Abstract
A homologue of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa, smeABC, was cloned from Stenotrophomonas maltophilia by using, as a probe, a PCR product amplified from this organism with primers based on the mexB sequence. The smeABC genes were hyperexpressed in a mutant strain displaying resistance to several antimicrobials, including aminoglycosides, β-lactams, and fluoroquinolones. Deletions in smeC but not smeB compromised this resistance, suggesting that SmeC contributed to the multidrug resistance of the mutant as part of another, as-yet-unidentified multidrug efflux system. Consistent with SmeC functioning independently of SmeAB, a promoter activity was identified upstream of smeC. Upstream of the smeABC genes, a putative two-gene operon, smeSR, encoding homologues of bacterial two-component regulatory systems was identified. The cloned smeR gene activated expression of a smeA-lacZ fusion, indicating that SmeR positively regulates expression of the smeABC genes. Consistent with this, the multidrug resistance of the SmeABC-hyperexpressing mutant was compromised by deletion of smeR. Intriguingly, SmeC expression in S. maltophilia paralleled a β-lactamase activity provided by a C-terminally truncated L2 enzyme, which was apparently responsible for the β-lactam resistance of the SmeABC-hyperexpressing mutant. This represents the first report of coregulation of an efflux resistance determinant and a β-lactamase.
Previously known as Pseudomonas maltophilia and Xanthomonas maltophilia, Stenotrophomonas maltophilia is an aerobic, nonfermentative gram-negative bacterium broadly distributed in nature (36). This organism has increasingly emerged as an important nosocomial pathogen, particularly for immunocompromised patients, although little is known regarding the virulence mechanisms of and risk factors for S. maltophilia (11, 28, 31, 42). S. maltophilia is characterized by its high-level intrinsic resistance to a variety of structurally unrelated antimicrobials, including β-lactams, quinolones, and aminoglycosides (7, 10, 11, 13, 14, 21). Constitutive production of the families of class B L1 metallo- and the class A L2 serine-β-lactamases is the major determinant for β-lactam (including carbapenem) resistance in this organism (37, 47, 51, 53). Aminoglycoside-modifying enzymes, including O-nucleotidyltransferases and N-acetyltransferases, in S. maltophilia are important contributors for aminoglycoside resistance (18, 20, 50).
Multiple antibiotic resistance in S. maltophilia, as in other gram-negative bacteria, is also attributable, in part, to limited outer membrane permeability (27) and active antibiotic extrusion (3, 4, 53), although these mechanisms are poorly characterized to date. Multidrug efflux pumps responsible for multiple antibiotic resistance of Pseudomonas aeruginosa, Escherichia coli, and Neisseria gonorrhoeae have, however, been well characterized (for a review, see reference 38). These gram-negative multidrug efflux systems are composed of three components—a cytoplasmic membrane transporter, a periplasmic but cytoplasmic membrane-associated fusion protein, and an outer membrane efflux channel protein (outer membrane factor [OMF])—which work together to extrude antimicrobials across both membranes simultaneously (35). The cytoplasmic membrane transporter belongs to the resistance-nodulation-cell division (RND) family of the secondary transporters, drug-proton antiporters that use proton motive force across the cytoplasmic membrane to actively extrude drugs from the cell (23, 34, 38, 45). Intriguingly, the various tripartite multidrug efflux systems in gram-negative bacteria share high identity and similarity at the levels of nucleotide and amino acid sequences, allowing ready identification of homologous multidrug efflux systems in the other bacterial species (see reference 45 for a review).
Multidrug efflux systems have also been described in S. maltophilia. Multidrug-resistant (MDR) strains have, for example, been reported that hyperexpress SmeM (53), a homologue of the outer membrane components of multidrug efflux systems in several gram-negative bacteria (38). Recently, MDR strains expressing the SmeDEF multidrug efflux pump have also been described in this organism (3, 4) and shown to play an important role in the resistance of clinical strains (5). Further study of the SmeDEF pump has demonstrated that this pump contributes to intrinsic multidrug resistance in S. maltophilia (53a). Here we report the identification and characterization of a novel multidrug efflux system of S. maltophilia, SmeABC. The SmeABC multidrug system is unique among the known multidrug efflux systems in gram-negative bacteria, being regulated by a two-component regulatory system encoded by the smeSR genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Two strains of S. maltophilia, ATCC 13637 and ULA-511, were used as parental wild-type strains (15, 53). Luria-Bertani (LB) broth (1% [wt/vol] Difco tryptone, 0.5% [wt/vol] Difco yeast extract, and 0.5% [wt/vol] NaCl) and agar (LB broth containing 1.5% [wt/vol] agar) were used as the growth media throughout, and bacterial cells were cultivated at 30 or 37°C as indicated. Plasmids were maintained in Escherichia coli with appropriate antibiotic selection (pBluescript II SK[+], pTZ19U, pAK1900, and pET21-d[+], 100 μg of ampicillin per ml; pLAFR3, pVLT31, pRK415, and pEX18Tc, 10 μg of tetracycline per ml; pCR-Blunt-TOPO, and pRK2103, 50 μg of kanamycin per ml; and pMP190, 30 μg of chlormphenicol per ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| S. maltophilia | ||
| ATCC 13637 | Wild-type | 53 |
| ULA-511 | Wild-type | 15 |
| K1385 | MDR derivative of ULA-511, overproducing SmeM | 53 |
| K1449 | ULA-511 ΔL1 bla ΔL2 bla | 53 |
| K1668 | MDR derivative of K1449 | This study |
| K1778 | ULA-511 ΔsmeBC | This study |
| K1779 | K1385 ΔsmeBC | This study |
| K1780 | K1449 ΔsmeBC | This study |
| K1781 | K1668 ΔsmeBC | This study |
| K1782 | K1449 ΔsmeB | This study |
| K1783 | K1668 ΔsmeB | This study |
| K1784 | K1449 ΔsmeC | This study |
| K1785 | K1668 ΔsmeC | This study |
| K1786 | K1449 ΔsmeR | This study |
| K1787 | K1668 ΔsmeR | This study |
| K1975 | K1668 L2 bla::pLZ736 (L2 bla gene of K1668 disrupted by insertion of pLZ736 via homologous recombination with L2 bla sequences on the vector) | This study |
| E. coli | ||
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR (φ80dlacΔ(lacZ)M15) | 46 |
| MM294 | supE44 λ−rfbD1 spoT thi-1 endA1 hsdR17 pro | 32a |
| S17-1 | thi pro hsdR recA Tra+ | 48 |
| P. aeruginosa | ||
| ML5087 | ilv-220 thr-9001 leu-9001 met-9011 pur-67 aphA | 48 |
| K1112 | ML5087 nalB | 48 |
| K1113 | K1112 ΔoprM | 48 |
| Plasmids | ||
| pAK1900 | E. coli-P. aeruginosa shuttle cloning vector, 4.75 kb, Apr Cbr | R. Sharp, this department |
| pBluescript II SK(+) | Phagemid cloning vector, 2.96 kb, MCS, Apr | Stratagene |
| pCR-Blunt-TOPO | Vector for directly cloning blunt-ended PCR product, MCS, 3.52 kb, Kmr | Invitrogen |
| pET-21d(+) | Polyhistidine tag vector, 5 kb, Apr | Novagen |
| pEX18Tc | Broad-host-range gene replacement vector, 6.35 kb, sacB Tcr | 16 |
| pLAFR3 | Cosmid vector, 22 kb, Tcr | 49 |
| pMP190 | Broad-host-range promoterless lacZ transcription fusion vector, Cmr | 54 |
| pRK415 | Broad-host-range cloning vector; plac, MCS, Tcr | 48 |
| pTZ19U | Phagemid cloning vector, 2.8 kb, Apr | Bio-Rad |
| pVLT31 | Broad-host-range cloning vector, MCS, 10 kb, Tcr | 48 |
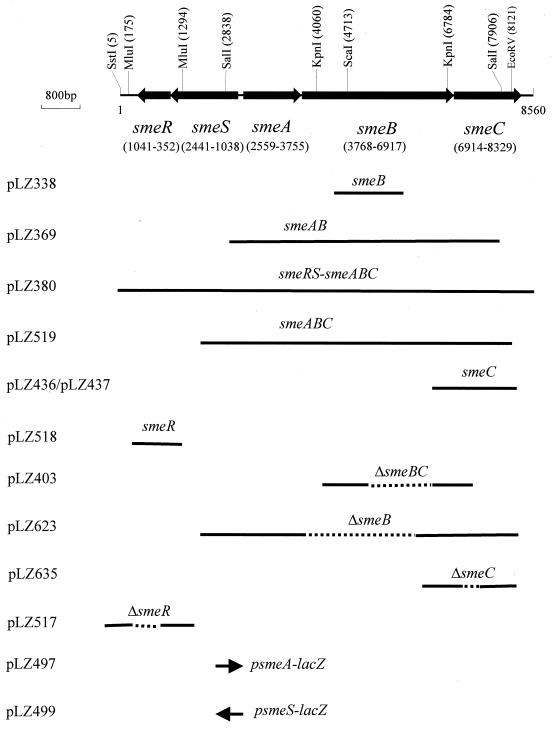
| pLZ338 | pBluescript II SK(+) derivative carrying a ca.-1.8-kb internal fragment of smeB | This study |
| pLZ370 | pLAFR3 derivative carrying the smeRSABC genes on a ca.-25-kb Sau3A fragment | This study |
| pLZ380 | pAK1900 derivative carrying the smeRSABC genes on a ca. 15-kb SstI fragment | This study |
| pLZ403 | pEX18Tc::ΔsmeBC; carries a 931-bp deletion in smeBC between bp 6205 and 7139 of the deposited smeRSABC sequence (GenBank accession no. AF173226) | This study |
| pLZ410 | pBluescript II SK(+) derivative carrying smeC on a ca.-3-kb KpnI fragment | This study |
| pLZ416 | pEX18Tc::L2 bla ΔStyI (L2 bla gene with a 4-bp deletion of an internal StyI site) | 53 |
| pLZ415A | pAK1900 derivative carrying the smeR gene on ca.-3-kb SphI fragment | This study |
| pLZ436 | pRK415 derivative carrying smeC on a ca.-3-kb KpnI fragment in same orientation as plac | This study |
| pLZ437 | pRK415 derivative carrying smeC on a ca.-3-kb KpnI fragment in opposite orientation to plac | This study |
| pLZ465 | pAK1900 derivative carrying the smeABC genes on a ca.-10-kb insert | This study |
| pLZ477 | pET21-d(+) derivative carrying smeC on a 1.4-kb EcoRI-HindIII fragment | This study |
| pLZ479 | pET21-d(+) derivative carrying smeR on a 0.7-kb EcoRI-HindIII fragment | This study |
| pLZ497 | pMP190 derivative carrying the smeABC promoter region (0.7-kb KpnI-XbaI fragment) upstream of lacZ | This study |
| pLZ499 | pMP190 derivative carrying the smeSR promoter region (0.7-kb XbaI-SalI fragment) upstream of lacZ | This study |
| pLZ515 | pBluescript II SK(+) derivative carrying the smeR gene on a 1.1-kb fragment | This study |
| pLZ517 | pEX18Tc::ΔsmeR; carries 349-bp deletion in smeR | This study |
| pLZ518 | pRK415 derivative carrying the smeR gene on a 1.1-kb HindIII-XbaI fragment in the same orientation as plac | This study |
| pLZ519 | pBluescript II SK(+) derivative carrying the smeABC gene on a ca.-7.2-kb SstI-HindIII fragment in same orientation as resident T3 promoter | This study |
| pLZ526 | pVLT31 derivative carrying the smeABC genes on a ca.-7.2-kb SstI-HindIII fragment in same orientation as resident lac promoter | This study |
| pLZ601 | pBluescript II SK(+) derivative carrying the smeABC genes on a ca.-7.2-kb insert in same orientation as resident T7 promoter | This study |
| pLZ605 | pRK415 derivative carrying the smeABC genes on a 7.2-kb HindIII-SstI fragment in same orientation as plac | This study |
| pLZ606 | pRK415 derivative carrying the smeS gene on a 2.2-kb HindIII-SstI fragment in same orientation as plac | This study |
| pLZ622 | pLZ605::ΔsmeB; carries an in-frame deletion of an internal 2.7-kb KpnI fragment | This study |
| pLZ623 | pEX18Tc::ΔsmeB; carries ΔsmeB of pLZ622 on a 4.5-kb HindIII-SstI fragment | This study |
| pLZ633 | pTZ19U derivative carrying the smeC gene on a 1.4-kb EcoRI-HindIII fragment | This study |
| pLZ634 | pTZ19U::ΔsmeC; pLZ633 derivative carrying a 130-bp deletion of internal BamHI-EcoRV fragment | This study |
| pLZ635 | pEX18Tc derivative carrying ΔsmeC of pLZ634 on a ca. 1.2-kb EcoRI-HindIII fragment | This study |
Abbreviations: Apr, ampicillin resistant; Cbr, carbenicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; MCS, multiple cloning site; plac, lac promoter.
DNA methodology.
Basic DNA procedures, including restriction endonuclease digestions, ligations, transformations, and agarose gel electrophoresis were performed as described previously (46). The alkaline lysis method (46) or a plasmid midi kit (Qiagen Inc., Mississauga, Ontario, Canada) was used to isolate plasmids from E. coli DH5α and S. maltophilia. The genomic DNA of S. maltophilia was extracted by the method of Barcak et al. (8). DNA fragments used in cloning were extracted from agarose gels using Prep-A-Gene (Bio-Rad Labs, Richmond, Calif.) as per the manufacturer’s instructions.
Selections of multiple-antibiotic-resistant strains.
Selection of multiple-antibiotic-resistant mutants was carried out as described previously (53). Briefly, 50 μl of an overnight culture of K1449 (an L1 and L2 β-lactamase-deficient strain derived from ULA-511) was plated onto antibiotic-containing LB agar (cefsulodin [16 μg/ml] and ciprofloxacin [4 μg/ml]) and incubated for 24 to 48 h at 37°C. Resistant colonies were subsequently tested for cross-resistance to additional antibiotics, and those exhibiting resistance to at least two structurally unrelated antibiotics were saved for further study.
Antimicrobial susceptibility assay.
Susceptibility testing was carried out by twofold serial dilution using LB broth, with an inoculum of 5 × 105 cells/ml. In some instances, this was carried out on LB agar plates containing twofold serial dilution of antibiotics with inoculum of 104 cells/spot (5 μl). Data were reported as MICs, which reflect the lowest concentrations of antibiotics inhibiting visible cell growth after an overnight incubation at 37°C.
Construction of a genomic library of S. maltophilia.
The genomic DNA of S. maltophilia ULA-511 was subjected to partial Sau3A digestion (46) and fractionated on a 10 to 40% (wt/vol) continuous sucrose gradient. Fractionations were examined by agarose gels, and those fractions enriched for DNA of ca. 20 kb in length were recovered, extensively dialyzed against 10 mM Tris-HCl (pH 8.0) containing 1 mM EDTA to remove the sucrose, and ligated to the broad-host-range cosmid cloning vector pLAFR3 (49), previously treated with BamHI and calf intestinal alkaline phosphatase (46). Following in vitro packaging of the ligated DNA with a Gigapack III Gold Package Extract Kit (Stratagene Cloning Systems, La Jolla, Calif.) and infection of E. coli DH5α cells according to instructions provided by the manufacturer, cosmid-containing DH5α clones were recovered on LB agar containing 10 μg of tetracycline per ml. Overall, more than 100,000 transductants were obtained, and these were pooled in groups and stored as 15% (vol/vol) glycerol stocks at −80°C for future use. Random screening of cosmids isolated from several transductants confirmed the presence of inserts of ca. 15 to 25 kb.
Amplification of a mexB homologue from S. maltophilia by PCR.
To identify multidrug efflux system-encoding genes in S. maltophilia, two PCR primers, mexb1xz (5′-GTGTTCCTGGTGATGTACCTGT-3′) and mexb3xz (5′-CGATGAGAATGGCGTTCTTCG-3′), derived from the internal sequences of the mexB gene of the P. aeruginosa mexAB-oprM operon (GenBank accession number L11616) (40, 41), were used to amplify a fragment of ca. 1.8 kb in a PCR using the genomic DNA of S. maltophilia (ATCC 13637 or ULA-511) as the template. The reaction mixture contained 0.1 μg of genomic DNA as the template, 40 pmol of each primer, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 2 mM MgSO4, 10% (vol/vol) dimethyl sulfoxide, and 2 U of Vent DNA polymerase (New England Biolabs, Mississauga, Ontario, Canada) in 1× Thermo reaction buffer and was heated for 2 min at 94°C followed by 30 cycles of 1 min at 94°C, 1 min at 52°C, and 1.5 min at 72°C. PCR products were purified using a QIAquick-spin PCR purification kit (Qiagen) and cloned into a SmaI-digested pBluescript II SK(+). The cloned insert in the resulting plasmid, dubbed pLZ338, was then sequenced using the universal T3 and T7 promoter primers (see below). Sequencing of pLZ338 identified the 1.8-kb insert as a mexB homologue. Thus, this gene was designated smeB (for Stenotrophomonas multidrug efflux).
Southern and colony hybridizations.
To obtain the intact smeB gene (and flanking DNA) directly from the genomic DNA of S. maltophilia or from a genomic library of S. maltophilia, Southern and colony hybridizations were carried out, respectively, with a 759-bp smeB-containing DNA fragment as the probe. This DNA fragment was obtained by PCR amplification of plasmid pLZ338 using primers smexb5 (5′-CCAGTTCTCGGTGACGAT-3′; anneals within smeB [bp 5171 to 5188 of GenBank accession no. AF173226]) and smexb6xz (5′-CGTAGCGCACGTTGACCA-3′; anneals within smeB [bp 5911 to 5928 of GenBank accession no. AF173226]) as described above. Following purification, the PCR product was labeled using a digoxygenin (DIG) High Prime DNA labeling kit (Boehringer Mannheim, Montreal, Quebec, Canada) as per the manufacturer’s instructions.
Southern hybridization was carried out in order to identify DNA fragments that were complementary to the 759-bp DIG-labeled smeB probe. Following digestion of S. maltophilia genomic DNA with a variety of restriction enzymes, the DNA fragments were separated on 0.8% (wt/vol) agarose gel and blotted onto a positively charged nylon membrane (Boehringer Mannheim), and smeB-complementary DNA was detected according to a protocol provided with the DIG High Prime DNA labeling and detection kit II (Boehringer Mannheim). A strong hybridization signal was obtained with a ca. 5-kb SalI fragment, and the corresponding region of an equivalent, unblotted gel of SalI-digested genomic DNA was excised, purified using Prep-A-Gene (Bio-Rad), and shotgun cloned into SalI-digested pBluescript II SK(+). One resultant plasmid, pLZ369, was shown by PCR and Southern hybridization to carry the 5-kb fragment. Nucleotide sequencing confirmed that the SalI-digested insert of pLZ369 carried the smeB gene in association with an upstream mexA-like gene (dubbed smeA) and a downstream oprM-like gene (smeC).
For colony hybridization, E. coli DH5α cells carrying cosmids with S. maltophilia genomic DNA were grown overnight on LB agar supplemented with tetracycline. Following the printing of the colonies onto a positively charged nylon membrane, the membrane was processed and screened with the aforementioned 759-bp DIG-labeled smeB probe according to instructions provided with DIG High Prime DNA labeling and detection kit II (Boehringer Mannheim). From the colony hybridization, a clone that positively reacted with the smeB probe was identified, and its cosmid, dubbed pLZ370, was further studied. PCR was used to amplify a product of 759 bp from pLZ370 using primers smexb5xz and smexb6xz (see above). Nucleotide sequencing indicated that this PCR fragment was identical to the aforementioned smeB gene (see above). The digestion of pLZ370 by SstI resulted in two DNA fragments (ca. 15 kb and ca. 10 kb), which were subcloned into an SstI-digested pAK1900, yielding plasmids pLZ380 and pLZ381, respectively. These plasmids were then subjected to nucleotide sequencing.
Cloning of sme genes.
Plasmid pLZ380 carries the intact smeABC genes as well as substantial unsequenced DNA beyond smeC. In order to better assess the contribution of smeABC alone to antimicrobial resistance, it was necessary to remove this downstream DNA. Suitable restriction sites for subcloning were not readily identified downstream of smeC, necessitating the use of a multistep cloning procedure. Initially, the intact smeABC genes and additional unknown sequence downstream of smeC were recovered on a ca. 10-kb MluI-XbaI fragment, which was cloned into SmaI-restricted pAK1900 following the polishing of its ends with T4 DNA polymerase. The resultant vector, pLZ465, was digested with SstI-EcoRV to release a ca.-7-kb fragment carrying the intact smeAB genes and the 5′ end of smeC. Next, the 3′ end of smeC (ca. 600 bp) with ca. 120 bp downstream sequence (i.e., ca. 720 bp total) was amplified from plasmid pLZ380 using primers soprm11xz (5′-AATACCCGGGATGGCAGCGTCGCATTGGC-3′; anneals within smeC; SmaI site underlined) and soprm12xz (5′-TATCAAGCTTCTGGTGGGTGCCGAC-3′; anneals ca. 120 bp downstream from smeC stop codon; HindIII site underlined). Following digestion with SmaI and HindIII, the PCR product was cloned into SmaI-HindIII-restricted pBluescript II SK(+) to produce pLZ509. Digestion of this plasmid with SstI-EcoRV generated an intact pBluescript II SK(+) fragment carrying the 3′ end of smeC, which was then ligated to the aforementioned SstI-EcoRV fragment of pLZ465 (see above) to produce pLZ519. The smeABC genes were then recovered on a ca.-8-kb SstI-HindIII fragment and cloned into pVLT31, yielding pLZ526.
The smeC gene was liberated from plasmid pLZ370 on a ca. 3-kb fragment following digestion with KpnI, as confirmed by Southern hybridization using a DIG-labeled smeC probe. The smeC probe was obtained by labeling of a PCR product, which was amplified from plasmid pLZ380 using primers soprm3xz (5′-ATGCTCTAGAACAACCGCGATCTGCGCGT-3′; anneals within smeC; XbaI site underlined) and soprm4xz (5′-TGAGAAGCTTGCCATCAACGTCGACAAC-3′; anneals within smeC; HindIII underlined). The PCR was formulated and processed as described above for smeB. The smeC-containing KpnI fragment was cloned into KpnI-digested pBluescript II SK(+), producing plasmid pLZ410. Subsequently, the smeC-containing fragment of pLZ410 was subcloned into KpnI-digested pRK415, yielding plasmids pLZ436 and pLZ437, which carried the smeC gene in the same and opposite orientation to the pRK415 lac promoter, respectively.
A blunt-ended ca.-1.1-kb fragment containing intact smeR gene was generated following digestion of pLZ415A with MluI and treatment with T4 DNA polymerase. This fragment was cloned into the SmaI-EcoRV-restricted pBluescript II SK(+) to produce pLZ515. The smeR gene was then released from pLZ515 by digestion with HindIII and XbaI, and the 1.1-kb fragment was subcloned into pRK415 to yield pLZ518, which permits transcription of the smeR gene under the control of plac.
Construction of sme deletion mutants.
To construct ΔsmeBC mutants, PCRs were performed to amplify sequences upstream (1.2 kb) and downstream (0.8 kb) of the intended deletion. The upstream fragment was amplified from pLZ369 using primers smexb23xz (5′-AGATGAATTCGCGCAGGTGGTGCTCAAG-3′; anneals upstream of smeB; EcoRI site underlined) and smexb24xz (5′-AGTATCTAGACTGGTCCAATGCGTGCTG-3′; anneals downstream of the smeB start codon; XbaI site underlined), while the downstream fragment was amplified with primers, soprm3xz (see above) and soprm4xz (see above). PCRs were formulated and processed as described above for smeB. The two PCR products were digested with EcoRI-XbaI or XbaI-HindIII, as appropriate, and cloned into the EcoRI-HindIII-digested gene replacement vector pEX18Tc via a three-piece ligation. The resultant plasmid, pLZ403, was introduced into E. coli S17-1 by transformation and mobilized into S. maltophilia strains ULA-511, K1385, K1449, and K1668 via conjugation (48). Transconjugants carrying pLZ403 in the chromosome were selected on LB agar containing tetracycline (40 μg/ml for ULA-511 and K1449; 80 μg/ml for K1385 and K1668) and norfloxacin (10 μg/ml; for counterselection). Transconjugants were then streaked onto LB agar containing 10% (wt/vol) sucrose, and sucrose-resistant colonies arising after overnight incubation at 37°C were screened for the presence of the smeBC deletion using PCR with three pairs of primers (soprm5xz-smexbxz8xz, soprm5xz-smexb27xz, and soprm5xz-smexb5xz [see above]).
To construct in-frame smeB deletion mutants, plasmid pLZ605 carrying the intact smeABC operon was digested by KpnI to remove two KpnI fragments that together contained smeA and the 5′ end of smeB. KpnI-restricted pLZ605 was then self-ligated to create pLZ613, which contained the 3′ end of smeB and the entirety of smeC. Subsequent digestion of pLZ519, which also carried the intact smeABC operon, with KpnI-ScaI released several fragments, including a 2.8-kb KpnI fragment carrying the entirety of smeA and the 5′ end of smeB. This fragment was then cloned into KpnI-restricted pLZ613. One of the resulting plasmids, pLZ622, in which the orientation of the smeA-smeB insert was the same as the smeB-smeC genes of pLZ613 (confirmed by PCR), carried a ca. 2.7-kb deletion in smeB. The smeB deletion was then recovered on a 4.5-kb fragment from pLZ622 and cloned into the HindIII-SstI restricted pEX18Tc to yield pLZ623, which was introduced to E. coli S17-1 by transformation. Plasmid pLZ623 was then mobilized into S. maltophilia strains K1449 and K1668 via conjugation (see above). Transconjugants carrying pLZ623 in the chromosome were selected on LB agar containing tetracycline (80 μg/ml) and norfloxacin (10 μg/ml; for counterselection). Transconjugants were then streaked onto LB agar containing 10% (wt/vol) sucrose, and sucrose-resistant colonies arising after overnight incubation at 37°C were screened for the presence of the smeB deletion using PCR with primers smebaxz and smebbxz.
To construct a ΔsmeC strain of S. maltophilia, an intact smeC gene was initially released from plasmid pLZ471 (see below) by digestion with EcoRI and HindIII. The 1.4-kb smeC-containing fragment was then cloned into EcoRI-HindIII-treated pTZ19U to yield pLZ633. Digestion of pLZ633 with BamHI-EcoRV removed a 130-bp DNA fragment from the middle of smeC. Following blunt ending of BamHI-EcoRV-digested pLZ633 with T4 DNA polymerase, the plasmid was recircularized via ligation to produce the ΔsmeC-carrying plasmid pLZ634. The ΔsmeC gene was then released from pLZ634 via EcoRI-HindIII digestion and cloned into pEX18Tc previously digested with EcoRI-HindIII, to produce pLZ635. This vector was introduced to E. coli S17-1 by transformation and mobilized into S. maltophilia strains K1449 and K1668 via conjugation (see above). Transconjugants carrying pLZ623 in the chromosome were selected on LB agar containing tetracycline (50 μg/ml) and norfloxacin (5 μg/ml; for counterselection). Transconjugants were then streaked onto LB agar containing 10% (wt/vol) sucrose, and sucrose-resistant colonies arising after overnight incubation at 37°C were screened for the presence of the smeC deletion using PCR with primers smexb17xz and soprm5xz.
To construct ΔsmeR mutants, two PCRs were performed to amplify DNA fragments 5′ (ca. 1 kb) and 3′ (ca. 0.6 kb) to the sequences to be deleted within smeR. Using plasmid pLZ370 as a template, sequences 5′ to the deletion were amplified with primers smer5xz (5′-TGACTCTAGACCGGTACATCGCTGCT-3′; anneals within smeS; XbaI site underlined) and smer6xz (5′-CTACAAGCTTCAACCGGATGGATGGCGCA-3′; anneals downstream of smeR start codon; HindIII underlined), while sequences 3′ to deletion were amplified with primers smer7xz (5′-CTCTGAATTCGAGCGCGCCATGTTG-3′; anneals downstream of smeR; EcoRI site underlined) and smer4xz (5′-TGAATCTAGACAGCCATGTGCGCAAC-3′; anneals upstream of smeR stop codon; XbaI site underlined). The PCR was formulated and processed as described above for smeB. The two PCR products were digested with EcoRI-XbaI or XbaI-HindIII, as appropriate, and cloned into EcoRI-HindIII-digested pEX18Tc via a three-piece ligation. The resultant plasmid, pLZ517, which contained an internal deletion of 340 bp in the smeR gene, was introduced into E. coli S17-1 and then mobilized into S. maltophilia strains K1449 and K1668 via conjugation. Transconjugants carrying pLZ517 in the chromosome were selected on LB agar containing tetracycline (80 μg/ml) and norfloxacin (10 μg/ml; for couterselection). Transconjugants were then streaked onto LB agar containing 10% (wt/vol) sucrose, and sucrose-resistant colonies arising after overnight incubation at 37°C were screened for the presence of the smeR deletion using PCR with primers smer6xz (see above) and smerbxz (see below).
Construction of an L2 bla insertion mutation.
Disruption of the L2 bla gene in K1668 was originally carried out by insertion of 4 bp near the middle of the gene, although this could still yield a truncated (and perhaps partially functional) L2 enzyme of 187 amino acids (c.f. 303 amino acids in the native enzyme). To create a null mutation, then, an internal fragment of the L2 bla gene was amplified by PCR and cloned into the gene replacement vector pEX18Tc. Following homologous recombination, the chromosomal L2 bla gene would be disrupted by insertion of pEX18Tc sequences. A ca.-400-bp fragment of the L2 bla gene (beginning immediately after the ATG start codon) was amplified with primers sml8xz (5′-ACTTGTCGACCGTCGCCGATTCCTGCAGTT-3′; SalI site underlined) and sml9xz (5′-AGATGGATCCTGATCGTGGCACGGCACAGA-3′; BamHI site underlined) using plasmid pLZ416 as a template. Reaction mixtures were formulated as described above and subjected to 30 cycles of 50 s at 94°C, 40 s at 56°C, and 30 s at 73°C before finishing with a 5-min incubation at 72°C. The resultant PCR product was restricted with SalI and BamHI and cloned into SalI-BamHI-restricted pEX18Tc, yielding pLZ736. Following introduction of this vector into E. coli S17-1, it was mobilized into S. maltophilia K1668 as described above, and transconjugants (e.g., K1975) were selected on tetracycline (25 μg/ml) and norfloxacin (2.5 μg/ml; for counterselection). K1795 would be effectively be L2−, containing as it would two incomplete versions of the L2 bla gene, one truncated at the 5′ end (immediately after the start codon) and one truncated at the 3′ end (400 bp into the gene).
RT-PCR.
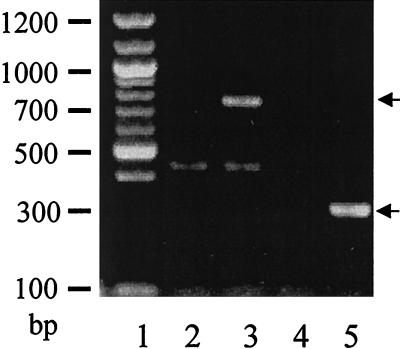
Total bacterial RNA was isolated from log-phase cultures (1 to 2 ml) of S. maltophilia strains using the Qiagen RNeasy Mini Kit (Qiagen), treated with RNase-free DNase (Promega, Madison, Wis.; 2 U of enzyme/μg of RNA for 60 min at 37°C), and repurified using the same kit. A 0.2-μg sample of DNase-treated RNA was used as a template for reverse transcription (RT)-PCR with the Qiagen OneStep RT-PCR kit according to a protocol supplied by the manufacturer. Primer pairs specific for and internal to smeA (smeaaxz, 5′-TACAGAATTCATGTCTCTCCTGCGCCCG-3′ [forward]; smexa2xz, 5′-ACCTTAACCTGTGCCTTG-3′ [reverse]; and smexa3xz, 5′-GTCGACCTGGTACAGCA-3′ [reverse]) were used to amplify and quantitate the corresponding mRNA, as a measure of smeABC expression. Thirty picomoles of each primer was used per reaction mixture (final volume of 50 μl), which involved a 30-min incubation at 50°C, followed by 15 min at 95°C and 40 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C before finishing with 10 min at 72°C. A 15-μl sample of each reaction product was analyzed by agarose (1.4% [wt/vol]) gel electrophoresis for the expected RT-PCR products (742 and 296 bp).
Expression of SmeC and SmeR.
To identify the products of the smeC and smeR genes, it was necessary to clone these genes into a suitable expression system. PCR was initially used to amplifying smeC and smeR from pLZ380 with two pairs of primers, smecaxz (5′-TCTGGAATTCATGAAGCCGATGCTGCTGC-3′; EcoRI site underlined) and smecbxz (5′-TCATAAGCTTCCGCGCGTCCGCGCCACCAC-3′; HindIII site underlined) for smeC and smeraxz (5′-TACAGAATTCATGAGCACGTCGCCCGCCAC-3′; EcoRI site underlined) and smerbxz (5′-TCATAAGCTTGGGCTCGTAGCTGTAGCCCAT-3′; HindIII site underlined) for smeR. The smeC-containing (1.4-kb) and smeR-containing (0.7-kb) PCR products were directly cloned into pCR-Blunt-II-TOPO vector as instructed by the manufacturer (Invitrogen, Carlsbad, Calif.), yielding plasmids pLZ471 and pLZ473, respectively. Cloned inserts were confirmed by DNA sequencing. Following digestion of pLZ471 and pLZ473 with EcoRI and HindIII, the smeC- and smeR-containing fragments were cloned into EcoRI-HindIII-restricted pET21-d(+) to yield pLZ477 and pLZ479, respectively. These plasmids were then transformed into E. coli BL21(DE3) carrying the pLysS plasmid, respectively, and expression of smeC and smeR was examined in whole-cell lysates of BL21(DE3). Briefly, cells grown overnight in LB broth were subcultured in 5 ml of LB broth to early exponential phase of growth (optical density at 600 nm, 0.2). The cells were then incubated with (to induce expression of the cloned genes) or without 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. One milliliter of cells was subsequently harvested, washed once with 50 mM sodium phosphate buffer (pH 7.5), and resuspended in the same buffer before being subjected to sodium dodecyl sulfate-polyacrylmide (11% [wt/vol]) gel electrophoresis (SDS–11% PAGE) as described previously (46).
Cloning of smeABC and smeSR promoter regions.
To evaluate expression of the smeABC and smeSR operons, the intergenic region encompassing sequences upstream of both was recovered on a ca.-0.7-kb PCR product produced using pLZ380 as the template and primers smexa6xz (5′-ACGGGTCGACCTGGTACAGC-3′; anneals within smeA) and smes1xz (5′-ACCGTCTAGATGCTCCCAGCCACCGT-3′; anneals within smeS). The PCR product was directly cloned into pCR-Blunt II-TOPO vector (Invitrogen) as per the manufacturer’s instructions, producing plasmid pLZ474. The insert DNA was then sequenced to ensure no error had been introduced during PCR. Digestion of pLZ474 with KpnI and XbaI released a 0.7-kb fragment whose cloning into KpnI-XbaI-restricted pMP190 (to yield pLZ497) placed the smeABC promoter region upstream of and in the proper orientation relative to the promoterless lacZ gene of this vector. Similarly, digestion of pLZ474 with SalI and XbaI released a 0.7-kb fragment whose cloning into SalI-XbaI-restricted pMP190 (to yield pLZ499) placed the putative smeSR promoter region upstream of and in the proper orientation relative to the promoterless lacZ gene of this vector.
Plasmid mobilization.
Introduction of plasmids pRK415 and pVLT31 and their derivatives from E. coli DH5α into S. maltophilia required a triparental mating procedure employing the helper vector pRK2013 (39), while introduction of plasmid pMP190 and its derivatives from E. coli S17-1 into S. maltophilia employed a biparental mating procedure. Briefly, in triparental mating, 100-μl aliquots of overnight cultures of plasmid-containing E. coli DH5α, pRK2013-containing E. coli MM294, and ULA-511 derivatives of S. maltophilia were mixed in a microcentrifuge tube, and in biparental mating, 100 μl of plasmid-containing E. coli S17-1 and S. maltophilia K1449 were mixed. The cell mixtures were subsequently pelleted by centrifugation, resuspended in 40 μl of LB broth, and spotted onto the center of an LB agar plate. Following incubation overnight at 37°C, bacterial cells were resuspended in 1 ml of LB broth and appropriate dilutions (10−2 to 10−4) were plated on LB agar containing norfloxacin (5 to 10 μg/ml; to counterselect the E. coli strains) and tetracycline (25 to 50 μg/ml; for pRK415 and pVLT31 and their derivatives) or chloramphenicol (20 μg/ml; for pMP190 and its derivatives). Plasmid DNA was prepared from S. maltophilia recipients using the miniprep procedure to confirm successful plasmid transfer.
β-Galactosidase assays.
Bacteria harboring plasmids pMP190 and pRK415 and their derivatives were cultured overnight at 37°C in LB broth supplemented with chloramphenicol (30 μg/ml) and tetracycline (25 μg/ml) and subsequently diluted 50-fold into fresh LB broth. Following growth to late log phase (optical density at 600 nm, 0.8 to 1.0), cultures were assayed for β-galactosidase activity as described previously (29)
β-Lactamase preparation and activity assays.
The extraction and assay of the S. maltophilia chromosomal β-lactamases were carried out as described previously (without imipenem induction) using nitrocefin as the chromogenic substrate (24). Large-scale extraction of β-lactamase from strain K1668 was carried out using a cold-osmotic shock method (33), followed by purification of the β-lactamase on a borate-affinity column (9). Six hundred milliliters of cells at the late-exponential phase of growth in LB broth was harvested and washed once with 100 ml of 30 mM Tris-HCl (pH 7.5). The cells were then resuspended in 30 ml of 30 mM Tris-HCl (pH 7.5), followed by the addition of 30 ml of 40% (wt/vol) sucrose (prepared in 30 mM Tris-HCl [pH 7.5]-2 mM EDTA). The cell suspension was centrifuged at 8,000 × g for 20 min, and the cell pellet resuspended in 25 ml of chilled H2O containing 1 mM MgCl2 before being centrifuged at 12,000 × g for 20 min. The β-lactamase-containing supernant recovered after centrifugation was mixed with an equal volume of 20 mM triethanolamine (pH 7.0) and then loaded onto an m-aminophenylboronic acid agarose (Sigma) column equilibrated with 20 mM triethanolamine (pH 7.0). The column was then washed with 50 ml of 20 mM triethanolamine (pH 7.0) containing 0.5 M NaCl, and the β-lactamase was eluted with 20 ml of 0.5 M sodium borate (pH 7.0), also containing 0.5 M NaCl. One-milliliter fractions were collected and those enriched for β-lactamase activity (assayed using the nitrocefin assay described above) and demonstrating high purity (assayed by SDS-PAGE with silver staining [46]) were pooled and dialyzed against 10 mM Tris-HCl (pH 7.5). The 22-kDa β-lactamase was then concentrated using a Centricon concentrator (Amicon, Inc., Beverly, Mass.) as previously described (53). Hydrolysis of β-lactams by the enzyme was determined spectrophotometrically at 230 nm (for penicillins) or at 260 nm (cephalosporins) using a 0.5 mM (penicillins) or 0.1 mM (cephalosporins) final substrate concentration. The enzyme activity was expressed in nanomoles of substrate hydrolyzed per minute per milligram of protein at room temperature. The rates of hydrolysis for each substrate were calculated relative to that of nitrocefin.
DNA sequencing.
Nucleotide sequencing of plasmid-borne DNA or PCR products was carried out by Cortec DNA Services Inc. (Queen’s University) using universal or custom primers. Compilation of DNA sequence data was performed using the PC Gene software package (version 2.1; Intelligenetics Inc., Mountain View, Calif.) and DNAMAN (version 4.11; Lynnon Biosoftware, Vaudreuil, Quebec, Canada). Promoter prediction was carried out using neural network promoter prediction software (http://www.fruitfly.org/seq_tools/promoter.html).
Nucleotide sequence accession number.
The nucleotide sequences of the smeABC and smeSR operons have been deposited with the GenBank sequence database under accession number AF173226.
RESULTS
Cloning and sequencing of the smeABC operon of S. maltophilia.
The RND components of the multidrug efflux systems of P. aeruginosa, Pseudomonas putida, and E. coli show a high degree of amino acid sequence identity. In an effort to recover a multidrug efflux system from S. maltophilia, then, primers from a highly conserved region of the mexB component of the mexAB-oprM multidrug efflux operon were designed and used to amplify an RND homologue from the chromosome of S. maltophilia. Using primers mexb1xz and mexb3xz, an expected 1.8-kb product was amplified from the genome of S. maltophilia strain ULA-511, and nucleotide sequencing revealed that it was highly homologous to mexB (data not shown). Primers based on the sequence of the PCR product were also successful in reamplifying appropriately sized fragments from the chromosomes of S. maltophilia ULA-511 and ATCC 13637. To recover the complete gene, then, a cosmid library of S. maltophilia ULA-511 genomic DNA was screened with a probe derived from the PCR product. One cosmid, pLZ370, was subsequently identified, and the mexB-like sequence was localized to a 15-kb SstI fragment, which was subcloned to produce plasmid pLZ380. Nucleotide sequencing of a portion of the insert DNA in this vector revealed five open reading frames, three of which were predicted to encode products with substantial homology to the MexAB-OprM (22, 40, 41), MexCD-OprJ (39), MexEF-OprN (19), and MexXY-OprM (1, 30, 52) multidrug efflux systems of P. aeruginosa; the TtgABC (44) and SrpABC (17) multidrug efflux systems of P. putida; and the AcrAB multidrug efflux system of E. coli (25) (Table 2). These were subsequently dubbed smeABC (for Stenotrophomas multidrug efflux) (Fig. 1). During this work a second multidrug efflux system was reported in S. maltophilia, encoded by the smeDEF genes (4). SmeDEF was also very similar to SmeABC (Table 2). The neural network prediction method (see Materials and Methods) identified a strong candidate promoter region upstream of smeA (−40 to −1 region, 5′CGCCAGCTGAGCCCGTCGCACAATCTCCATATTCTCTGCA-3′ [consensus hexamers underlined]) with transcription predicted to initiate 55 bp upstream of the efflux genes.
TABLE 2.
Characteristics of the putative proteins encoded by smeABC and smeSR operons
| Protein | No. of amino acid residues | Molecular mass (kDa) | Putative function | Homologuesa (% identity) |
|---|---|---|---|---|
| SmeA | 398 | 42 | Inner-membrane fusion lipoprotein | MexAPa (43), MexCPa (40), MexEPa (30), MexXPa (31), TtgAPp (40), SrpAPp (42), AcrAEc (44), YegMEc (23), SmeDSm (44) |
| SmeB | 1,049 | 112 | RND transporter | MexBPa (52), MexDPa (44), MexFPa (38), MexYPa (42), TtgBPp (53), SrpBPp (47), AcrBEc (55), YegNEc (29), YegOEc (29), SmeFSm (53) |
| SmeC | 471 | 50 | Outer-membrane efflux lipoprotein | OprMPa (50), OprJPa (39), OprNPa (25), TtgCPp (45), SrpCPp (42), TolCEc (17), YegBEc (15), SmeDSm (44) |
| SmeS | 467 | 51 | Two-component system sensory kinase | CpxAEc (26), CreCEc (24), CreCPa (25), SrpSPp (8), BaeSEc (39) |
| SmeR | 229 | 26 | Two-component system response regulator | CpxREc (36), CreBEc (36), CreBPa (43), SrpRPp (10), BaeREc (54) |
The organisms are indicated in subscripts after their proteins (Pa, P. aeruginosa; Pp, P. putida; Ec, E. coli; Sm, S. maltophilia) and the identity of the homologues with respect to SmeABC and SmeSR is indicated in parentheses. The sources (GenBank accession numbers of the homologues compared are indicated in parentheses: MexAB-OprM (L11616); MexCD-OprJ (U57969), MexEF-OprN (X99514), MexXY (AB015853), TtgABC (AF031417), SrpABC (AF029405)-SrpSR (AF061937), AcrAB (U00734)-TolC (AE000385), YegMNOB (AE000297)-BaeSR (D14054; AE000297), SmeDEF (AJ252200), CpxAR (AE000466), CreBC of E. coli (AE000510) and of P. aeruginosa (AE004484). The putative operon yegMNOB (including two RND transporter genes, yegN and yegO) is located upstream of the baeSR operon, but both operons, yegMNOB and baeSR, are transcribed in the same orientation.
FIG. 1.
Physical map of the smeSR and smeABC operons. Restriction fragments cloned into appropriate plasmids are featured. See the text for a detailed description of the plasmids.
Expression of SmeABC in MDR mutants of S. maltophilia.
To assess whether SmeABC contributed to antimicrobial resistance in wild-type S. maltophilia, the smeBC genes were deleted from the chromosome of S. maltophilia ULA-511 and its L1-L2 β-lactamase-deficient mutant K1449. The resultant strains, K1778 (data not shown) and K1780 (Table 3), failed to demonstrate enhanced susceptibility to a variety of antimicrobial agents (Table 3) or heavy metal salts (data not shown). RT-PCR subsequently confirmed, however, that the efflux system was not expressed in wild-type S. maltophilia (Fig. 2, lanes 2 and 4). To assess whether this efflux system was overexpressed in and responsible for the multidrug resistance of a number of previously reported MDR mutants (e.g., K1385) (53), the smeBC deletion was engineered into these strains as well. Again, however, no impact on resistance was observed (data not shown). Subsequently, one of several mutants of S. maltophilia K1449 selected on ciprofloxacin and cefsulodin and displaying resistance to aminoglycosides, β-lactams, and fluoroquinolones was shown to express smeABC (e.g., K1668 [Fig. 2, lanes 3 and 5; Table 3]). Interestingly, this mutant showed increased susceptibly to trimethoprim, perhaps due to decreased expression of a trimethoprim resistance determinant as a result of smeABC hyperexpression. Deletion of smeBC in strain K1668 compromised resistance to β-lactams, aminoglycosides, and fluoroquinolones (see K1781 in Table 3), consistent with this efflux system playing a role in the multidrug resistance of K1668. Interestingly, however, an in-frame deletion of smeB failed to alter the resistance of K1668 (see K1783 in Table 3), although deletion of smeC from this strain once again compromised the multidrug resistance of this mutant (see K1785 in Table 3). Moreover, the cloned smeABC genes (e.g., plasmid pLZ526) collectively failed to promote resistance to any antimicrobial in wild-type S. maltophilia strains (data not shown). This suggested that SmeABC does not function as a multidrug efflux system but rather that SmeC plays a role in antimicrobial resistance independent of SmeAB, possibly as the OMF component of a yet-to-be-identified multidrug efflux system.
TABLE 3.
Influence of SmeABC and SmeSR on antibiotic susceptibility of S. maltophilia
| Antibiotic | MIC (μg/ml) fora:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| K1449 (wild type) | K1780 (ΔsmeBC) | K1782 (ΔsmeB) | K1784 (ΔsmeC) | K1786 (ΔsmeR) | K1668 (MDR) | K1781 (ΔsmeBC) | K1783 (ΔsmeB) | K1785 (ΔsmeC) | K1787 (ΔsmeR) | |
| β-Lactams | ||||||||||
| Penicillin | 2–4 | 1–2 | 2 | 2 | 2 | 32 | 2 | 64 | 2 | 8 |
| Carbenicillin | 2–4 | 2 | 2 | 2 | 2 | 128 | 2 | 128 | 4 | 32 |
| Ampicillin | 1 | 1 | 1 | 1 | 1 | 32–64 | 0.5 | 64 | 1 | 16 |
| Cefsulodin | 4–8 | 4 | 8 | 8 | 4 | 128 | 4 | 256 | 4 | 32 |
| Cefotaxime | 2 | 2 | 2 | 2 | 2 | 16 | 2 | 32 | 2 | 4 |
| Cefoperazone | 4 | 2 | 2 | 2 | 4 | 8 | 2 | 8 | 2 | 4–8 |
| Cefepime | 1–2 | 1 | 2 | 2 | 2 | 4 | 1 | 4 | 2 | 4 |
| Cefpirome | 2 | 2 | 4 | 2 | 2 | 8 | 4 | 8 | 2 | 4 |
| Imipenem | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Aminoglycosides | ||||||||||
| Amikacin | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 | >2,048 | 1,024 | >2,048 | 1,024 | 2,048 |
| Gentamicin | 512–1,024 | 1,024 | 1,024 | 1,024 | 1,024 | >2,048 | 512 | >2,048 | 1,024 | 2,048 |
| Kanamycin | 512–1,024 | 512 | 1,024 | 1,024 | 512 | 4,096 | 512 | 4,096 | 1,024 | 1,024 |
| Streptomycin | 512–1,024 | 512–1,024 | 1,024 | 1,024 | 512 | >4,096 | 512 | 4,096 | 1,024 | 2,048 |
| Tobramycin | 2,048 | 2,048 | 2,048 | 2,048 | 2,048 | >2,048 | 2,048 | >2,048 | 2,048 | >2,048 |
| Quinolones | ||||||||||
| Nalidixic acid | 4 (8) | 4 (8) | 8 | 4 | 4 | 8 (32) | 4 (8) | 4 | 4 | 4 |
| Ciprofloxacin | 2–4 (4) | 2–4 (4) | 4 | 4 | 4 | 8–16 (16) | 4 (4) | 16 | 4 | 4–8 |
| Norfloxacin | 8 (32) | 8 (32) | 8 | 8 | 8 | 16 (128) | 8 (32) | 8 | 8 | 8 |
| Trovafloxacin | (1) | (1) | ND | ND | ND | (4) | (1) | ND | ND | ND |
| Other | ||||||||||
| Chloramphenicol | 8–16 (16) | 8 (16) | 16 | 8 | 8 | 2–4 (4) | 8 (16) | 4 | 16 | 8 |
| Tetracycline | 8 (32) | 8 (32) | 8 | 8 | 8 | 4 (32) | 8 (32) | 8 | 8 | 8 |
| Trimethoprim | 512–1,024 | 256 | 512–1,024 | 128 | 256 | 64 | 128 | 128 | 512 | 256 |
| Erythromycin | 512 | 512 | 512 | 512 | 512 | 512 | 512 | 512 | 512 | 512 |
Susceptibilities of K1449 and its deletion derivatives (K1780, K1782, K1784, and K1786) and the MDR derivative of K1449 (K1668) and its deletion derivatives (K1781, K1783, K1785, and K1787) to the indicated antibiotics were assessed in LB broth or on LB agar plates as outlined in Materials and Methods. The MICs in parentheses were obtained using LB agar plate assays. ND, not determined.
FIG. 2.
smeA expression in S. maltophilia measured by RT-PCR of RNA isolated from strains K1449 (lanes 2 and 4) and K1668 (lanes 3 and 5) using smeA-specific primers, smeaaxz and smexa2xz (lanes 2 and 3) or smeaaxz and smexa3xz (lanes 4 and 5). The expected smeA products are indicated by arrows. DNA size markers are shown on the left (lane 1).
SmeC functionally replaces OprM.
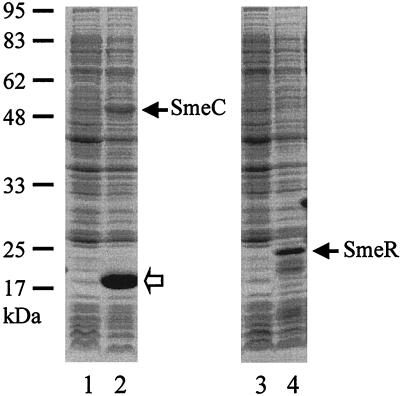
To further assess the possibility of SmeC functioning as the OMF of a multidrug efflux system in S. maltophilia, its ability to functionally replace OprM of the MexAB-OprM multidrug efflux system of P. aeruginosa was examined. To this end, the cloned gene, which was shown to be active and to direct the expression of the expected ca.-50-kDa protein (Fig. 3, lane 2) was introduced into P. aeruginosa strain K1113, a nalB derivative rendered antibiotic hypersusceptible through deletion of the oprM gene. When smeC was introduced into K1113 on vector pRK415 in the same orientation as the resident lac promoter of this vector (see pLZ436) substantial restoration of antibiotic resistance was observed (Table 4), although not to levels seen for the OprM+ parent strain of K1113 (see K1112 in Table 4). When smeC was introduced into K1113 such that it was in the opposite orientation with respect to the pRK415 lac promoter (see pLZ437), resistance levels were much lower but still higher (with the exception of carbenicillin) than that seen for the vector control lacking smeC (Table 4). These data are consistent with SmeC functioning, apparently as part of a MexAB-SmeC multidrug efflux system, in efflux-mediated multidrug resistance in P. aeruginosa, suggesting that it likely plays a similar role in S. maltophilia. Moreover, they also indicate that a weak promoter must exist upstream of smeC, providing for the expression and resistance seen for K1113 harboring pLZ437. Indeed, using the neural network promoter prediction method (see Materials and Methods) a weak promoter was identified upstream of this gene, with a predicted transcription start site 90 bp upstream of smeC (data not shown).
FIG. 3.
Analysis of SmeC and SmeR expression in E. coli BL21(DE3) harboring smeC-containing plasmid pLZ477 (lanes 1 and 2) or smeR-containing plasmid pLZ479 (lanes 3 and 4) by SDS-PAGE of cell lysates prepared from the cells without (lanes 1 and 3) and with (lanes 2 and 4) induction by IPTG. The SmeC and SmeR proteins are indicated by arrows, and the molecular mass standards are shown on the left. A possible SmeC degradation product is indicated by the open arrow.
TABLE 4.
Functional replacement of P. aeruginosa OprM outer membrane efflux protein by SmeC
| P. aeruginosa strain | Relevent genotype | Plasmid | MIC (μg/ml) ofc:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CAR | CPZ | CIP | NOR | CAM | ERY | NOV | |||
| ML5087 | Wild type | None | 64 | 8 | 0.4 | 1 | 128 | 256 | 128 |
| K1112 | nalB | None | 512 | 32 | 1.6 | 4 | 512 | 512 | >512 |
| K1113 | nalB ΔoprM | None | 0.5 | 0.25 | 0.006 | 0.015 | 2 | 32 | 8 |
| K1113 | nalB ΔoprM | pRK415 (vector) | 0.5 | 0.25 | 0.006 | 0.015 | 2 | 32 | 8 |
| K1113 | nalB ΔoprM | pLZ436 (pRK415::smeCa) | 8 | 4 | 3.2 | 2 | 64 | 512 | 128 |
| K1113 | nalB ΔoprM | pLZ437 (pRK415::smeCb) | 0.5 | 2 | 1.6 | 1 | 32 | 128 | 32 |
smeC was cloned in the same orientation as plac.
smeC was cloned in the opposite orientation to plac.
Abbreviations: CAR, carbenicillin; CPZ, cefoperazone; CIP, ciprofloxacin; NOR, norfloxacin; CAM, chloramphenicol; ERY, erythromycin; NOV, novobiocin.
A two-component regulatory system regulates smeABC expression.
Two open reading frames (dubbed smeSR) organized into a probable operon and transcribed divergently from the efflux genes were identified immediately upstream of smeABC (Fig. 1). The predicted products of these genes showed substantial similarity to a family of sensor kinases (SmeS) and transcriptional activators (SmeR) that work together to promote target gene expression in response to stress (Table 2) (12, 32, 43). Again, a strong candidate promoter was identified upstream of smeS (−40 to −1 region, 5′-GCCTTGCGTTGACCCGTGCACGCTGCCGTGAAACTGTGCA-3′, consensus hexamers underlined) with a predicted transcription start site occurring 69 bp upstream of the smeS start codon. To assess the significance of smeSR vis-à-vis regulation of smeABC expression, a deletion of the smeR gene was engineered into the chromosome of S. maltophilia strains K1449 and K1668 and the impact on antimicrobial susceptibility was examined. As expected, elimination of smeR in the SmeABC-hyperexpressing mutant K1668 compromised, to some extent, the resistance of this mutant to several antibiotics (see K1787 in Table 3), consistent with its playing a positive role in expression of the efflux genes. In contrast, elimination of this gene from the susceptible parent strain of K1668 (i.e., K1449) had no impact on resistance (see K1786 in Table 3), since this strain fails to express smeABC in the first place (see above). Interestingly, deletion of smeR, while reducing MICs, failed to compromise resistance to the same extent that deletion of smeC did (compare K1787 with K1781 and K1785 in Table 3). To assess the influence of SmeR on smeABC expression directly, the promoter region upstream of smeA was cloned into the lacZ fusion vector pMP190 and the impact of the cloned smeR gene on β-galactosidase activity was measured. As expected, the cloned smeR gene was shown to produce a protein of ca. 25 kDa (Fig. 3, lane 4). As seen in Table 5, smeR promoted a ca.-25-fold increase in expression of the smeA-lacZ fusion relative to a vector control lacking the smeR gene. Similarly, the cloned smeR gene also activated expression of a smeS-lacZ fusion and did so to a similar extent (Table 5), indicating that SmeR positively regulates both smeABC and its own smeSR operon. Nonetheless, the smeABC hyperexpression seen in MDR strain K1668 did not result from a mutation within smeRS or the smeS-smeA intergenic region (data now shown).
TABLE 5.
Influence of SmeR on smeA and smeSa
| Promoterb | SmeRc | β-Galactosidase activity (Miller unit)d |
|---|---|---|
| — | − | 15 ± 5 |
| — | + | 19 ± 4 |
| smeA | − | 15 ± 2 |
| smeA | + | 374 ± 34 |
| smeS | − | 16 ± 2 |
| smeS | + | 396 ± 8 |
Assessed in S. maltophilia K1449 harboring appropriate plasmids (below).
The promoters of the smeA and smeS genes were carried on plasmids pLZ497 (smeA) and pLZ499 (smeS), respectively. —, promoterless lacZ transcription fusion vector pMP190.
The smeR gene was carried on plasmid pLZ518. Symbols: −, pRK415 (vector); +, pLZ518.
The data shown are the averages ± standard deviations of two independent assays in which triplicate samples were determined.
Elevated β-lactamase activity in a SmeABC-hyperexpressing MDR mutant.
The SmeABC-hyperexpressing strain K1668 exhibits increased resistance to several β-lactam antibiotics despite the absence of both the L1 and L2 enzymes. While the implication is that SmeC contributes directly to β-lactam resistance, presumably as part of a multidrug efflux system capable of accommodating β-lactams, it was also possible that this resistance was attributable to a previously unidentified β-lactamase. To assess this, then, the β-lactamase activities of the MDR strain K1668 and its ΔL1 L2 parent strain K1449 were measured. Unexpectedly, the MDR strain demonstrated a marked increase in β-lactamase activity relative to K1449, though still well below that seen for the L1-L2-producing strain (Table 6 and reference 53). Moreover, elimination of smeBC (K1781) or smeC (K1785) reduced the β-lactamase activity to levels seen for the K1449 parent strain, while a smeB deletion had no effect (Table 6). Deletion of smeR from K1668 (K1787), on the other hand, had an intermediate effect on β-lactamase activity, reducing it to a level below that of the MDR strain but above that seen for its parent (Table 6). Indeed, the relative β-lactamase activities exactly paralleled the MICs seen for β-lactams, indicating that the β-lactam resistance of K1668 was likely explained not by any efflux activity provided by a pump that includes SmeC but by this β-lactamase.
TABLE 6.
β-Lactamase activity of smeABC and smeR mutants of S. maltophilia
| Strain | Relevent phenotype | β-Lactamase activitya
|
|
|---|---|---|---|
| Hydrolysis rate (nmol/mg/min) | Relative rate (%) | ||
| ULA-511 | Wild type (L1+ L2+) | 47,000 | 260,000 |
| K1449 | ΔL1 ΔL2 parent | 18 | 100 |
| K1780 | ΔsmeBC | 21 | 110 |
| K1782 | ΔsmeB | 22 | 120 |
| K1784 | ΔsmeC | 23 | 130 |
| K1786 | ΔsmeR | 29 | 160 |
| K1668 | ΔL1 ΔL2 MDR | 1,400 | 7,700 |
| K1781 | ΔsmeBC | 16 | 90 |
| K1783 | ΔsmeB | 1,300 | 7,200 |
| K1785 | ΔsmeC | 16 | 87 |
| K1787 | ΔsmeR | 260 | 1,500 |
The enzyme activity was determined in 50 mM sodium phosphate buffer (pH 7.2). Hydrolysis rate is presented as nanomoles of nitrocefin hydrolyzed per milligram of protein per minute, and the values are averages of three to four assays. The relative rate of hydrolysis is expressed using the hydrolysis rate from the enzyme of L1- and L2-deficient parent strain K1449 as 100%.
While this might represent a novel activity, it was possible, too, that it was attributable to the L2 β-lactamase, perhaps as a result of suppression of the 4-bp insertion used to produce the L2 knockout in K1449 (53). PCR amplification and sequencing of the L2 gene confirmed, however, that the mutation was conserved in all strains being used. The enzyme was subsequently purified although attempts at obtaining an N-terminal amino acid sequence failed. Finally, we were unable to obtain evidence (using RT-PCR) that expression of a truncated version of the 303-amino-acid L2 enzyme was being induced in K1668 (the 4-bp insertion occurs within the middle of the gene, such that a protein of 187 amino acids could be produced), although a disruption of this gene by insertion of pEX18Tc sequences (yielding strain K1975) did eliminate the β-lactamase activity and, thus, β-lactam resistance of K1668 (data not shown). Resistance to other agents was not affected. Thus, coexpression (or activation) of a truncated L2 enzyme with SmeABC was apparently responsible for the β-lactam resistance of MDR strain K1668.
DISCUSSION
Despite the hyperexpression of SmeABC in the MDR strain K1668, it is clear that resistance is dependent only upon the SmeC OMF component of this multidrug efflux system homologue. So while it is likely that SmeABC functions in efflux, the identity of the substrates is unknown and does not include antimicrobials. Nonetheless, the overproduction of SmeC does provide for multidrug resistance in S. maltophilia, suggesting that it may contribute to efflux-mediated multidrug resistance as the OMF of another, as-yet-unidentified efflux system. The fact, however, that the cloned gene (as part of a complete smeABC operon) was unable to promote resistance to antimicrobials in a wild-type strain of S. maltophilia suggests that such a system is not expressed in wild-type cells. Upregulation of this system, together with SmeC, is likely necessary for resistance and occurs, in fact, in the SmeABC-hyperexpressing multidrug-resistant strain K1668.
The observation that the smeC gene cloned in the opposite orientation relative to the resident lac promoter of the pRK415 cloning vector was able to complement, to some extent, the antibiotic susceptibility of an OprM-deficient mutant of P. aeruginosa is consistent with the smeC gene possessing its own, albeit weak, promoter. Similarly, the oprM gene of the mexAB-oprM multidrug efflux operon of P. aeruginosa was also shown to possess a weak promoter, which permitted its expression independent of mexAB (54). This was likely necessary since this protein functions as the OMF of several multidrug efflux systems in this organism (1, 30, 52), and thus, its expression would likely be needed even under circumstances when MexAB-OprM expression is not. The fact that smeC but not smeAB contributes to antimicrobial resistance and can be expressed independently of these genes suggests that SmeC also functions as part of an additional, as-yet-unidentified efflux system(s) which does accommodate antimicrobials. Similarly, a three-component RND-type efflux system, AmeABC, was recently described in Agrobacterium tumefaciens, where loss of the inner-membrane-associated components (AmeAB, homologues of SmeAB) did not impact antimicrobial susceptibility while loss of the OMF component, AmeC, did (37a). The putative AmeABC pump was implicated in export of intermediates of purine biosynthesis, while AmeC was proposed to also function in drug efflux as part of an as-yet-unidentified efflux system (37a). That smeC expression is driven by a promoter upstream of smeA as well as its own promoter is also supported by the demonstration that inactivation of smeR, which encodes a positive regulator of smeABC expression, has only an intermediate effect on resistance levels. While loss of smeR will negatively impact the expression of smeC from the smeA upstream promoter (which is the target for SmeR [see Table 5]) smeC expression is still possible from its own promoter in these mutants. Given the correlation between SmeC production and antimicrobial resistance in K1668, then, smeR knockouts are expected to provide at least some resistance while smeC knockouts are not, and this is, in fact, what is seen.
The discovery that the β-lactam resistance of a SmeABC (i.e., SmeC)-overexpressing strain is due to increased β-lactamase activity and not efflux is interesting. While it might reflect coregulation of the smeC and β-lactamase genes, it appears more likely that the status of SmeC impacts, e.g., cell physiology (perhaps via accumulation or release of certain substrates) in such a way as to induce (or activate) the enzyme. This is supported by the observation that deletion of smeC itself produces a decline in β-lactamase activity which is more striking than the loss provided by the smeR knockout (i.e., it is likely that the ΔsmeR effect on β-lactamase activity results from its impact on SmeC levels and not loss of, e.g., SmeR-dependent activation of the β-lactamase gene). Certainly the presence or absence of SmeC has a major impact on the cell, inasmuch as the SmeABC-hyperexpressing strain grows markedly more slowly than its parent or smeC deletion derivatives, forms smaller cells, and is stained much less intensely with Gram stain (data not shown). Nonetheless, given the homology between SmeR and other regulators of β-lactamase gene expression (e.g., BlrAB [2] and CreB [6]) (Table 2), the possibility of SmeR regulating β-lactamase gene expression is intriguing. A connection between expression of multidrug efflux pumps and β-lactamase was made previously in P. aeruginosa, where it was noted that increased expression of the MexCD-OprJ multidrug efflux system in an nfxB mutant was accompanied by a decrease in expression of the organism’s AmpC β-lactamase (26). Thus, efflux systems can both positively and negatively influence β-lactamase production.
The nature of the connection between the L2 β-lactamase and SmeABC is unknown at this point. While it would be worthwhile to study this in a wild-type background, SmeABC-expressing derivatives of, e.g., ULA-511 are not currently available, and the nature of the mutation(s) leading to SmeABC expression in MDR strain K1668 is unknown (precluding construction of a SmeABC-expressing derivative of ULA-511). Still, L2 activity is readily measurable in wild-type S. maltophilia (which does not express SmeABC), indicating that expression of these two resistance determinants is not intimately linked, though SmeABC (or SmeC) expression may necessitate production of some L2 enzyme.
Acknowledgments
This work was supported by funding from the Canadian Bacterial Diseases Network (one of the Networks of Centers of Excellence). K.P. is a Canadian Cystic Fibrosis Foundation Scholar.
REFERENCES
- 1.Aires, J. R., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alksne, L. E., and B. A. Rasmussen. 1997. Expression of the AsbA1, OXA-12, and AsbM1 β-lactamases in Aeromonas jandaei AER 14 is coordinated by a two-component regulon. J. Bacteriol. 179:2006–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, A., and J. L. Martinez. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso, A., and J. L. Martinez. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44:3079–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso, A., and J. L. Martinez. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:1879–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avison, M. B., P. Niumsup, T. R. Walsh, and P. M. Bennett. 2000. Aeromonas hydrophila AmpH and CepH β-lactamases: derepressed expression in mutants of Escherichia coli lacking creB. J. Antimicrob. Chemother. 46:695–702. [DOI] [PubMed] [Google Scholar]
- 7.Baltch, A. L., R. P. Smith, and W. Ritz. 1994. Comparative antimicrobial activity of FK037, cefpirome, ceftazidime and cefepime against aminoglycoside-sensitive and aminoglycoside-resistant Pseudomonas aeruginosa and Pseudomonas spp. Chemotherapy 40:391–398. [DOI] [PubMed] [Google Scholar]
- 8.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J.-F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321–337. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright, S. J., and S. G. Waley. 1984. Purification of β-lactamases by affinity chromatography on phenylboronic acid-agarose. Biochem. J. 221:505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denton, M., N. J. Todd, K. G. Kerr, P. M. Hawkey, and J. M. Littlewood. 1998. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J. Clin. Microbiol. 36:1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denton, M., and K. G. Kerr. 1988. Microbiological and clinical aspects of infections associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Wulf, P., B. J. Akerley, and E. C. Lin. 2000. Presence of the Cpx system in bacteria. Microbiology 146:247–248. [DOI] [PubMed] [Google Scholar]
- 13.Fass, R. J., J. Barnishan, M. C. Solomon, and L. W. Ayers. 1996. In vitro activities of quinolones, β-lactams, tobramycin, and trimethoprim-sulfamethoxazole against nonfermentative gram-negative bacilli. Antimicrob. Agents Chemother. 40:1412–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felegie, T. P, V. L. Yu, W. Rumans, and R. B. Yee. 1979. Susceptibility of Pseudomonas maltophilia to antimicrobial agents, singly and in combination. Antimicrob. Agents Chemother. 16:833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felici, A., G. Amicosante, A. Oratore, R. Strom, P. Ledent, B. Joris, L. Fanuel, and J.-M. Frere. 1993. An overview of the kinetic parameters of class B β-lactamases. Biochem. J. 291:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
- 17.Kieboom, J., J. J. Dennis, J. A. M. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85–91. [DOI] [PubMed] [Google Scholar]
- 18.King, B. A., K. P. Shannon, and I. Phillips. 1978. Aminoglycoside 6′-N acetyltransferase production by an isolate of Pseudomonas maltophilia. J. Antimicrob. Chemother. 4:467–468. [DOI] [PubMed] [Google Scholar]
- 19.Köhler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J.-C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345–354. [DOI] [PubMed] [Google Scholar]
- 20.Lambert, T., M.-C. Ploy, F. O. Denis, and P. Courvalin. 1999. Characterization of the chromosomal aac(6′)-lz gene of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 43:2366–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesco Bornet, M., J. Pierre, D. Sarkis Karam, S. Lubera, and E. Bergogne. 1992. Susceptibility of Xanthamonas maltophilia to six quinolones and study of outer membrane proteins in resistant mutants selected in vitro. Antimicrob. Agents Chemother. 36:669–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X.-Z., and K. Poole. 2001. Mutational analysis of the OprM outer membrane component of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 183:12–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. 1998. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda, N, E. Sakagawa, S. Ohya, N. Gotoh, and T. Nishino. 2001. Hypersusceptibility of the Pseudomonas aeruginosa nfxB mutant to β-lactams due to reduced expression of the AmpC β-lactamase. Antimicrob. Agents Chemother. 45:1284–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mett, H., S. Rosta, B. Schacher, and R. Frei. 1988. Outer membrane permeability and β-lactamase content in Pseudomonas maltophilia clinical isolates and laboratory mutants. Rev. Infect. Dis. 10:765–769. [DOI] [PubMed] [Google Scholar]
- 28.Micozzi, A., M. Venditti, M. Monaco, A. Friedrich, F. Taglietti, S. Santilli, and P. Martino. 2000. Bacteremia due to Stenotrophomonas maltophilia in patients with hematologic malignancies. Clin. Infect. Dis. 31:705–711. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. F. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Mine, T, Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muder, R. R., A. P. Harris, and S. Muller. 1996. Bacteremia due to Stenotrophomonas (Xanthomonas) maltophilia: a prospective, multicenter study of 91 episodes. Clin. Infect. Dis. 22:508–512. [DOI] [PubMed] [Google Scholar]
- 32.Nagasawa, S., K. Ishige, and T. Mizuno. 1993. Novel members of the two-component signal transduction genes in Escherichia coli. J. Biochem. (Tokyo) 114:350–357. [DOI] [PubMed] [Google Scholar]
- 32a.Neidhardt, F. C. 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 33.Neu, H. C., and J. Chou. 1967. Release of surface enzymes in Enterobacteriaceae by osmotic shock. J. Bacteriol. 94:1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516–523. [DOI] [PubMed] [Google Scholar]
- 35.Nikaido, H. 2000. How do exported proteins and antibiotics bypass the periplasm in gram-negative bacterial cells? Trends Microbiol. 8:481–483. [DOI] [PubMed] [Google Scholar]
- 36.Palleroni, N. J., and J. F. Bradbury. 1993. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int. J. Syst. Bacteriol. 43:606–609. [DOI] [PubMed] [Google Scholar]
- 37.Payne, D. J., R. Cramp, J. H. Batson, J. Neal, and D. Knowles. 1994. Rapid identification of metallo- and serine β-lactamses. Antimicrob. Agents Chemother. 38:991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Peng, W. T., and E. W. Nester. 2001. Characterization of a putative RND-type efflux system in Agrobacterium tumefaciens. Gene 270:245–252. [DOI] [PubMed] [Google Scholar]
- 38.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J.-I. Yamagishi, X.-Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713–724. [DOI] [PubMed] [Google Scholar]
- 40.Poole, K., D. E. Heinrishs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529–544. [DOI] [PubMed] [Google Scholar]
- 41.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinn, J. P. 1998. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin. Infect. Dis. 27:S117–S124. [DOI] [PubMed] [Google Scholar]
- 43.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-TIE. J. Bacteriol. 180:3323–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Sanschagrin, F., J. Dufresne, and R. C. Levesque. 1998. Molecular heterogeneity of the l-1 metallo-β-lactamase family from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 42:1245–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srikumar, R., X.-Z. Li, and K. Poole. 1997. The inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanhoof, R., P. Sonck, and E. Hannecart-Pokorni. 1995. The role of lipopolysaccharide anionic binding sites in aminoglycoside uptake in Stenotrophomonas (Xanthomonas) maltophilia. J. Antimicrob. Chemother. 35:167–171. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, T. R., A. P. MacGowan, and P. M. Bennett. 1997. Sequence analysis and enzyme kinetics of the L2 serine β-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1460–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., X.-Z. Li, and K. Poole. 2000. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob. Agents Chemother. 44:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Zhang, L., X.-Z. Li, and K. Poole. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, Q., X.-Z. Li, R. Srikumar, and K. Poole. 1998. Contribution of outer membrane efflux protein OprM on antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob. Agents Chemother. 42:1682–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]