ACA AAG AGA GTG CAA CGG-3′ and 5′-ATG ATT CTC GCC GCT GAA GCC-3′, and the conditions were 35 cycles of 94°C for 45 s, 62°C for 45 s, and 72 for 45 s, with a final extension of 72°C for 10 min (41). The PCR products were separated in 0.8% agarose gels and were visualized under UV light after staining with ethidium bromide. SHV-specific PCR products obtained from genomic DNA from wild-type K. pneumoniae isolates were digested with NheI (New England Biolabs, Beverly, Mass.) to distinguish the SHV-type ESBLs from broad-spectrum SHV-type ESBLs (31).
NheI RFLP analysis of SHV-specific PCR products.
We used a combination of IEF and NheI restriction fragment length polymorphism (RFLP) analysis of SHV-specific PCR products to identify multiple SHV-type β-lactamases per isolate. Some SHV-type ESBLs with pIs of 8.2 and 7.6 present a glycine-to-serine substitution at position 238. This point mutation is due to the change of guanosine to adenosine (in boldface) in the sequence G′CTAGC, which creates a new NheI restriction site (31). This restriction site is absent from the structural genes of the SHV-6 or the SHV-8 ESBL and in the structural genes of the SHV-1 and SHV-11 broad-spectrum β-lactamases (http://www.lahey.org/studies/webt.htm). The presence of an NheI site in the SHV-specific product will give two bands of 768 and 162 bp, and its absence will result in no cleavage and, thus, in a full-length fragment of 930 bp (31, 37).
Sequencing of blaTEM, blaSHV, and blaCTX specific PCR products.
PCR products containing blaTEM, blaSHV, or blaCTX-M were purified with a QIAquick PCR purification kit (QIagen) and were subjected to direct sequencing on an ABI PRISM 377 automated sequencer (Perkin Elmer Biosystems, Foster City, Calif.). The identity of the gene encoding TEM β-lactamases was resolved by sequencing of the blaTEM-specific PCR products with primers 5′-TTA CCA ATG CTT AAT CA-3′ (position 1069), 5′-CTC GTC GTT TGG TAT GGC-3′ (position 733), 5′-TTA CTG TCA TGC CAT CC-3′ (position 560), and 5′-CCT GCA GCA ATG GCA ACA-3′(position 749) (25). Primers 5′-GGG TTA TTC TTA TTT GTC GC-3′ (position 58) and 5′-TTA GCG TTG CCA GTG CTC-3 (position 988) were used to sequence the blaSHV genes (37; http://www.lahey.org/studies/webt.htm). Identification of the gene coding for the CTX-M-10 enzyme was performed with primers CTX-M-F8 (5′-CCG CGC TAC ACT TTG TGG C-3′; position 86 upstream from the start codon), CTX-M-F3 (5′-GCT GAT GAG CGC TTT GCG-3′; position 193), and CTX-M-R3 (5′-TTA CAA ACC GTT GGT GAC G-3′; position 858) (33).
PFGE.
Chromosomal DNA was prepared for pulsed-field gel electrophoresis (PFGE) as described previously (22). After digestion with XbaI (Roche Diagnostics), the DNA fragments were separated by electrophoresis in 1.2% agarose gels (pulsed-field agarose certified; Bio-Rad) and 0.5× Tris-borate-EDTA buffer by using a contour-clamped homogeneous electric field (CHEF-DRIII system; Bio-Rad). Electrophoresis conditions were 12°C at 6 V/cm for 27 h with pulse times ranging from 1 to 15 s for 7 h and 15 to 35 s for 17 h. The results were analyzed by following the criteria established by Tenover et al. (43).
RESULTS
Epidemiological background.
The first ESBL-producing K. pneumoniae strain isolated in our institution was obtained in September 1989. Since then and until December 2000, a total of 159 K. pneumoniae isolates recovered from 58 patients were identified as ESBL producers. These isolates represented 4.8% of all K. pneumoniae clinical isolates recovered in our institution during the period studied. Sixty-two ESBL-producing K. pneumoniae isolates from the 58 patients were selected for further studies. Among these 62 isolates, 31 distinct strains or clonal types were identified (see below).
The sites of isolation of the 62 clinical isolates were the respiratory tract (n = 22; 35.5%), urine (n = 14; 22.6%), blood (n = 11; 17.7%), rectal swabs (n = 5; 8.1%), wounds (n = 4; 6.5%), catheters (n = 3; 4.8%), and other sites (n = 3; 4.8%). Rectal swabs were taken from all potential reservoirs in the cardiopediatric ward for screening of ESBL-producing K. pneumoniae when the hospital experienced an outbreak in this ward in 1997 and 1998 (see below). Patient origins and the dates of isolation of the ESBL-producing K. pneumoniae isolates are expressed in Table 1. Of the 58 patients, 41% were in intensive care facilities, 36% were in surgical wards, 16% were in medical wards, and 7% were outpatients. The prevalence of ESBL-producing K. pneumoniae isolates from 1989 to 1996 ranged from 0.8 to 3.6%. Increased rates were observed during 1997 and 1998 (12.1 and 18.2%, respectively) due to an outbreak in the cardiopediatric unit involving 11 patients (2). In 1999 and 2000, the prevalence was similar to that in previous years, once the outbreak was controlled after the adoption of infection control measures (2).
TABLE 1.
Clinical data, PFGE profiles, IEF results, and β-lactamase characterization for ESBL-producing K. pneumoniae isolates recovered at the Hospital Ramón y Cajal (1989 to 2000)
| ESBL and PFGE types | Ward | Mo/yr | No. of isolates | No. of patients | β-Lactamase pI by IEF | Source (no. of isolates) | Conjugation frequency | Estimated plasmid size (kb) | Plasmid typea | ESBL type identified by PCR | ESBL identified by sequencingb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TEM type | |||||||||||
| KP8T | Cardiopediatric ICU | 9/95 | 2 | 2 | 7.6 + 5.9c | Blood (1) Respiratory (1) | 1 × 10−2 | 85 | pRYCE11 | TEM | TEM-4 |
| KP20T | Cardiopediatric ICU | 7/97 | 1 | 1 | 7.6 + 5.9 | Respiratory (1) | 1 × 10−2 | 85 | pRYCE11 | TEM | |
| KP21T | Cardiopediatric ICU | 7/97 | 1 | 1d,e | 7.6 + 5.9 | Blood (1) | NTf | TEM | |||
| 1 | 5.9 | Respiratory (1) | NT | ||||||||
| KP6T | Cardiopediatric ICU | 5/97–2/98 | 11 | 11e | 7.6 + 5.9 | Respiratory (5), rectal swabs (3), blood (2), other (1) | 1 × 10−2 | 85 | pRYCE11.1 | TEM | TEM-4 |
| KP27T | Cardiopediatric ICU | 5/99 | 1 | 1 | 5.9 | Respiratory (1) | 1 × 10−2 | 85 | pRYCE11 | TEM | TEM-4 |
| SHV type | |||||||||||
| KP1S | Internal medicine ICU | 9/89 | 1 | 1 | 7.6 + 5.4 | Urine (2) | 1 × 10−5 | FEg | SHV | SHV-2 | |
| Cardiopediatric ICU | 11–12/90 | 3 | 3 | Respiratory (2) | FE | ||||||
| KP11S | Cardiopediatric ICU | 7/90 | 1 | 1 | 7.6 + 5.4 | Blood (1) | 1 × 10−6 | NDh | SHV | SHV-2 | |
| KP2S | Cardiopediatric ICU | 6–11/91 | 4 | 4e,i | 7.6 + 5.4 | Wound (2)Respiratory (2) | 1 × 10−6 | 147 | pRYCE12 | SHV | SHV-2a |
| KP2aS | Cardiopediatric ICU | 11/91 | 1 | 7.6 + 5.4 | Blood (1) | 1 × 10−6 | 147 | pRYCE12 | SHV | SHV-2a | |
| KP3S | Cardiopediatric Cardiopediatric ICU | 1–11/92 | 4 | 2e | 7.6 + 5.4 | Blood (2) Respiratory (2) | 2 × 10−6 | 63 | pRYCE13 | SHV | SHV-2a |
| KP9S | Cardiopediatric ICU | 2–3/92 | 2 | 2 | 7.6 + 5.4 | Respiratory (2) | 2 × 10−6 | 75 | pRYCE14 | SHV | SHV-2a |
| KP14S | Cardiopediatric ICU | 7/92 | 1 | 1 | 7.6 + 5.4 | Respiratory (1) | 1 × 10−5 | 147 | pRYCE12 | SHV | SHV-2a |
| KP17S | Cystic fibrosis | 6/94 | 1 | 1 | 7.6 + 8.2 | Respiratory (1) | 3 × 10−7 | pRYCE15 | SHV | SHV-5 | |
| KP18S | Pediatric ICU | 8/95 | 1 | 1 | 7.6 | Respiratory (1) | ND | ND | ND | ND | ND |
| KP23S | Internal medicine | 7/98 | 1 | 1 | 7.6 | Urine (1) | 3 × 10−8 | 50 | pRYCE16 | SHV | SHV-2 |
| KP25S | Internal medicine | 7/98 | 1 | 1 | 7.6 | Respiratory (1) | 6 × 10−7 | 3 | pRYCE17 | SHV | SHV-2 |
| KP7S | Nephrology | 12/98 | 1 | 1 | 7.6 | Urine (1) | NT | SHV | SHV-2 | ||
| KP34S | Cardiopediatric ICU | 8–9/00 | 1 | 1 | 7.6 | Rectal swab (1) | 1.4 × 10−8 | >147 | FE | SHV | SHV-2 |
| KP34aS | Cardiopediatric ICU | 8/00 | 1 | 1 | 7.6 | Blood (1) | 1.4 × 10−8 | >147 | FE | SHV | SHV-2 |
| KP38S | Cardiopediatric ICU | 8/00 | 1 | 1 | 7.6 | Urine (1) | 1.4 × 10−8 | 70 | pRYCE18 | SHV | SHV-2 |
| CTX-M type | |||||||||||
| KP12C | Urology | 10/90 | 1 | 1 | 7.6 + 8.1 | Urine (1) | 3 × 10−7 | >147 | FE | CTX-M-10 | CTX-M-10 |
| KP13C | Outpatient | 11/91 | 1 | 1 | 8.1 | Rectal (1) | 3 × 10−7 | 147 | pRYCE19 | CTX-M-10 | CTX-M-10 |
| KP15C | Cardiopediatric ICU | 7/93 | 1 | 1 | 8.1 | Blood (1) | 5 × 10−8 | 147 | pRYCE20 | CTX-M-10 | CTX-M-10 |
| KP4C | Cardiovascular ICU | 8–9/93 | 1 | 1 | 8.1 | Blood (1) | NT | CTX-M-10 | |||
| Endocrinology | 1 | 1 | Wound (1) | ||||||||
| KP4aC | Urology | 7/98 | 1 | 1d | 8.1 | Urine (1) | 1 × 10−7 | 60 | pRYCE21 | CTX-M-10 | CTX-M-10 |
| Urology | 6/98 | 1 | 8.1 + 7.6 | Urine (1) | 1 × 10−7 | 60 | CTX-M-10 | CTX-M-10 | |||
| KP39C | Internal medicine | 4/96 | 2 | 2 | 8.1 | Urine (1), others (1) | NT | CTX-M-10 | |||
| KP22C | Pneumology | 9/97 | 1 | 1 | 8.1 | Respiratory (1) | 1.5 × 10−7 | 80 | pRYCE22 | CTX-M-10 | |
| KP26C | Pneumology | 7/98 | 1 | 1 | 8.1 | Respiratory (1) | 1 × 10−8 | 40 | pRYCE23 | CTX-M-10 | CTX-M-10 |
| KP26aC | Pneumology | 5/99 | 1 | 1 | 8.1 | Respiratory (1) | 1 × 10−8 | 40 | CTX-M-10 | CTX-M-10 | |
| Gastroenterology | 9/99 | 1 | 1 | 8.1 | Respiratory (1) | 1 × 10−8 | 40 | CTX-M-10 | |||
| KP30C | Endocrinology | 4/99 | 1 | 1 | 8.1 | Urine (1) | 3 × 10−7 | 63 | pRYCE24 | CTX-M-10 | |
| KP29C | Outpatient | 4/99 | 1 | 1 | 8.1 | Wound (1) | NT | CTX-M-10 | |||
| KP40C | Gastroenterology | 11/99 | 1 | 1 | 8.1 | Others (1) | 5 × 10−7 | 70 | CTX-M-9 | CTX-M-9 | |
| KP31C | Traumatology | 2/00 | 1 | 1 | 8.1 | Urine (1) | 1.5 × 10−7 | 60 | pRYCE24.1 | CTX-M-10 | |
| KP32C | Nefrology | 4/00 | 1 | 1 | 8.1 | Urine (1) | NT | CTX-M-10 | |||
| KP32Ca | Urology | 2-5/00 | 1 | 1 | 8.1 + 7.6 | Urine (1) | NT | CTX-M-10 | |||
| KP36C | Outpatient | 7/00 | 1 | 1 | 8.1 | Urine (1) | 5 × 10−7 | 40 | pRYCE25 | CTX-M-10 | CTX-M-10 |
EcoRI restriction enzyme analysis profile.
An empty field indicates that sequencing of the bla gene was not performed.
β-Lactamases produced by transconjugants are underlined.
A single patient had two isolates that belonged to the same clonal type but that had different pI profiles.
Patients carrying different clonal types: one patient carried isolates corresponding to clones KP21T and KP6T, and one patient carried isolates corresponding to clones KP2S and KP3S
NT, no transfer.
FE, failed extraction.
ND, not done.
One patient carried two variants of the same clonal type.
β-Lactam susceptibility profile and associated resistance.
The MICs for 62 clinical isolates representing each distinct clonal type or β-lactamase pattern and their corresponding transconjugants are shown in Table 2. All isolates were uniformly resistant to piperacillin (MICs, ≥128 μg/ml). In all but four isolates, this resistance was reduced by tazobactam to a susceptible range (MICs, 4 to 16 μg/ml). Moreover, all isolates were susceptible or intermediate to amoxicillin-clavulanate (MICs, 4/2 to 16/8 μg/ml) and susceptible to cefoxitin (MICs, <32 μg/ml). Isolates that produced ESBLs of the TEM type were more resistant to extended-spectrum cephalosporins and monobactams than those that produced ESBLs of the SHV type. Cefotaxime MICs ranged from 16 to 256 μg/ml for TEM-type ESBL-producing isolates and from 4 to 128 μg/ml for SHV-type ESBL-producing isolates. The corresponding ranges of ceftazidime MICs were 8 to 64 and 1 to 32 μg/ml, and the corresponding ranges of aztreonam MICs were 8 to 512 and 0.5 to 256 μg/ml. In both groups, cefepime was slightly more active (MICs, 2 to 32 μg/ml). For isolates with ESBLs of the CTX-M type, cefotaxime MICs (16 to 512 μg/ml) were higher than ceftazidime MICs (0.5 to 16 μg/ml), whereas cefepime and aztreonam MICs were between those of cefotaxime and ceftazidime (Table 2).
TABLE 2.
Antimicrobial susceptibilities of ESBL-producing K. pneumoniae isolates and their corresponding E. coli transconjugants at the Hospital Ramón y Cajal (1989 to 2000)a
| ESBL type by PFGE and clone | MIC (μg/ml)
|
Resistance phenotypea | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | PIP | TZP | CFZ | CXM | FOX | CTX | CAZ | FEP | ATM | ||
| TEM type | |||||||||||
| KP6T | 8 | 512 | 4 | 64 | 512 | 4 | 16 | 8 | 4 | 8 | G, T |
| KP8T | 8 | 1,024 | 8 | 32 | 512 | 8 | 128 | 32 | 8 | 16 | G, T |
| KP20T | 16 | 2,048 | 16 | 128 | 512 | 4 | 256 | 64 | 32 | 512 | G, T |
| KP21T | 4 | 2,048 | 8 | 128 | 256 | 16 | 64 | 64 | 2 | 32 | G, T |
| KP27T | 16 | 512 | 16 | 128 | 512 | 4 | 256 | 64 | 16 | 32 | G, L T, S, Nt, Te, C, Su, KI |
| KP6T R+ | 8 | 256 | 2 | 16 | 32 | 2 | 8 | 4 | 0.5 | 4 | G, T |
| KP8T R+ | 8 | 128 | 2 | 8 | 128 | 2 | 8 | 16 | 2 | 16 | G, T |
| KP20T R+ | 8 | 512 | 2 | 128 | 512 | 2 | 32 | 32 | 8 | 32 | G, T |
| KP27T R+ | 8 | 256 | 2 | 128 | 32 | 2 | 8 | 16 | 2 | 16 | G, T, Su |
| SHV type | |||||||||||
| KP1S | 8 | >1,024 | 256 | 128 | 32 | 4 | 128 | 32 | 16 | 32 | G, K, T, A, S, Nt, N, Te, C, Su |
| KP11S | 16 | 512 | 16 | 32 | 32 | 4 | 16 | 16 | 16 | 32 | Te, C, Na, Su, S |
| KP2S | 16 | 256 | 8 | 128 | 32 | 8 | 4 | 4 | 1 | 1 | G, T, NtI, Te |
| KP2aS | 16 | 512 | 8 | >128 | 256 | 8 | 32 | 32 | 16 | 16 | G, T, NtI, Te |
| KP3S | 16 | 1,024 | 16 | 128 | 512 | 8 | 16 | 8 | 16 | 8 | G, K, T, N, NtI |
| KP9S | 8 | 512 | 4 | >128 | 512 | 8 | 128 | 16 | 8 | 16 | G, K, TI, SI, Te, C, Su |
| KP14S | 8 | 512 | 4 | >128 | 512 | 2 | 128 | 32 | 8 | 32 | G, TI |
| KP17S | 8 | >1,024 | 128 | >128 | 256 | 4 | 128 | 512 | 8 | 256 | G, TI, C, Su, Tp, S, Na |
| KP18S | 8 | >1,024 | 64 | >128 | 128 | 4 | 64 | 32 | ND | ND | G, T |
| KP23S | 8 | 1,024 | 8 | >128 | 512 | 4 | 16 | 4 | 4 | 64 | G, T, K, NI, SI, Te, C, Su, Tp, Na, |
| KP25S | 8 | >1,024 | >1,024 | >128 | 128 | 4 | 16 | 32 | 8 | 256 | G, K, T, S, Su, CI, NI, ApI, TpI |
| KP7S | 8 | 128 | 4 | >128 | 16 | 4 | 2 | 4 | 2 | 1 | G, T, K, S, N, Te, C, Su, Tp, Na |
| KP34S | 16 | 512 | 2 | >128 | 16 | 2 | 8 | 1 | 1 | 0.5 | G, T, K, S, Te, C, Su |
| KP34aS | 16 | 512 | 2 | >128 | 32 | 2 | 8 | 2 | 1 | 1 | G, T, K, S, C, Su |
| KP38S | 8 | 256 | 2 | >128 | 128 | 2 | 16 | 4 | 8 | 2 | G, T, NtI |
| KP1S R+ | 8 | 128 | 2 | 128 | 32 | 2 | 8 | 4 | 2 | 4 | G, K, T, A, S, Nt, Te |
| KP11S R+ | 4 | 128 | 8 | 32 | 32 | 2 | 8 | 1 | 2 | 4 | Te, C, Na, Su, SI |
| KP2S R+ | 16 | 256 | 8 | 32 | 64 | 2 | 2 | 4 | 1 | 1 | G, T, NtI, Te |
| KP2aS R+ | 16 | 512 | 2 | >128 | 64 | 2 | 8 | 16 | 4 | 8 | G, T, NtI, Te |
| KP3 S R+ | 16 | 512 | 2 | 128 | 32 | 2 | 4 | 4 | 2 | 2 | G, K, T, N, NtI |
| KP9S R+ | 8 | 256 | 2 | 64 | 128 | 2 | 8 | 16 | 2 | 16 | G, TI |
| KP14S R+ | 8 | 128 | 2 | 32 | 128 | 2 | 8 | 16 | 1 | 4 | G, TI |
| KP17S R+ | 4 | 128 | 1 | 64 | 64 | 2 | 4 | 2 | 2 | 2 | G, TI, C, S |
| KP23S R+ | 8 | 128 | 1 | 64 | 16 | 2 | 4 | 4 | 1 | 2 | K, N, C |
| KP25S R+ | 4 | 128 | 1 | 64 | 32 | 4 | 4 | 32 | 4 | 4 | G, TI, S, CI |
| KP34S R+ | 8 | 128 | 2 | 8 | 32 | 2 | 8 | 2 | 2 | 8 | G, T, K, S, C |
| KP34aS R+ | 8 | 256 | 2 | 8 | 32 | 2 | 8 | 2 | 2 | 8 | G, T, K, S, C |
| KP38S R+ | 4 | 128 | 2 | 8 | 64 | 2 | 16 | 8 | 8 | 16 | G, T, NtI |
| CTX-M type | |||||||||||
| KP12C | 8 | 256 | 1 | >128 | 128 | 4 | 64 | 16 | 1 | 64 | Su, S |
| KP13C | 16 | 256 | 4 | >128 | 256 | 2 | 64 | 2 | 8 | 8 | |
| KP15C | 8 | 128 | 1 | >128 | 512 | 4 | 16 | 1 | 8 | 16 | |
| KP4C | 8 | 512 | 4 | >128 | 512 | 4 | 16 | 1 | 2 | 1 | |
| KP4aC | 8 | 512 | 4 | >128 | 512 | 4 | 64 | 2 | 16 | 2 | |
| KP22C | 8 | 1024 | 8 | >128 | 512 | 4 | 128 | 2 | 4 | 8 | |
| KP26C | 8 | 256 | 4 | >128 | 512 | 4 | 32 | 1 | 2 | 4 | |
| KP26aC | 8 | 256 | 1 | >128 | 512 | 4 | 16 | 1 | 2 | 4 | |
| KP30C | 8 | 1,024 | 8 | >128 | 512 | 4 | 512 | 8 | 32 | 64 | |
| KP29C | 8 | 512 | 8 | >128 | 512 | 2 | 128 | 4 | 8 | 64 | |
| KP31C | 4 | 512 | 2 | >128 | 512 | 2 | 128 | 0.5 | 1 | 2 | |
| KP40C | 4 | 256 | 8 | >128 | 256 | 2 | 8 | 0.5 | 2 | 2 | K, S, N, Te, Su, Tp, Nor, Spec |
| KP32C | 4 | 256 | 2 | >128 | 512 | 4 | 64 | 0.5 | 4 | 2 | |
| KP32aC | 4 | 512 | 4 | >128 | 512 | 4 | 128 | 1 | 4 | 4 | |
| KP36C | 8 | 512 | 2 | >128 | 512 | 2 | 512 | 1 | 8 | 4 | |
| KP39C | 8 | 512 | 8 | >128 | 256 | 4 | 32 | 2 | 4 | 8 | |
| KP12C R+ | 8 | 64 | 2 | >128 | 128 | 4 | 32 | 16 | 0.5 | 16 | |
| KP13C R+ | 8 | 64 | 2 | >128 | 256 | 2 | 8 | 2 | 2 | 4 | |
| KP15C R+ | 8 | 64 | 4 | 64 | 64 | 2 | 8 | 2 | 1 | 1 | |
| KP4aC R+ | 8 | 512 | 4 | >128 | 512 | 4 | 64 | 2 | 2 | 2 | |
| KP22C R+ | 8 | 128 | 2 | 64 | 128 | 4 | 32 | 4 | 2 | 2 | |
| KP26C R+ | 8 | 256 | 2 | >128 | 512 | 4 | 32 | 2 | 1 | 2 | |
| KP30C R+ | 8 | 64 | 2 | >128 | 32 | 2 | 8 | 0.5 | 2 | 4 | |
| KP31C R+ | 4 | >64 | 2 | >128 | 256 | 2 | 8 | 0.5 | 8 | 4 | |
| KP36C R+ | 4 | >64 | 2 | >128 | 512 | 2 | >8 | 2 | 8 | 16 | |
| KP40C R+ | 8 | 256 | 4 | >128 | 256 | 2 | 8 | 4 | 4 | 4 | K, S, N, Te, Su, Tp, Spec |
Abbreviations: AMC, amoxicillin-clavulanate; PIP, piperacillin; TZP, piperacillin-tazobactam; CFZ, cefazolin; CXM, cefuroxime; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; S, streptomycin; G, gentamicin; T, tobramycin; Te, tetracycline; C, chloramphenicol; A, amikacin; Nt, netilmicin; N, neomycin; Su, sulfisoxazole; Tp, trimethoprim; Spec, spectinomycin; Ap, apramycin; Na, nalidixic acid; ND, not done. I, intermediate; R+, E. coli transconjugants of ESBL-producing K. pneumoniae isolates.
All isolates with ESBLs of the TEM type and all but one of the isolates with ESBLs of the SHV type expressed transferable resistance to gentamicin and tobramycin. In contrast, none of the isolates with a CTX-M-10 β-lactamase were resistant to aminoglycosides. The latter isolates were also susceptible to tetracycline, chloramphenicol, or trimethoprim, whereas 60% of the isolates with ESBLs of the SHV type were resistant to tetracycline, chloramphenicol, and sulfonamides; but resistance to these drugs was not always cotransferred with the ESBL resistance phenotype (Table 2). It is noteworthy that 19% of the isolates studied were resistant to nalidixic acid and that all of them exhibited reduced susceptibilities to ciprofloxacin.
Phenotypic characterization of β-lactamases: the problem of multiplicity within phenotypic groups.
Four different ESBL phenotypic groups were identified by their antimicrobial susceptibility profiles; these groups were then classified by analytical IEF, as follows: (i) isolates with a TEM-type ESBL with a pI of 5.9 (17 of 62 isolates; 27%), (ii) isolates with an SHV-type ESBL with a pI of 7.6 (23 of 62 isolates; 37%), (iii) an isolate with an SHV-type ESBL with a pI of 8.2 (1 of 62 isolates; 1.5%), and (iv) isolates with a CTX-M-type ESBL with a pI of 8.1 (20 of 62 isolates; 32%). A common problem in the characterization of ESBLs is the frequent association of an ESBL with broad-spectrum β-lactamases. Thus, PCR analysis was used to identify the simultaneous presence of both broad- and extended-spectrum β-lactamases. In our work, 27% of the 62 isolates studied harbored a β-lactamase with a pI of 5.4; this β-lactamase probably corresponded to the TEM-1 enzyme. All isolates that exhibited ESBLs of the TEM and CTX-M types could be amplified with SHV-specific primers, although no β-lactamase band with a pI of 7.6 was always detected (Table 1). When the NheI restriction method (see Materials and Methods) was applied to the SHV-specific PCR products from wild-type K. pneumoniae isolates that produced a β-lactamase of the SHV type, a noncleaved 930-bp fragment was obtained in all cases. This fragment was never obtained from the DNA of the transconjugants, suggesting that it contains part of the sequence of a chromosomally encoded SHV-1 enzyme, a frequent finding in K. pneumoniae isolates (8; G. S. Babini and D. M. Livermore, Letter, Antimicrob. Agents Chemother. 44:2230, 2000).
Genetic characterization of β-lactamases: DNA amplification and sequencing.
TEM, SHV, and CTX-M β-lactamase genes from the E. coli transconjugants were amplified and sequenced with the primers mentioned in Materials and Methods. A 1,076-bp amplification product was obtained from all five isolates of different clonal types harboring an ESBL of the TEM type. The DNA sequences of strains corresponding to the four clonal types had the same deduced amino acid sequence, which contained the L21F, E104K, G238S, and T265 M amino acid substitutions (Ambler numbering); this sequence corresponds to that of the TEM-4 β-lactamase (42). They also presented silent nucleotide substitutions A346G, C436T, T682C, and G925A and the substitution C32T, which creates the Pa and Pb overlapping promoters described by Chen and Clowes (9). According to the recently proposed nomenclature for TEM-type β-lactamases (23, 24), this β-lactamase would be designated TEM-4f (Pa/Pb).
For 12 of 13 isolates of different clonal types with the SHV-type β-lactamase, a 930-bp amplification product was obtained. The sequence of this product covers the region of blaSHV that corresponds to the part of the SHV protein located between L35 and G283. Analysis of these sequences in all strains revealed the substitution G238S with respect to the sequence for the SHV-1 enzyme (residue numbering of Ambler). Six of these isolates had L35, A140, and T142 sequences, which indicates the presence of a typical SHV-2 sequence. Five isolates had G35, A140, and T142 sequences plus two silent nucleotide substitutions. This pattern is indicative of the SHV-2a ESBL. The blaSHV gene of the remaining isolate was identified as that for SHV-5.
Seventeen isolates with the CTX-M-type β-lactamase corresponding to 12 different clonal types had positive results by PCR with primers specific for CTX-M-10. Eight of the isolates (corresponding to isolates obtained in 1990, 1991, 1993, 1998, 1999, and 2000) were sequenced. All eight sequences studied presented the same amino acid changes (A27V and R38Q) and the 15 nucleotide substitutions (at positions 92, 125, 141, 165, 294, 342, 375, 453, 531, 576, 636, 654, 669, 732, and 807) that are specific for CTX-M-10 in the CTX-M-1 cluster constituted by the CTX-M-1, CTX-M-3, CTX-M-10, CTX-M-11, and CTX-M-12 enzymes (21, 33, 46; this study). These sequences were compared with the sequences available for this enzyme cluster, including the unpublished CTX-M-11 sequence (GenBank accession no. AY005110). Finally, one isolate with a β-lactamase with a pI of 8.1 could be amplified with primers specific for CTX-M-9 but not with those used to identify CTX-M-10. This strain was considered to harbor the CTX-M-9 enzyme, recently described in different areas of Spain (38, 41).
Plasmids encoding ESBLs.
In order to detect the presence of common plasmids among different strains, plasmid DNA was obtained from K. pneumoniae isolates that were representatives of isolates with the different PFGE patterns or whose β-lactamases had different IEF profiles (35 of the 62 isolates included in the study). Plasmid DNA was also isolated from the recipient strain when transconjugants were available. All 35 wild-type isolates had one or more plasmids (one to six per organism) (data not shown) with molecular sizes ranging from 5 to 147 kb. Analysis of E. coli transconjugants expressing ESBLs revealed the presence of large plasmids, with estimated molecular sizes of 147, 85, 65, and 40 kb. Thirty-one different whole-plasmid patterns corresponding to the 31 clonal types observed as described above were identified. Most ESBL enzymes (81%; 28 of 35 isolates) were encoded by transferable plasmids, with the frequency of transferability being similar for all β-lactamase types: 83% for TEM-type β-lactamases, 86% for SHV-type β-lactamases, and 73% for CTX-M-type β-lactamases.
Transconjugants from four of the five clones harboring the TEM-type ESBL have a common plasmid of approximately 85 kb which was transferred at a very high frequency (10−2 transconjugants/donor). In contrast, a high degree of diversity of plasmids coding for SHV- or CTX-M-type enzymes was observed (Table 1; Fig. 1). The molecular sizes of these genetic elements ranged from 50 to 147 kb. The frequency of transfer was much lower for these plasmids than for plasmids encoding ESBLs of the TEM type (10−6 to 10−8)
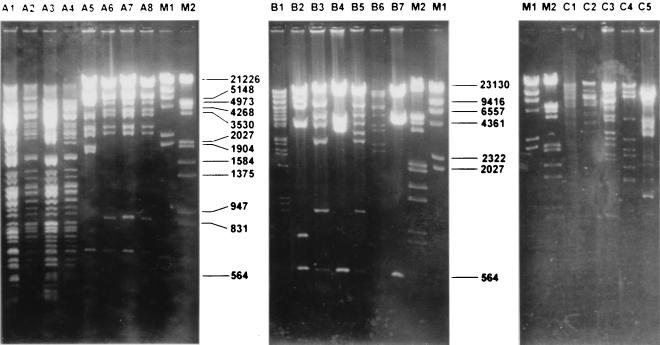
FIG. 1.
Fingerprinting analysis of plasmids from E. coli transconjugants of ESBL-producing K. pneumoniae strains. (A) EcoRI and PstI digests of plasmid DNAs obtained from TEM-4-producing isolates. Lane A1, KP6T R+ digested with PstI; lane A2, KP8T R+ digested with PstI; lane A3, KP20T R+ digested with PstI; lane A4, KP27T R+ digested with PstI; lane A5, KP6T R+ digested with EcoRI; lane A6, KP8T R+ digested with EcoRI; lane A7, KP20T R+ digested with EcoRI; lane A8, KP27T R+ digested with EcoRI; lane M1, DNA molecular size marker II (Roche Diagnostics); lane M2, DNA molecular size marker III (Roche Diagnostics). (B) EcoRI digest of plasmid DNA obtained from CTX-M-10-producing isolates. Lane B1, KP13C R+ digested with EcoRI; lane B2, KP4aC R+ digested with EcoRI; lane B3, KP31C R+ digested with EcoRI; lane B4, KP26C R+ digested with EcoRI; lane B5, KP30C R+ digested with EcoRI; lane B6, KP22C R+ digested with EcoRI; lane B7, KP36C R+ digested with EcoRI; lane M2, DNA molecular size marker III (Roche Diagnostics); lane M1, DNA molecular size marker II (Roche Diagnostics). (C) EcoRI digest of plasmid DNA obtained from SHV-producing isolates. Lane M1, DNA molecular size marker II (Roche Diagnostics); lane M2, DNA molecular size marker III (Roche Diagnostics); lane C1, KP2S R+ digested with EcoRI; lane C2, KP14S R+ digested with EcoRI; lane C3, KP9S R+ digested with EcoRI; lane C4, KP38S R+ digested with EcoRI; lane C5, KP23S R+ digested with EcoRI.
Population structure.
RFLP analysis by PFGE with XbaI with all 62 ESBL-producing K. pneumoniae isolates revealed 31 clones or karyotypes, with banding patterns giving fragments in a range from 30 to 600 kb (Table 1; Fig. 2). Among these K. pneumoniae clones, 5 clones harbored an TEM-type ESBL (TEM-4), 13 clones harbored an SHV-type ESBL (SHV-2 [n= 7], SHV-2a [n = 4], SHV-5 [n = 1], and noncharacterized at a molecular level [n = 1]), and 13 clones harbored a CTX-M-type ESBL (CTX-M-9 [n = 1] or CTX-M-10 [n = 12]). Not a single clone contained two different types of ESBLs. The evolution of these clones in space and time is detailed below.
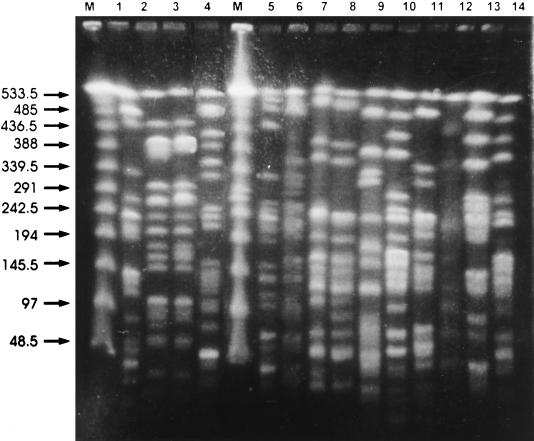
FIG. 2.
Patterns generated by PFGE of SmaI-digested chromosomal DNA obtained from CTX-M-10 producing K. pneumoniae isolates. Lane M, bacteriophage lambda ladder PFGE marker (New England Biolabs); lane 1, K. pneumoniae KP31C; lane 2, K. pneumoniae KP32C; lane 3, K. pneumoniae KP32aC; lane 4, K. pneumoniae KP36C; lane 5, K. pneumoniae KP13C; lane 6, K. pneumoniae KP15C; lane 7, K. pneumoniae KP4C; lane 8, K. pneumoniae KP4aC; lane 9, K. pneumoniae KP22C; lane 10, K. pneumoniae KP26C; lane 11, K. pneumoniae KP30C; lane 12, K. pneumoniae KP29C; lane 13, K. pneumoniae KP12C; lane 14, K. pneumoniae KP39C. The numbers to the left of the gel are in kilobases.
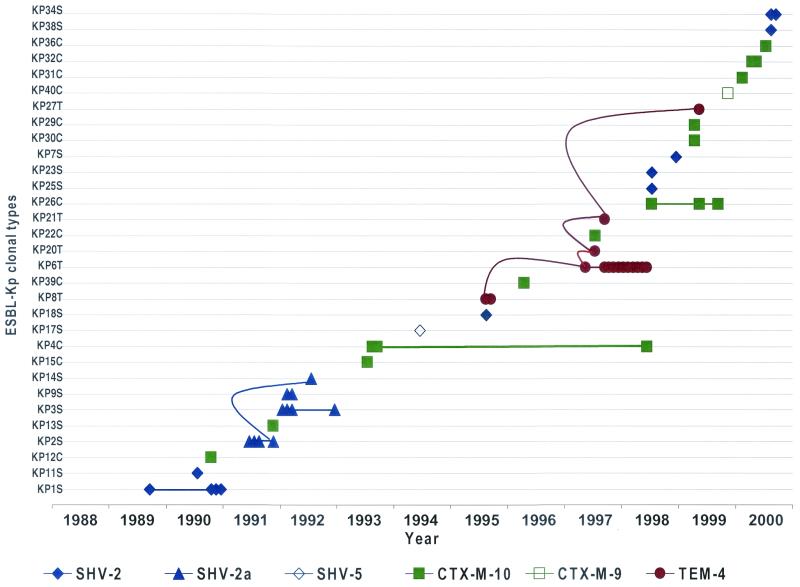
In September 1995, clone KP8T (which contains a plasmid that codes for the TEM-4 ESBL) was isolated from two patients in the cardiopediatric intensive care unit (ICU). In July 1997, two other TEM-4 producing clones, KP20T and KP21T, were each isolated from a single patient in the same unit; one of them (clone KP20T) harbored a plasmid identical to that harbored by clone KP8T (pRYC11). At the end of 1997, a new TEM-4-producing clone (clone KP6T) carrying a plasmid (pRYC11.1) closely related to one harbored by clone KP8T (the PstI and EcoRI restriction patterns were very similar, and thus plasmid pRYC11.1 was considered a variant of pRyC11) KP8T (Fig. 1). KP6T was responsible for a 1-year-long epidemic in the same unit that involved 11 patients. One year later, another clone (clone KP27T) with the same plasmid (pRYC11) and the same ESBL was isolated from a single patient of the unit. These observations indicate not only the successful maintenance of a single plasmid type in the cardiopediatric ICU during 4 years (Fig. 1 and 3) but also the variable epidemiological success of the five TEM-4-producing clones since only one clone (clone KP6T) caused a substantial number of cases of infection during an extended period of time.
FIG. 3.
Emergence and stability of ESBL-producing K. pneumoniae (Kp) clonal types during the period studied (1988 to 2000) in the Hospital Ramón y Cajal. Each symbol represents a single patient (n = 58). The straight lines represent the persistence of a specific clone over the indicated time period. The curved lines represent the spread of a specific plasmid among different clonal types over the indicated time period.
The first SHV-2-producing K. pneumoniae isolate (clone KP1S) was recovered from a single patient in the cardiopediatric ICU in 1989. One year later, a small outbreak involving three patients in the same ICU was caused by the same clone. Another SHV-2-producing strain (clone KP11S) was isolated from a single patient in this unit during the time of the small outbreak. In 1991, clone KP2S and a related clone, clone KP2aS (both of which share an identical SHV-2a-encoding plasmid), were isolated from four patients in the same ward. Eight months later, another SHV-2a-producing clone, clone KP14S, with the same plasmid as clone KP2S, appeared in a single patient in the same unit. In January 1992, a small outbreak (four patients) in the cardiopediatric ICU was caused by a clone with a different SHV-2a karyotype, clone KP3S (which harbored a plasmid distinct from that of clone KP2S). Again, a new SHV-2a-producing clone, clone KP9S, was isolated from two patients in the same unit. After an approximately 7-year period free of β-lactamase-producing clones, new SHV-2-producing clones (clones KP34S and KP38S) emerged in the same unit, but these two clones harbored different ESBL-encoding plasmids. In 1998, new strains (clones KP23S, KP25S, and KP7S) with different SHV-2-encoding plasmids were recovered from single patients in the internal medicine and nephrology wards. These observations document the sporadic emergence of different SHV-2- or SHV-2a-producing K. pneumoniae clones generally unable to produce outbreaks.
The first K. pneumoniae clone (clone KP12C) harboring a plasmid coding for a CTX-M-type ESBL (CTX-M-10) was isolated very early in our institution from a single patient in the urology ward (October 1990). During the following 2 years, the same enzyme was identified in different clones (clones KP13C, KP4C, and KP15C) carrying different plasmids and was found in isolates from quite different areas (including in an outpatient). A variant clone of KP4C, KP4aC, was isolated from a patient in the urology ward 5 years later (June 1998). From 1996 to 1998, isolates with new CTX-M-10 karyotypes (clones KP26C and, probably, clones KP39C and KP22C) sporadically appeared in single patients in the internal medicine, pneumology, and gastroenterology wards. In 1999 and 2000, two isolates of variant clone KP26aC were found in the pneumology and gastroenterology wards, respectively, and other CTX-M-10-producing clones were detected in single patients from the endocrinology, trauma, nephrology, and urology wards (clones KP30C, KP31C, and KP32C) and even from an outpatient (clones KP29C, KP36C). One of the clones, clone KP40C, harbored the CTX-M-9 enzyme.
DISCUSSION
To our knowledge, this is the first study to document the natural history of K. pneumoniae strains harboring ESBLs in a single institution from its time of emergence in 1989 to 2000. The overall prevalence of ESBL-producing K. pneumoniae (4.8%) found in our hospital is lower than those reported in other European countries and the United States (7 to 44%) (10, 39, 48). A reduced variety of β-lactamases was found: essentially, TEM-4, SHV-2 (and SHV-2a), and CTX-M-10 were found. On the contrary, an unexpected clonal diversity was detected among the ESBL-producing isolates, showing that most strains have been unable to be maintained or to spread in the hospital setting. This observation challenges many conventional thoughts about the nosocomial epidemiology of antibiotic resistance.
A group of five clones harbored the β-lactamase TEM-4. This enzyme has infrequently been found among ESBLs, and to our knowledge, its production is restricted to E. coli and Salmonella isolates from France and Spain (10, 12, 14, 42). Only two of the five clones were able to produce more than a single clinical case of infection, but one of them (clone KP6T) spread during 9 months among patients in the cardiopediatric ICU. Globally, TEM-4-producing strains were detected in our hospital over a period of 4 years, and no other TEM-type ESBL appeared during the entire observation period. Indeed, TEM-type ESBLs have scarcely been found in Spain and were only described in the early 1990s, in contrast to the situation in other European countries, where TEM-3, TEM-10, and new TEM derivatives (TEM-24, TEM-56, TEM-47 to TEM-49 TEM-52, and TEM-68) have recently been isolated in France, England, Portugal, Italy, and Eastern Europe (4, 10, 15–17, 29, 34). The same TEM-4-encoding plasmid was found in all strains studied, indicating plasmid-mediated direct or indirect transclonal transfer of TEM-4 production. Considering previous observations from studies with other enzymes (16, 35), we could eventually suspect a situation in which the TEM-4-producing plasmid (pRYCE11) is endemic in the hospital, in another hospital, or in the community, with plasmids circulating among noninfective strains (probably involving different species) and occasionally entering different K. pneumoniae clones. In our case, the extremely high frequency of transfer (10−2) of pRYCE11 may have facilitated plasmid selection and dissemination. In summary, for the strains harboring TEM-4, we have confined K. pneumoniae nonepidemic clones or epidemic and, probably, endemic or epidemic TEM-4-encoding plasmids.
Eight different K. pneumoniae clones harbored the SHV-2-type ESBL, and three more, isolated only during the period from 1991 to 1993, harbored SHV-2a (which differs from SHV-2 by the G35L substitution). The first ESBL obtained in our institution was of the SHV-2 type and corresponded to the first Spanish epidemic of β-lactamase-producing Enterobacteriaceae, which occurred in 1988 (14). Only two of seven K. pneumoniae clones harboring SHV-2 were detected in more than a single patient, but three of four clones harboring SHV-2a were able to spread between patients in the cardiopediatric ICU, causing limited epidemics of infections. The distribution of the SHV-2 and SHV-2 ESBLs in so many different clones cannot be explained by the conjugative dissemination of a single or a limited number of plasmids, as was case for the interclonal spread of the TEM-4 ESBL. With a single exception (the same plasmid in two different clones, clones KP2S and KP14S), the plasmids obtained from the SHV-2- or SHV-2a-producing isolates were considered unrelated by restriction enzyme digestion. To explain this puzzling clonal heterogeneity, some hypotheses may be considered. Our hospital may have been subjected to the multiple and continuous introduction of different SHV-2-producing K. pneumoniae clones. Eventually, by the introduction of other SHV-2-producing organisms, this may have served as a source from which the ESBLs were occasionally acquired by different K. pneumoniae clones. Having ruled out significant plasmid dissemination, blaSHV-2 gene spread may have been facilitated by the interspecies or interclonal transfer of transposable elements. The role of transposable elements in SHV-2 dissemination can be suggested, as the ancestor gene blaSHV-1 possibly exists as part of a mobile element (32). Moreover, blaSHV-2a has been found to be integrated into the chromosome of a Pseudomonas aeruginosa isolate, with investigators suggesting that the flanking sequences are of enterobacterial origin (26, 45). Finally, we cannot rule out the possibility of an independent intraclonal evolution of the very common SHV-1 non-ESBL from K. pneumoniae (found in all of our isolates) to SHV-2 and SHV-2a. Note that the SHV enzymes, including SHV-1, are intrinsically more efficient than the TEM enzymes against broad-spectrum cephalosporins (45). In reaction to the selection pressure that results from treatment with these antibiotics, a strain with blaSHV-1 may have first evolved by gene duplication, and then a single mutation (at position 238) could have produced a derivative that simultaneously harbors blaSHV-1 and blaSHV-2, which occurred in all strains in our series (8). The fact that silent mutations were absent from the SHV-2 sequence studied in our strains suggests that this evolution may recently have occurred from a particular SHV-1 gene. Another SHV variant, SHV-5, was detected in only a single clone (from a single patient) in our hospital during the observation period. SHV-2 and SHV-5 have been described worldwide, being more commonly found in countries of the Mediterranean area, including Spain (36, 45). Both have been detected in a wide variety of enterobacterial species (14, 18, 45). However, the variant of SHV-2, SHV-2a, has been described only in clinical isolates of K. pneumoniae and E. coli from Germany, Switzerland, and Korea (45) and, more recently, in a French P. aeruginosa isolate (26).
A group of 13 different clones harbored the same CTX-M-10 ESBL and were isolated from 1990 to 2000. Only 3 of 13 of these clones were able to produce more than a single case of infection, but 2 of these clones (clones KP4C and KP26C) were recovered at very distant periods of time (5 and 2 years, respectively). This is the first report of a CTX-M-producing K. pneumoniae isolate in a Mediterranean country and the first to document that a K. pneumoniae isolate harbors CTX-M-10, originally identified in one E. coli isolate from our hospital (33). Note that the first K. pneumoniae isolate to produce CTX-M-10 (now identified) was isolated in our institution in 1990, at approximately the same time that other CTX-M-type ESBL-producing strains were detected in geographically different areas: CTX-M-1 in Germany in 1989, MEN-1 in France from an Italian patient in 1989, and CTX-M-2 in Argentina in 1989 (46). We were certainly challenged by the overwhelming clonal diversity of the CTX-M-10-producing K. pneumoniae clones in our institution. As in the previous case of the SHV-2 ESBL and different from what was observed for strains harboring the TEM-4 ESBL, we were unable to document the presence in these clones of a common transmissible genetic element that could explain the polyclonal structure of our ESBL-producing K. pneumoniae isolates. This result suggests that blaCTX-M-10 may have been transferred to plasmids of different origins and may have been able to disseminate in different K. pneumoniae clones, and the presumptive involvement of a transposable element is being investigated. Supporting this view, all blaCTX-M-10 sequences that we studied carried an identical array of 15 noncoding genetic polymorphisms. On the other hand, all CTX-M-producing isolates lacked the usual pattern of resistance to aminoglycoside antibiotics frequently linked to the presence of other ESBLs (14, 19, 36). This pattern of greater susceptibility may suggest a community or environmental source of the genetic element carrying blaCTX-M-10. Indeed, we isolated the CTX-M-10 ESBL from two outpatients who came to the hospital 10 years apart, and we have recently identified blaCTX-M-10 in a typical environmental organism, Enterobacter gergoviae (R. Cantón, submitted for publication). If the environment is a relevant source of ESBLs, as it has been suggested, the hypothesis that ESBLs are introduced from multiple sources should be considered to explain the observed clonal diversity. Note that clone KP4C was initially isolated in 1993 in two different medical wards and reemerged in 1998 (in the urology ward). During this period, no other K. pneumoniae isolates producing CTX-M enzymes were recovered in our institution, although this type of ESBL was detected in E. coli and Enterobacter sp. isolates from patients staying in the same wards (33; T. M. Coque et al., 41st Intersci. Conf. Antimicrob. Agents Chemother, abstr. 298, 2001). These data would indicate that blaCTX-M-10 has the ability to be maintained as a minority gene population in the hospital microbial metagenome during extended periods of time.
The present study indicates that the epidemiology of ESBL-producing organisms in hospitals may differ from what was considered the conventional wisdom. In our case, except in a very limited number of well-defined cases (frequently associated with the same high-risk areas of the hospital), there was no real epidemic (caused by single bacterial clones) or endemic (maintenance of a single clone during extended periods of time). Interestingly, most ESBL-producing K. pneumoniae clones were extremely ephemeral, being poorly selected and maintained in the hospital setting. The reasons for such frequent clonal turnover remain obscure. We could suggest the frequent acquisition of ESBL genes by different clones in the absence of antibiotic selection. Eventually, clonal competition with susceptible K. pneumoniae strains may occur; this may involve a reduction in the fitness of a resistant clone related to the maintenance of a plasmid harboring an ESBL. On the contrary, small numbers of ESBL genes have successfully persisted for more than a decade in the institution. This may indicate that the unit of selection is not the bacterial clone but the ESBL gene itself (which may emerge by mutation) or its genetic vector. Indeed, phenotypic evolution is caused by the selection of genes as replicators, and this is particularly clear in nonchromosomally domesticated genes, which make up infective genetic elements, such as plasmids or transposons (5). We can hypothesize that the maintenance of the ESBL genes in the environment and particularly in the hospital provides a general advantage for the entire bacterial community (not only for particular clones). These genes, whose numbers in different bacterial populations are amplified by antibiotic selection pressure, may occasionally enter different K. pneumoniae clones. Even more infrequently, these clones are able to spread in the hospital, causing conventional epidemics. Previous reports have pointed out the responsibility of particular clones or plasmids in epidemics of K. pneumoniae harboring ESBLs in hospitals (1, 4, 7, 15, 16, 35, 47, 49), although a more complex situation involving multiple different enzymes and strains has also been demonstrated (11, 17). It is our hope that our study, based on a very long observational period during which all isolates were collected and characterized, will contribute to a more complete understanding of the spread and evolution of ESBL genes in hospitals.
Acknowledgments
We are grateful to María Isabel García (Faculty of Pharmacy, Complutensis University, Madrid, Spain) for excellent technical assistance with the sequence analysis and to Jorge Mata for computer assistance and Juan-Carlos Galán for helpful discussions (Servicio de Microbiología, Hospital Ramón y Cajal). We also thank Encarna Simarro and the Spanish Culture Type Collection for kindly providing us with strains S. enterica serovar Virchow harboring blaCTX-M-9 and strains E. coli V517 and E. coli NCTC 50192, respectively.
This work was supported in part by research grants from the Comunidad Autónoma de Madrid (grant 08.20017/2000.1) and the Fondo de Investigaciones Sanitarias (grant FIS 01/412).
REFERENCES
- 1.Arlet, G., M. Rouveau, I. Casin, P. J. M. Bouvet, P. H. Lagrange, and P. Philippon. 1994. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 β-lactamase and which were isolated in 14 French hospitals. J. Clin. Microbiol. 32:2553–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asensio, A., A. Oliver, P. González-Diego, F. Baquero, J. C. Pérez-Díaz, P. Ros, J. Cobo, M. Palacios, D. Lasheras, and R. Cantón. 2000. Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin. Infect. Dis. 30:55–60. [DOI] [PubMed] [Google Scholar]
- 3.Baquero, F., D. Bouanchaud, M. C. Martínez-Pérez, and C. Fernández. 1978. Microcin plasmids: a group of extrachromosomal elements for low-molecular-weight antibiotics in Escherichia coli. J. Bacteriol. 135:342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barroso, H., A. Freitas-Vieira, L. M. Lito, J. Melo Cristino, M. J. Salgado, H. Ferreira Neto, J. C. Sousa, G. Soveral, T. Moura, and A. Duarte. 2000. Survey of Klebsiella pneumoniae producing extended-spectrum β-lactamases at a Portuguese hospital: TEM-10 as endemic enzyme. J. Antimicrob. Chemother. 45:611–616. [DOI] [PubMed] [Google Scholar]
- 5.Bell, G. 1997. Phenotypic evolution is caused by the selection of genes as replicators, p.35–36. In G. Bell (ed.), Selection, the mechanism of evolution. Chapman & Hall, New York, N.Y.
- 6.Bonnet, R., C. Dutour, J. L. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240-Gly. Antimicrob. Agents Chemother. 45:2269–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, P. A., C. E. Cherubin, V. Idemyor, B. A. Rasmussen, and K. Bush. 1994. Multiply resistant Klebsiella pneumoniae from two Chicago hospitals: identification of the extended-spectrum β-lactamase TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob. Agents Chemother. 38:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaves, J., M. G. Ladona, C. Segura, A. Coira, R. Reig, and C. Ampurdanés. 2001. SHV-1 β-lactamase is mainly a chromosomally encoded species-specific enzyme in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. T., and R. C. Clowes. 1984. Two improved promoter sequences for the beta-lactamase expression arising from a single base-pair substitution. Nucleic Acids Res. 11:3219–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and the French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essack, E. S., L. M. C. Hall, D. G. Pillay, M. L. McFadyen, and D. M. Livermore. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Aránguiz, A., R. Alonso, K. Colom, L. Gallego, A. Morla, J. Garaizar, and R. Cisterna. 1991. Estudio multicéntrico de la resistencia a cefotaxima durante el año 1991: detección y caracterización de nuevas β-lactamasas de espectro ampliado. Rev. Esp. Quimioter. 4:137–144. [Google Scholar]
- 13.Fernández-Rodríguez, A., R. Cantón, J. C. Pérez-Díaz, J. Martínez-Beltrán, J. J. Picazo, and F. Baquero. 1992. Aminoglycoside-modifying enzymes in clinical isolates harboring extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 36:2536–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Rodríguez, A., J. Reguera, J. C. Pérez-Díaz, J. J. Picazo, and F. Baquero. 1992. Primera epidemia española de resistencia plasmídica a cefalosporinas de tercera generación: implicación de SHV-2. Enf. Infec. Med. Clin. 10:458–461. [PubMed] [Google Scholar]
- 15.Fiett, J., A. Palucha, B. Miaczynska, M. Stankiewicz, H. Przondo-Mordaska, W. Hryniewicz, and M. Gniadkowski. 2000. A novel complex mutant β-lactamase, TEM-68, identified in a Klebsiella pneumoniae isolate from an outbreak of extended spectrum β-lactamase-producing klebsiellae. Antimicrob. Agents Chemother. 44:1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gniadkowski, M., A. Palucha, P. Grzesiowski, and W. Hryniewicz. 1998. Outbreak of ceftazidime-resistant Klebsiella pneumoniae in Warsaw, Poland: clonal spread of the TEM-47 extended-spectrum β-lactamase (ESBL)-producing strain and transfer of a plasmid carrying the SHV-5 like ESBL-encoding gene. Antimicrob. Agents Chemother. 42:3079–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gniadkowski, M., I. Schneider, R. Jungwirth, W. Hryniewicz, and A. Bauernfeind. 1998. Ceftazidime-resistant Enterobacteriaceae isolates from three Polish hospitals: identification of three novel TEM- and SHV-5-type extended-spectrum β-lactamase-producing klebsiellae. Antimicrob. Agents Chemother. 42:514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heritage, J., F. H. M’Zali, D. M. Gascoyne-Binzi, and P. M. Hawkey. 1999. Evolution and spread of SHV extended-spectrum β-lactamases in gram negative bacteria. J. Antimicrob. Chemother. 44:309–318. [DOI] [PubMed] [Google Scholar]
- 19.Jacoby, G. A., and L. Sutton. 1991. Properties of plasmids responsible for production of extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarlier, V., M. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867–878. [DOI] [PubMed] [Google Scholar]
- 21.Kariuki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis. Methods Mol. Med. 15:17–31. [DOI] [PubMed] [Google Scholar]
- 23.Leflon-Guibout, V., B. Heym, and M. Nicolas-Chanoine. 2000. Updated sequence information and proposed nomenclature for blaTEM genes and their promoters. Antimicrob. Agents Chemother. 44:3232–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leflon-Guibout, V., V. Speldooren, B. Heym, and M. Nicolas-Chanoine. 2000. Epidemiological survey of amoxicillin-clavulanate resistance and corresponding molecular mechanisms in Escherichia coli isolates in France: new genetic features of blaTEM genes. Antimicrob. Agents Chemother. 44:2709–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mabilat, C., and S. Goussard. 1995. PCR detection and identification of genes for extended-spectrum β-lactamases, p.553–557. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 26.Naas, T., L. Philippon, L. Poirel, E. Ronco, and P. Nordmann. 1999. An SHV-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1281–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.National Committee for Clinical Laboratory Standards. 2000. Methods for diffusion disk antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Neuwirth, C., R. Labia, E. Siebor, A. Pechinot, S. Madec, E. B. Chaibi, and A. Kazmierczak. 2000. Characterization of TEM-56, a novel β-lactamase produced by a Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 44:453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordmann, P. 1998. Trends in β-lactam resistance among Enterobacteriaceae. Clin. Infect. Dis. 27(Suppl. 1):S1000–S1006. [DOI] [PubMed] [Google Scholar]
- 31.Nüesch-Inderbinen, M. T., H. Hächler, and F. H. Kayser. 1996. Detection of the genes coding for extended spectrum SHV β-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 15:398–402. [DOI] [PubMed] [Google Scholar]
- 32.Nugent, M. E., and R. W. Hedges. 1979. The nature of the genetic determinant for the SHV-1 beta-lactamase. Mol. Gen. Genet. 175:239–243. [DOI] [PubMed] [Google Scholar]
- 33.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagani, L., M. Perilli, R. Migliavacca, F. Luzzaro, and G. Amicosante. 2000. Extended-spectrum TEM and SHV-type β-lactamase-producing Klebsiella pneumoniae strains causing outbreaks in intensive care units in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 19:765–772. [DOI] [PubMed] [Google Scholar]
- 35.Palucha, A., B. Mikiewicz, W. Hryniewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of extended spectrum β-lactamase-producing organism of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44:489–499. [DOI] [PubMed] [Google Scholar]
- 36.Philippon, A., G. Arlet, and P. H. Lagrange. 1994. Origin and impact of plasmid-mediated of extended spectrum β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):S17–S29. [DOI] [PubMed] [Google Scholar]
- 37.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) in Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saurina, G., J. M. Quale, V. M. Manikal, E. Oydna, and D. Landman. 2000. Antimicrobial resistance in Enterobacteriaceae in Brooklyn, NY: epidemiology and relation to antibiotic resistance patterns. J. Antimicrob. Chemother. 45:895–898. [DOI] [PubMed] [Google Scholar]
- 40.Shannon, K., P. Stapleton, X. Xiang, A. Johnson, H. Beattie, F. El Bakri, B. Cookson, and G. French. 1998. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains causing nosocomial outbreaks of infection in the United Kingdom. J. Clin. Microbiol. 36:3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simarro, E., F. Navarro, J. Ruíz, E. Miró, J. Gómez, and B. Mirelis. 2000. Salmonella enterica serovar Virchow with CTX-M-like β-lactamase in Spain. J. Clin. Microbiol. 38:4676–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sougakoff, W., A. Petit, S. Goussard, D. Sirot, A. Bure, and P. Courvalin. 1989. Characterization of the plasmid genes blaT-4 and blaT-5 which encode the broad-spectrum β-lactamases TEM-4 and TEM-5 in Enterobacteriaceae. Gene 23:339–348. [DOI] [PubMed] [Google Scholar]
- 43.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Threlfall, E. J., and N. Woodford. Plasmid profile typing and plasmid fingerprinting. Methods Mol. Biol. 46:225–236. [DOI] [PubMed]
- 45.Tzouvelekis, L. S., and R.A. Bonomo. 1999. SHV-type β-lactamases. Curr. Pharm. Design 5:847–865. [PubMed] [Google Scholar]
- 46.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137–142. [DOI] [PubMed] [Google Scholar]
- 47.Wiener, J., J. P. Quinn, P. A. Bradford, R. V. Goering, C. Nathan, K. Bush, and R. A. Weinstein. 1999. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 281:517–523. [DOI] [PubMed] [Google Scholar]
- 48.Winokur, P. L., R. Cantón, J.-M. Casellas, and N. Legakis. 2001. Variations in the prevalence of strains expressing an extended-spectrum β-lactamases phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific Region. Clin. Infect. Dis. 32(Suppl. 2):S94–S103. [DOI] [PubMed] [Google Scholar]
- 49.Yuan, M., H. Aucken, L. M. C. Hall, T.L. Pitt, and D. M. Livermore. 1998. Epidemiological typing of klebsiellae with extended-spectrum β-lactamases from European intensive care units. J. Antimicrob. Chemother. 41:527–539. [DOI] [PubMed] [Google Scholar]