Abstract
We have previously reported that 2,5,6-trichloro-1-(β-d-ribofuranosyl)benzimidazole (TCRB) and its 2-bromo analog (2-bromo-5,6-dichloro-1-(β-d-ribofuranosy)benzimidazole [BDCRB]) are potent and selective inhibitors of human cytomegalovirus (HCMV) replication that block viral DNA maturation via HCMV gene products UL89 and UL56. To determine if phosphorylation is required for antiviral activity, the in vitro metabolism of BDCRB was examined and the antiviral activities of nonphosphorylatable 5′-deoxy analogs were determined. Reverse-phase high-performance liquid chromatography (HPLC) analysis of extracts from uninfected and HCMV-infected cells incubated with [3H]BDCRB revealed two major metabolites. Both were less polar than naturally occurring nucleoside monophosphates, but one peak coeluted with a BDCRB-5′-monophosphate (BDCRB-5′-MP) standard. Further analysis revealed, however, that neither metabolite partitioned with BDCRB-5′-MP on anion-exchange HPLC. Their retention patterns were not affected by incubation with alkaline phosphatase, thereby establishing that the compounds were not nucleoside 5′-monophosphates. Both compounds were detected in uninfected and HCMV-infected cells and in mouse live extracts, but neither has been identified. Like TCRB and BDCRB, the nonphosphorylatable 5′-deoxy analogs were potent and selective inhibitors of HCMV replication. The 5′-deoxy analogs maintained inhibition of HCMV replication upon removal of BDCRB, whereas an inhibitor of DNA synthesis did not. Similar to TCRB, its 5′-deoxy analog (5′-dTCRB) did not affect viral DNA synthesis, but 5′-dTCRB did inhibit viral DNA maturation to genome-length units. Additionally, virus isolates resistant to TCRB were also resistant to 5′-dTCRB and the 5′-deoxy analog of BDCRB. Taken together, these results confirm that TCRB, BDCRB, and their 5′-deoxy analogs have common mechanisms of action and establish that these benzimidazole ribonucleosides, unlike other antiviral nucleosides, do not require phosphorylation at the 5′ position for antiviral activity.
HCMV is one of eight identified human herpesviruses. In an immunocompetent host, HCMV infection is typically asymptomatic; however, in an immunocompromised patient, infection can result in significant morbidity and mortality (2). HCMV is a common opportunistic pathogen among immunocompromised individuals and is often a contributory cause of death in AIDS patients (28). HCMV infection has also been implicated in an increased risk of organ rejection following heart (18) and kidney (3) transplants and in the restenosis of diseased coronary arteries following angioplasty (27).
Drugs currently approved for the treatment of HCMV fall into two categories: an antisense oligonucleotide and small-molecule inhibitors. Fomivirsen is a 21-base phosphorothioate-linked oligonucleotide complementary to HCMV immediate-early 2 mRNA (33). This drug inhibits HCMV replication in a sequence-dependent manner; however, resistance to fomivirsen does not map to the complementary locus, suggesting a nonantisense mechanism of action (30). All three small-molecule drugs approved for the treatment of HCMV infections inhibit the viral DNA polymerase. GCV is a nucleoside analog and must be phosphorylated on the equivalent of the 5′ position by an HCMV-encoded protein kinase, UL97 (21, 40, 41). Two additional phosphate residues are added by cellular enzymes, and the resulting triphosphate of GCV inhibits the HCMV-encoded DNA polymerase (26). Likewise, cidofovir, a nucleotide analog, is phosphorylated to its diphosphate form, which is similar to a triphosphate and which is the active DNA polymerase inhibitor (22). Finally, the pyrophosphate analog foscarnet is known to inhibit a wide variety of DNA polymerases including that of HCMV (14).
The need for new drugs for the treatment of HCMV infections is apparent due to the toxicities of the drugs mentioned above, their low bioavailabilities, and the development of resistance to the currently available drugs (7, 8, 22, 34). Resistance to one class of drugs can be overcome and toxicity can be ameliorated by the development of new drugs with novel mechanisms of action. In 1995, we reported on the antiviral activities of benzimidazole nucleosides, a novel class of inhibitors of cytomegalovirus replication (Fig. 1) (43). The lead compound, TCRB, is a potent and selective inhibitor of HCMV replication; its 2-bromo analog (BDCRB) is even more potent. These compounds do not inhibit DNA synthesis but instead act late in the viral replication cycle by disrupting the cleavage of newly formed, polygenomic DNA concatemers into monomeric, genomic-length pieces (46).
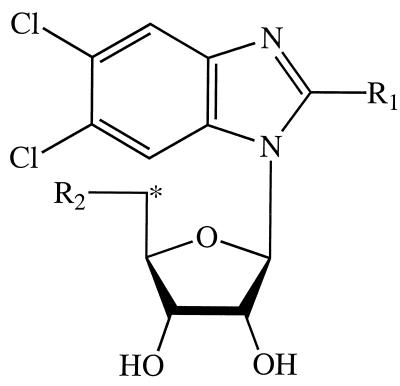
FIG. 1.
Structures of benzimidazole ribonucleosides. The asterisk indicates the position of the tritium label in [3H]BDCRB. The moieties at positions R1 and R2 were as follows: in DRB, H and OH, respectively; in TCRB, Cl and OH, respectively; in BDCRB, Br and OH, respectively; in 5′-TCRB, Cl and H, respectively; and in 5′-BDCRB, Br and H, respectively.
HCMV that is resistant to TCRB and BDCRB has been isolated, and the mutations responsible for resistance have been mapped to HCMV genes UL89 and UL56 (25, 46). Consequently, we have proposed that the benzimidazole nucleoside analogs inhibit the products of these genes. Consistent with this proposal, UL89 is homologous to the bacteriophage T4 gene for protein gp17 (9), which is a terminase responsible for the processing of concatemeric T4 DNA during particle assembly (38). UL56 is homologous to HSV UL28, which is required for efficient DNA cleavage and packaging (1, 42). HCMV UL56 may have intrinsic nuclease capabilities, supporting a role in DNA cleavage (6). It is not known how TCRB interacts with either of these proteins.
Inhibition of HCMV DNA processing is a novel and poorly understood mechanism of action. Since most antiviral nucleoside analogs must be activated by kinases to the corresponding 5′-phosphates, the studies reported herein were undertaken to determine if TCRB is phosphorylated and if phosphorylation is required for activity against HCMV. Two approaches have been used: (i) a search for phosphorylated metabolites of [3H]BDCRB in cell lysates and (ii) a comparison of the activities and mechanisms of action of the 5′-deoxy analogs of TCRB and BDCRB, which cannot be phosphorylated, to those of TCRB.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used throughout the report: DRB, 5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole; TCRB, 2,5,6-trichloro-1-(β-d-ribofuranosyl)benzimidazole; BDCRB, 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole, the 2-bromo analog of TCRB; 5′-dTCRB and 5′-dBDCRB, the 5′-deoxy analogs of TCRB and BDCRB, respectively (Fig. 1); BDCRB-5′-MP, the 5′-monophosphate of BDCRB; [3H]BDCRB, 2-bromo-5,6-dichloro-1-(β-d-[5′-3H]ribofuranosyl)benzimidazole; GCV, ganciclovir; FBS, fetal bovine serum; HCMV, human cytomegalovirus; HFFs, human foreskin fibroblasts; HPLC, high-performance liquid chromatography; HSV, herpes simplex virus; MEM(E), minimal essential medium with Earle’s salts; MOI, multiplicity of infection; ORF, open reading frame; 20× SSC, 3 M sodium chloride plus 0.3 M sodium citrate (pH 7.0).
Compounds.
TCRB and BDCRB were synthesized and characterized as described by Townsend et al. (43). 5′-dTCRB and 5′-dBDCRB were synthesized and characterized by similar methods (44). [3H]BDCRB, BDCRB-5′-MP, and BDCRB-5′-diphosphate were synthesized by and provided through the courtesy of Burroughs Wellcome Co. The label in [3H]BDCRB was located on the 5′ position of the ribose moiety (Fig. 1). The compound had a specific activity of 1.1 Ci/mmol and a radiochemical purity of >98.9% as determined by HPLC.
Cell culture procedures.
The routine growth of HFF and MRC-5 cells was performed in MEM(E) with 10% FBS. Cells were routinely passaged at dilutions of 1:2 by conventional procedures by use of 0.05% trypsin with 0.02% EDTA in HEPES-buffered saline (39). CEM cells were grown in RPMI with 10% FBS. All cell lines were screened periodically for mycoplasma contamination and were found to be negative.
Virological procedures.
The Towne strain, plaque-purified isolate Po, of HCMV was kindly provided by Mark Stinski, University of Iowa. The AD169 strain of HCMV was obtained from the American Type Culture Collection and was plaque purified in K. K. Biron’s laboratory. HCMV strains B11, D10, and C4 were selected for resistance to TCRB as previously described by us (25). Stocks of HCMV were prepared by infecting HFF cells at 0.01 PFU per cell by a previously described procedure (45). Virus titers were determined by using monolayer cultures of HFF cells, also as described previously (36). Viral assays, but not the preparation of viral stocks, were done in the presence of 100 U of penicillin G per ml and 100 μg of streptomycin per ml.
Metabolism of [3H]BDCRB in uninfected CEM cells.
CEM cells (5 × 105) were diluted into RPMI with 10% FBS and 10 μCi of (9.1 μM) [3H]BDCRB per ml. The cells were incubated for 48 h at 37°C and were harvested by centrifugation (250 × g for 5 min at room temperature). The resulting pellets were resuspended in 10 ml of cold Saline G described by Puck et al. (37) and repelleted. This pellet was resuspended in 0.6 N trichloroacetic acid at 4°C and kept on ice for 15 min. The suspensions were clarified by centrifugation at 12,000 × g for 30 s at 4°C. The supernatant was removed and extracted with an equal volume of ice-cold Freon containing 0.5 M trioctylamine (35). The mixture was centrifuged at 12,000 × g for 30 s to separate the layers, and the lower layer was removed by aspiration and discarded. The aqueous layer was frozen at −76°C for subsequent HPLC analysis.
Incorporation of [3H]BDCRB into cellular RNA and DNA was examined by incubation of CEM cells with 1 μM (1.1 μCi/ml) [3H]BDCRB for 48 h. Parallel incubations were performed under identical conditions with 1 μCi of [3H]Urd or [3H]dThd per ml as the substrate. The cells were lysed, and DNA was separated from RNA by isopycnic centrifugation in CsSO4 gradients as we have described previously (32). Following centrifugation, the gradients were separated into approximately 50 fractions, and acid-insoluble material was determined by liquid scintillation spectrometry as described previously (32).
Metabolism of [3H]BDCRB in HCMV-infected and uninfected HFF cells.
HFF cells (≈2 × 106 cells) were planted in 25-cm2 flasks and grown overnight. They were either mock infected or infected with HCMV at 2 × 105 PFU per flask. After 1 h, the inoculum was removed and replaced with fresh medium containing 10 μCi of (9.1 μM) [3H]BDCRB per ml (see the legend to Fig. 4). The comparative data mentioned in Results were obtained in two separate experiments in which HCMV-infected and uninfected HFF cells were incubated with [3H]BDCRB for the following time intervals: 0 to 48, 0 to 72, 0 to 120, 48 to 72, or 48 to 120 h. At 120 h the cells were harvested by trypsinization, pelleted, and extracted as described above. Extracts were stored at −76°C for subsequent analysis by HPLC.
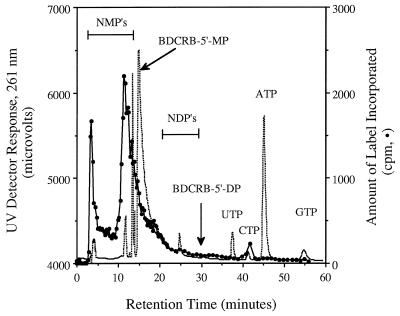
FIG. 4.
Anion-exchange HPLC analysis of an extract from HCMV-infected HFF cells incubated with [3H]BDCRB. Following infection at an MOI of 0.15, the cells were incubated with 10 μCi of (9.1 μM) [3H]BDCRB per ml for 5 days. Nucleosides and nucleotides were extracted and separated by anion-exchange HPLC on an Ultrasil-AX 10-μm column (4.6 by 250 mm), as detailed in the text. The dotted line is the response from the UV detector; the line defined by solid circles is the counts per minute from labeled metabolites. Horizontal bars indicate the ranges in which naturally occurring nucleoside monophosphates (NMP’s) and nucleoside diphosphates (NDP’s) eluted. The retention time of BDCRB-5′-MP was determined by the addition of a nonradioactive standard to the infected cell extract. That of BDCRB-5′-diphosphate (BDCRB-5′-DP) was determined in a separate run.
Alkaline phosphatase digestion.
In a separate series of experiments, the metabolism of BDCRB was studied with a mouse liver extract. The extract was prepared by Dounce homogenization of mouse liver in physiological saline plus dithiothreitol, followed by centrifugation at 9,000 × g for 30 min. The supernatant was separated, incubated with 10 μCi of [3H]BDCRB per ml for 1 h, and then extracted as described above. Two types of analyses were performed. (i) An aliquot of an extract was evaporated under a flow of nitrogen to 35% of its original volume. This was mixed with 5 volumes of buffer (50 mM glycine [pH 9.1]) containing 25 μM each 5′-AMP and 3′-AMP, and the mixture was digested with bovine intestinal mucosal alkaline phosphatase (1 U; Sigma) for 1 h at 37°C. The reaction was stopped by the addition of ice-cold methanol to a final concentration of 60%, and the products were analyzed by reverse-phase HPLC as described below. (ii) An aliquot of the liver extract was chromatographed by reverse-phase HPLC, and fractions containing the metabolite with a peak at 21 min (see below and Fig. 2) were pooled and lyophilized. The pellet was resuspended in buffer plus 5′-AMP and 3′-AMP and treated with alkaline phosphatase as described for the first type of analysis. A sample of synthetic BDCRB-5′-MP was prepared and treated with alkaline phosphatase in a similar manner.
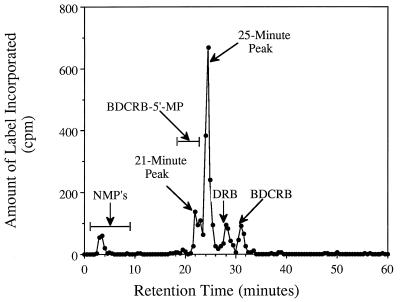
FIG. 2.
Reverse-phase HPLC of extracts from uninfected CEM cells incubated with [3H]BDCRB. The cells were incubated with 10 μCi of (9.1 μM) [3H]BDCRB per ml for 48 h, after which nucleosides and nucleotides were isolated by trichloroacetic acid precipitation and Freon extraction. Reverse-phase separations were performed on a μBondapak C18 column (3.9 by 300 mm), as detailed in the text. The retention times of naturally occurring nucleoside monophosphates (NMP’s) and BDCRB-5′-MP were determined by addition of standards to the extract and are indicated by horizontal bars.
HPLC analysis.
HPLC was performed on a Spectra-Physics chromatography system consisting of a model SP8800 ternary pump, an SP8500 dynamic mixer, an SP8780 autosampler, and an SP8490 variable-length detector. Peaks were integrated on a model SP4270 integrator. System and data management was achieved with a Compaq model 386 computer with WINner 386 software (Spectra-Physics). When radioactive compounds were separated, fractions were collected and analyzed with a Beckman LS 8100 series liquid scintillation system.
Anion-exchange separations were performed on an Ultrasil-AX 10-μm column (4.6 by 250 mm; Beckman) with a guard column (3.2 by 4.5 mm; Beckman) of the same material. The flow rate was 1 ml/min. Following equilibration with 0.002 M KH2PO4 (pH 4.65), cell extracts were injected and a linear gradient to 0.5 M KH2PO4 (pH 4.65) was applied over 40 min. After a 20-min isocratic period with the high-salt buffer, the column was reequilibrated for 20 min with 0.002 M KH2PO4 (pH 4.65) prior to the next injection. The UV absorbance at 261 nm of the effluent was monitored; fractions were collected for quantitation of the radiolabel by liquid scintillation spectrometry.
Reverse-phase separations were performed on a μBondapak C18 column (3.9 by 300 mm; Waters) at a flow rate of 1 ml/min. The column was equilibrated in a buffer of 50 mM ammonium acetate-0.1% triethylamine (pH 5.5), and cell extracts were injected. A linear gradient to 80% acetonitrile in water was applied over 60 min, and this concentration was maintained for 5 min. The UV absorbance and radioactivity of the effluent were monitored as described above.
Antiviral assays with HCMV.
For plaque reduction assays, HFF cells at 85,000 cells per well in 24-well cluster plates were infected with HCMV at 100 PFU/well in MEM(E) with 5% FBS, 100 U of penicillin G per ml, and 100 μg of streptomycin per ml. The inoculum was removed after 1 h and was replaced with medium containing selected drug dilutions and 0.5% methylcellulose. All drug concentrations were tested in duplicate by using four to seven one-half log10 dilutions. The plates were incubated at 37°C for 10 days and stained with crystal violet. The plaques were counted under 40-fold magnification. Drug effects were determined by calculation of the number of plaques in the presence of drug as a percentage of the number of plaques in the absence of drug.
For yield reduction assays, we used a procedure that we devised previously (36). Briefly, HFF cells were planted in 24- or 96-well cluster dishes, incubated overnight, and infected with HCMV at an MOI of 0.5. After virus adsorption, the inoculum was replaced with fresh medium containing test compounds in duplicate or triplicate in a manner which provided drug concentrations from 100 to 0.14 μM by using one-half log10 dilutions. Plates were incubated at 37°C for 7 days and subjected to one cycle of freezing and thawing. Aliquots from each of the wells were transferred to a fresh 96-well monolayer culture of HFF cells and were serially diluted across the plate. Cultures were incubated, cells were stained, plaques were enumerated at 40-fold magnification, and titers were calculated as described above.
For both plaque and yield assays, dose-response relationships were constructed by plotting the percent inhibition of plaque number or the log10 of the percent inhibition of virus titer against log10 drug concentrations. Fifty or 90% inhibitory concentrations were interpolated from the resulting dose-response linear regression lines. Samples containing GCV as a positive control were used in all assays.
DNA synthesis analysis.
Detection of intracellular viral DNA was performed as described in the package insert for the cytomegalovirus antiviral susceptibility test kit (Diagnostic Hybrids). Briefly, HFF cells in 24-well cell culture dishes were infected with HCMV at 3.0 PFU per cell. After 2 h, the inoculum was removed and replaced with medium containing selected drug concentrations. The plates were incubated for 4 days, and supernatants were removed for determination of the virus titer by limiting dilution as described previously (36). The cells were lysed by the addition of the DNA wicking agent included with the kit. Lysates were absorbed to Hybridwix and probed. Wicks were counted in a Beckman LS 8100 series liquid scintillation system in Ecolume (ICN) scintillation cocktail. All points are the averages for duplicate samples.
Contour-clamped homogeneous electric field separation.
MRC-5 cells were infected with HCMV at 3.0 PFU/cell in MEM(E) with 5% FBS, 100 U of penicillin G per ml, and 100 μg of streptomycin per ml. The inoculum was removed after 2 h and was replaced with fresh medium containing selected drug concentrations tested in duplicate. The plates were incubated for 3 days at 37°C, and then the cells were trypsinized, pelleted, and washed once with phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.4]). The pelleted cells were suspended in 0.8% low-melting-point agarose (Sigma) in phosphate-buffered saline and cast into plug molds. The plugs were digested in 0.45 M EDTA (pH 9.0)-1% sarkosine-0.5 mg of proteinase K per ml at 45°C for 48 h. Aliquots were loaded into 1% agarose gels and electrophoresed as described previously (46). Upon completion, the DNA was stained with ethidium bromide, nicked with 1,500 mJ in a Stratagene Stratalinker, and denatured for 30 min in 0.5 M NaOH-1.5 M NaCl. DNA was transferred to a Nytran membrane (Schleicher & Schuell) using a positive pressure blot transfer apparatus (Stratagene Posiblot) for 16 h. The membranes were neutralized for 10 min in 0.5 M Tris (pH 7.4) with 3 M NaCl and washed for 10 min in 2× SSC. They were incubated overnight at 42°C in hybridization solution (3× SSC, 50% formamide, 5× Denhardt’s solution, and 20 mM Na3PO4 [pH 6.5]) with 1% glycine. A 1:1 mixture of two cosmids (pC7S31 [4] and pC7S37 [24]), each of which encode ≈0.15 map unit from within the unique long arm of the HCMV genome was labeled by nick translation (0.2 μg) and diluted with 100 μg of salmon sperm DNA per ml. An equal volume of formamide was added, and the DNA was denatured at 65°C for 15 min. This was diluted into hybridization solution with 5% dextran sulfate. The membranes were probed overnight at 42°C and briefly rinsed with 2× SSC. Stringency washes were as follows: twice with 0.1× SSC-0.1% sodium dodecyl sulfate at 42°C for 30 min and twice with 0.1× SSC-0.1% sodium dodecyl sulfate at 56°C for 30 min. The blots were analyzed with a Molecular Dynamics PhosphorImager; the amount of monomeric, genomic-length DNA was calculated as a percentage of the total DNA in the lane.
Maturational assay.
The effect of the compounds on the cleavage and maturation of HCMV DNA was measured by an indirect, virological approach devised by one of us (K. K. Biron). The assay is based upon the observation that inhibition of HCMV DNA cleavage by BDCRB results in the accumulation of concatemeric DNA (46) which is reversible upon removal of the drug (P. M. Krosky, K. Z. Borysko, and J. C. Drach, unpublished data). MRC-5 cells were planted in 12-well cluster plates and were infected with the AD169 strain of HCMV at an MOI of ≈1 PFU/cell. BDCRB (20 μM) was added to all cultures, and the plates were incubated at 37°C for 96 h to permit accumulation of concatemeric DNA. The BDCRB-containing medium was then decanted, and the cell sheets were rinsed with phosphate-buffered saline. Drug-free medium or medium containing selected concentrations of the compounds listed in Table 2 was added, and the medium was changed after 1 h to remove any residual BDCRB. Incubation was continued for an additional 24 h, and the cultures were frozen at −80°C. At a later time, the samples were thawed, sonicated, and clarified by centrifugation; and the HCMV titer was determined by limiting dilution across 96-well cluster plates. The virus titer was calculated on the basis of the number of plaques observed per aliquot of supernatant multiplied by the dilution factor.
TABLE 2.
Mode-of-action comparisons among benzimidazole nucleosides
| Test compound | IC90a (μM) by:
|
IC50 (μM) for DNA synthesisd | |
|---|---|---|---|
| Maturation assayb | Yield assayc | ||
| BDCRB | 0.08 | 0.29 ± 0.15 | >25e |
| 5′-dBDCRB | <0.04e | 0.018 ± 0.003 | >5 |
| 5′-dTCRB | 0.03 | 0.36 ± 0.26 | >25 |
| 1263W94 | 27 | 0.55 ± 0.06 | 0.3 |
IC90, 90% inhibitory concentration.
The maturation assay was a single-cycle yield reduction experiment in which 20 μM BDCRB was removed from HCMV-infected cultures and replaced at 96 h postinfection by the test compound.
Data are from two or three single-cycle yield reduction experiments in which the test compound was present for the entire experiment.
The assay was a single-cycle yield experiment in which the amount of DNA was measured by a hybridization assay. IC50, 50% inhibitory concentration.
The 50 or 90% inhibitory concentration was not reached at the noted highest or lowest concentration tested.
RESULTS
Metabolism of BDCRB in uninfected CEM cells.
Most antiviral nucleoside analogs require phosphorylation for activation. Some analogs such as GCV and acyclovir are selectively phosphorylated by virally encoded kinases (15, 16, 41). To determine if the antiviral activities of the benzimidazole ribonucleosides involve a phosphorylated product, the metabolism of [3H]BDCRB (Fig. 1) was examined in uninfected and HCMV-infected cells.
In an initial investigation of the metabolic stability of BDCRB, uninfected CEM cells were incubated with 10 μCi of (9.1 μM) [3H]BDCRB per ml, and extracts from these cells were analyzed by reverse-phase HPLC. Two products that eluted after the naturally occurring nucleoside monophosphates were observed in the chromatogram (Fig. 2). The first peak (at 21 min) coeluted with a BDCRB-5′-MP standard, suggesting the identity of the labeled metabolite. A second major peak was retained longer, indicating that it was less polar. A very small radioactive peak was observed in the monophosphate region of the chromatogram (NMPs in Fig. 2), but this coeluted with ribose. Because [3H]BDCRB was labeled in the 5′ position of ribose and because ribose is a catabolite of BDCRB in vivo (17), we hypothesized that this peak was labeled ribose and we did not pursue further identification. Two small peaks corresponding to unchanged BDCRB and to another known catabolite, DRB, were also seen (Fig. 2).
Lack of incorporation of BDCRB into cellular RNA and DNA.
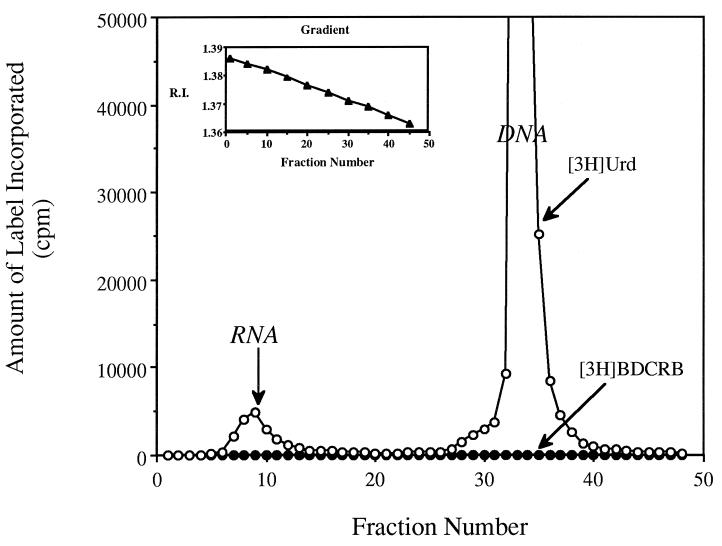
As a part of the study described above, the level of incorporation of [3H]BDCRB into the nucleic acids of CEM cells was determined. Incorporation could occur only if the tentatively identified BDCRB-5′-MP was converted to its triphosphate and this was a substrate for a DNA or RNA polymerase. Following incubation of CEM cells with 1 μM [3H]BDCRB for 48 h, cells were lysed and DNA was separated from RNA by isopycnic centrifugation in CsSO4 gradients as described previously (32). Data in Fig. 3 show that no incorporation of label above the background was detected in either RNA or DNA, but [3H]Urd was readily incorporated into RNA and DNA in a parallel experiment. The detection limit (calculated by using the specific activity of [3H]BDCRB, the amount of DNA per cell, and the number of cells) established that <1 molecule of BDCRB-MP was incorporated into DNA per 6 × 105 deoxynucleoside monophosphate molecules. In contrast, our previous studies showed that 615 molecules of arabinosyl-AMP are incorporated per 6 × 105 deoxynucleoside monophosphate molecules when 1 μM vidarabine is incubated with uninfected KB cells (32).
FIG. 3.
Cesium sulfate isopycnic gradient separation of cellular RNA and DNA. CEM cells were incubated with [3H]BDCRB (•) at 1 μM (1.1 μCi/ml) for 48 h, and the amount of labeled acid-precipitable material was determined. Parallel incubations were performed under identical conditions with 1 μCi of [3H]Urd per ml (○) as a marker for incorporation into RNA and DNA (following metabolism to dTTP). (Inset) Nature of gradient determined by refractive index (R.I.).
Metabolism of BDCRB in HCMV-infected cells.
More detailed studies of the metabolism of [3H]BDCRB were performed with uninfected and HCMV-infected HFF cells to determine if BDCRB metabolism was affected by viral infection and to further characterize the putative BDCRB-5′-MP. HCMV-infected HFF cells were incubated with 10 μCi of (9.1 μM) [3H]BDCRB per ml, and extracts were analyzed by reverse-phase HPLC (data not shown). A retention pattern virtually identical to that seen in uninfected CEM cells (Fig. 2) was observed, except that the quantities of the two major metabolites were greater.
In order to verify that the labeled metabolite with the peak at 21 min was BDCRB-5′-MP, an extract identical to that analyzed by reverse-phase HPLC was analyzed by anion-exchange HPLC, which separates compounds in the opposite order from that into which they are separated by reverse-phase HPLC. Under these conditions, both labeled metabolites eluted before the BDCRB-5′-MP standard but eluted near the naturally occurring nucleoside monophosphates (Fig. 4). On the basis of these data, neither metabolite could be BDCRB-5′-MP. As a further check of metabolite identity, fractions from this chromatogram that eluted at between 10 and 13 min were pooled, lyophilized, and rechromatographed on a reverse-phase column. Under these conditions the labeled peak and a BDCRB-5′-MP standard were retained for identical times, giving a pattern very similar to that shown in Fig. 2 (data not shown). Together these experiments established that the metabolite with the peak at 21 min from the reverse-phase HPLC was not BDCRB-5′-MP and that the process of anion-exchange chromatography had not changed this labeled metabolite, leading to a false conclusion about its identity. Furthermore, the long retention times of the metabolites on reverse-phase HPLC, especially that with the peak at 25 min, suggested that the BDCRB metabolites are not highly polar and therefore cannot be nucleoside phosphates.
Additional metabolite characterization.
As a more direct test of this hypothesis, the metabolites were incubated with alkaline phosphatase. Separate experiments were performed in which [3H]BDCRB was incubated with a supernatant (obtained by centrifugation at 9,000 × g) from mouse liver and extracts were analyzed by reverse-phase HPLC. A pattern of retention of labeled material nearly identical to that shown in Fig. 2 was observed (data not shown). Fractions of the eluted labeled peaks were pooled, lyophilized, and incubated with alkaline phosphatase. Extracts prepared from incubation reactions that were not chromatographed were treated in an identical manner. Reverse-phase HPLC analysis revealed no change in chromatographic patterns between samples incubated with or without alkaline phosphatase. To establish that BDCRB-5′-MP was a substrate for alkaline phosphatase, separate incubations were run. Under identical incubation conditions, 3′-AMP, 5′-AMP, and BDCRB-5′-MP were rapidly dephosphorylated (data not shown). Therefore, it is unlikely that the metabolites were any type of nucleoside phosphates.
The lack of evidence for nucleoside phosphates as the unknown BDCRB metabolites prompted an additional study that compared the retention times of the unknown metabolites to the retention times for known analogs of BDCRB that could be metabolites. Retention times were determined by reverse-phase HPLC for the following analogs and potential metabolites of BDCRB: the 2,5,6-dehalogenated analog of BDCRB (14.0 min), BDCRB-5′-MP (23.7 min), the 2-hydroxy analog of BDCRB (25.6 min), 2′-deoxy-DRB (27.0 min), the 2-amino analog of BDCRB (27.2 min), 2′-deoxy-BDCRB (29.9 min), the 2-methoxy analog of BDCRB (30.8 min), BDCRB (32.5 min), 5′-dTCRB (35.1 min), and the heterocyclic base of BDCRB (38.9 min). Of these, only the 2-hydroxy analog had a retention time and the chemical structure consistent with those of a BDCRB metabolite. Consequently, an extract from uninfected cells (described below) was chromatographed with standards of BDCRB, DRB, and the 2-hydroxy analog as described in the legend to Fig. 2. Retention times for both standard compounds and labeled metabolites were nearly identical to those shown in Fig. 2. The 2-hydroxy analog eluted separately from and approximately 1 min after the metabolite with the peak at 25 min, thereby establishing that none of the analogs listed above could be the labeled BDCRB metabolites.
Comparison of BDCRB metabolism in uninfected and HCMV-infected cells.
A comparison of the foregoing data showed that the amounts of the two major metabolites—hereafter referred to as the 21-min and the 25-min peaks (Fig. 2)—were greater in HCMV-infected cells. These differences were examined more thoroughly by incubating uninfected and HCMV-infected HFF cells with 10 μCi of [3H]BDCRB per ml at selected times postinfection and analyzing the metabolites by reverse-phase HPLC. The two major metabolites were present in both infected and uninfected cells, regardless of the conditions used. Infected cells consistently showed higher concentrations of both metabolites, especially after long incubation times (data not shown), raising the possibility of slow hydrolysis of the labeled compound with efficient salvage of the resulting [3H]ribose to produce the unknown metabolites. Even though we could not identify these metabolites, we can conclude that BDCRB was not converted to a 5′-nucleoside phosphate in either uninfected or HCMV-infected cells, and it is unlikely that viral proteins were involved in the metabolism.
Activity of 5′-deoxy analogs of TCRB and BDCRB.
As a direct test of the hypothesis that phosphorylation at the 5′ position of BDCRB is not needed for antiviral activity, the 5′-deoxy analogs 5′-dTCRB and 5′-dBDCRB were synthesized and evaluated. These compounds cannot be phosphorylated at the 5′ position, yet they were more potent inhibitors of HCMV replication than the parent ribosyl nucleosides TCRB and BDCRB (Table 1). These results are nearly identical to those we obtained previously with closely related erythrosyl benzimidazoles (20). The erythrosyl analogs lack the entire 5′-hydroxymethyl moiety, not just the 5′-hydroxy group. Like 5′-dTCRB and 5′-dBDCRB, they cannot be 5′ phosphorylated, yet they are more active than TCRB and BDCRB. Together these data clearly establish that, unlike most nucleoside antiviral drugs, phosphorylation of benzimidzole nucleosides at the 5′ position is not necessary for activity against HCMV.
TABLE 1.
Activities of benzimidazole nucleosides against HCMV isolates sensitive and resistant to TCRB and BDCRB
| Compound | Mean ± SD IC50 or IC90 (μM)a for virus isolateb:
|
|||
|---|---|---|---|---|
| Towne (wild type) | B11 (UL89) | D10 (UL89) | C4 (UL56 + UL89) | |
| TCRB | 2.8 ± 1.1 | 16 ± 4.9 | 17 ± 7.6 | 56 ± 31 |
| BDCRB | 1.2 ± 0.78 | 12 ± 6.7 | 7.3 ± 5.8 | 28 ± 7.4 |
| 5′-dTCRB | 0.58 ± 0.46c | 2.9 ± 0.5c | 3.1 ± 0.14c | 13d |
| 5′-dBDCRB | 0.15 ± 0.001c | |||
| 5′-dBDCRBe | 0.06 ± 0.01 | 3.0 ± 0.8 | ||
| GCV | 1.5 ± 0.6 | 3.1 ± 0.2c | 4.6 ± 2.1 | 5.5 ± 3.2 |
Results are from plaque reduction assays performed in duplicate with the indicated isolates of HCMV selected for resistance to TCRB, as described in the text. The 50% inhibitory concentration (IC50s) were calculated from three to six separate experiments with at least four drug concentrations each, unless marked otherwise.
The genotype of each isolate is given in parentheses. The UL number specifies the gene with the mutation that confers resistance to TCRB; the UL89 mutation is D344E, and the UL56 mutation is Q204R (25). B11 and D10 are different isolates with the same mutation that results in resistance to TCRB, which we have described previously (25).
The experiment was performed twice, in duplicate, by using at least four drug concentrations.
The experiment was performed once, in duplicate, by using five drug concentrations.
The results are presented as 90% inhibitory concentrations (IC90s) from two yield reduction experiments performed in duplicate or triplicate wells by using at least five drug concentrations.
HCMV resistant to TCRB and BDCRB.
In an initial attempt to understand the mechanism of action of these new analogs and to determine if they were similar to the mechanism of action of the parent compounds, the 5′-deoxy analogs were evaluated against HCMV strains selected for resistance to TCRB. The isolates designated B11 and D10 both encode a point mutation in UL89 (Asp344Glu); isolate C4 has the same mutation in UL89 plus an additional mutation in UL56 (Gln204Arg) (25). The data in Table 1 show that isolates D10 and B11 were 5- to 10-fold resistant to TCRB, BDCRB, and 5′-dTCRB and that isolate C4 was 20- to 50-fold resistant to these three drugs and to 5′-dBDCRB compared with the level of sensitivity of the wild type. GCV had similar potency against the three resistant viruses. From this viral cross-resistance to the benzimidazoles, we hypothesize that the 5′-deoxy analogs inhibit HCMV replication in a way analogous to that for TCRB via the proteins encoded by HCMV UL89 and UL56.
Mode of action of 5′-dTCRB.
In order to explore this hypothesis more closely, 5′-dTCRB was evaluated for its capacity to inhibit viral DNA synthesis and processing. Prior studies established that TCRB and BDCRB do not inhibit HCMV DNA synthesis but, instead, inhibit the processing of viral DNA (25, 46). In an assay that measured intracellular viral DNA levels, neither TCRB nor 5′-dTCRB inhibited the accumulation of HCMV DNA at drug concentrations sufficient to cause a decrease in the supernatant viral load of several orders of magnitude (Fig. 5). The decrease in viral DNA synthesis seen with 5′-dTCRB at 30 μM most likely was due to toxicity (31). Additionally, both TCRB and 5′-dTCRB inhibited the conversion of the high-molecular-weight HCMV DNA to monomeric units in a concentration-dependent manner, as determined by pulsed-field gel electrophoresis (Fig. 6). In contrast, 1263W94, a known inhibitor of HCMV DNA synthesis that acts via UL97 (C. Talarico et al., Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. H-90, 1998), was essentially inactive. This is consistent with earlier studies indicating that the activity of TCRB is mediated through HCMV gene products UL89 and UL56 (25, 46). These proteins have been proposed to be involved in the cleavage of concatemeric DNA to monomers during viral maturation (6, 9). Because viral DNA is not processed in the presence of 5′-dTCRB, we postulate that this compound also interferes with the activity of the proteins encoded by UL89 and UL56.
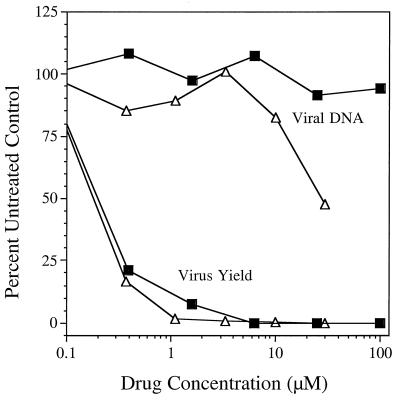
FIG. 5.
Benzimidazole nucleosides inhibited HCMV replication but not viral DNA synthesis. Selected concentrations of TCRB (▪) and 5′-dTCRB (▵) were incubated in duplicate wells with HFF cells infected with HCMV at 3 PFU/cell. Viral DNA was detected by DNA-DNA hybridization, and yield was quantified by serial dilution of supernatants across 96-well culture plates seeded with HFF cells.
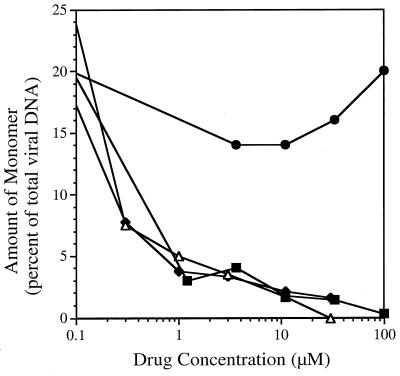
FIG. 6.
Inhibition of cleavage of concatemeric DNA to monomeric genomic units by benzimidazole nucleoside analogs. Selected concentrations of TCRB (▪), BDCRB (◊), 5′-dTCRB (▵), and 1263W94 (•) were incubated in duplicate wells with HFF cells infected with HCMV at 3 PFU/cell. After 3 days, the cells were suspended in agarose and digested to release DNA. The DNA was separated by contour-clamped homogeneous electric field separation, transferred to a nylon membrane, and probed for HCMV-specific sequences. The amount of genomic-length monomeric DNA is expressed as a percentage of the total amount of viral DNA in the well. In the absence of test compounds, monomeric DNA was 25% ± 5% of the total DNA in three determinations.
On the basis of our prior observation that the activity of BDCRB could be reversed by removal of the drug at any time in the viral replication cycle prior to DNA processing (P. M. Krosky, K. Z. Borysko, J. C. Drach, and K. K. Biron, unpublished data), an assay was devised to determine if the modes of action of the new compounds involved effects on DNA synthesis or DNA processing. Cells were incubated in the presence of 20 μM BDCRB for 96 h so that the HCMV replication cycle could proceed through viral DNA synthesis but then be halted prior to viral DNA processing. The subsequent removal of BDCRB permitted DNA processing to occur, resulting in the production of infectious virus. If a second (test) compound was added after removal of BDCRB, then the effect of the second compound on DNA processing was observed simply by the quantification of infectious virus produced during the next 24 h. Thus, a second compound that inhibited DNA synthesis would have no effect on virus production because it was added after the completion of viral DNA synthesis. In contrast, a compound with an effect on DNA processing would continue to have an inhibitory effect on viral replication. Table 2 presents data from such experiments and shows that BDCRB, 5′-dTCRB, and 5′-dBDCRB were highly active in this assay. In contrast, 1263W94 was essentially inactive since it is an inhibitor of HCMV DNA synthesis. All compounds had a marked effect on viral replication (yield) when they were present for the entire replication cycle. Table 2 also presents data that show directly the effect of 1263W94 on viral DNA synthesis and the lack of an effect by BDCRB, 5′-dTCRB, and 5′-dBDCRB. Taken together with the foregoing, these observations demonstrate that the 5′-deoxy analogs of TCRB and BDCRB act by a mechanism very similar or identical to that for the parent compounds, namely, inhibition of DNA processing.
DISCUSSION
The need for new drugs for the treatment of HCMV infections is apparent due to the multiple deficiencies of the currently available therapies. The trihalogenated benzimidazole ribonucleosides have limited toxicities and are potent and selective inhibitors of HCMV replication. They inhibit HCMV through a unique mechanism of action involving an interference with cleavage of concatemeric viral DNA (25, 46). In the present study, we have been unable to detect phosphorylated metabolites of BDCRB, but we have established that the 5′-deoxy analogs of TCRB and/or BDCRB are potent inhibitors of HCMV replication and DNA processing. These results establish that this novel mechanism of action does not involve a 5′-phosphorylated intermediate.
Other antiviral nucleosides actually are prodrugs that require activation by phosphorylation, but the benzimidazole ribonucleosides have proved to be quite different. HPLC analysis of extracts from cells treated with BDCRB provided no evidence of phosphorylated products. Two radioactive metabolites were observed under all chromatographic conditions and were seen in both infected and uninfected cells. We could find no evidence that these compounds are 5′-monophosphorylated metabolites of BDCRB because HPLC readily separated the labeled metabolites from other nucleoside monophosphates. These results indicate that TCRB and BDCRB are not prodrugs in the sense that many nucleoside analogs such as acyclovir and GCV are, as acyclovir and GCV require conversion to their triphosphates to be active (4, 5).
That phosphorylated metabolites of BDCRB were not identified does not totally nullify the possibility of their existence. Although we did not detect BDCRB-5′-MP, we cannot totally eliminate the possibility that it was formed at concentrations undetectable by the methods that we used, that its half-life was quite short, or that it was destroyed in the extraction process. However, there is little support for any of these hypotheses. The existence of minute amounts of an “activated” metabolite would require a compound with very great and unusual potency. In addition, the phosphorylated metabolites of a wide variety of antiviral nucleosides are readily formed and easily detected, including those of GCV (5) and cidofovir (23). Furthermore, the naturally occurring nucleotides and a standard of BDCRB-5′-MP were stable throughout the extraction process. Consequently, we have no reason to hypothesize that BDCRB-5′-MP was formed or that it is the active species.
Likewise, we have no evidence to suggest that BDCRB-2′- or -3′-monophosphates were formed. Because we did not have these compounds as standards, we cannot eliminate the possibility that one or the other was formed. This possibility would require, however, that BDCRB-2′- and -3′-monophosphates chromatographed differently from BDCRB-5′-MP on an anion-exchange column and that they were not susceptible to hydrolysis by alkaline phosphatase. Both seem unlikely. In contrast, the related RNA synthesis inhibitor, DRB, is phosphorylated at the 2′ and 5′ positions (11, 13). However, this phosphorylation is not needed for inhibitory activity (10); in fact, the presence of a nonphosphorylated 3′-hydroxyl group is required for activity (12).
HCMV-infected cells consistently showed larger amounts of the two metabolites than uninfected cells. A likely explanation for the increase in metabolism is a nonspecific increase in the level of cellular protein expression. It is well documented that HCMV infection stimulates the expression of many cellular proteins including transcription factors, enzymes involved in DNA metabolism, proto-oncogene products, and cell surface adhesion proteins (29). Although we have few data beyond those in Fig. 2, a slow hydrolysis of the labeled compound and efficient salvage of the resulting [3H]ribose to produce the unknown metabolites is consistent with an HCMV-induced increase in cellular enzyme levels.
The comparison of the modes of action between TCRB and 5′-dTCRB presented herein gave no indication that the activity of 5′-dTCRB was different in any way from the activity of TCRB. Both compounds were selective inhibitors of HCMV and had little or no effect against HSV type 1 (J. C. Drach et al., Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. H112, 1995). Neither TCRB nor the corresponding 5′-deoxy analog inhibited DNA synthesis, both compounds extended the effects of BDCRB, and both compounds inhibited concatemer cleavage. Furthermore, HCMV strains resistant to TCRB were cross resistant to 5′-dTCRB, demonstrating that the mutations that provided resistance to TCRB also provided resistance to the 5′-deoxy analog.
The increased potencies of the 5′-deoxy analogs compared to those of TCRB and BDCRB have not been fully explored. Analysis of structure-activity data obtained for other analogs of TCRB containing changes in the 5′ position has shown that the viral target tolerates a wide variety of modifications without a loss of activity. In general, analogs with small substituents in the 5′ position are more active than those with large substituents in that position (19). The substitution of hydrogen in 5′-dTCRB for 5′-hydroxyl in TCRB is consistent with this pattern. In addition, benzimidazole erythronucleosides are more potent than benzimidazole ribonucleosides (20). The potent activities against HCMV of these analogs that lack the entire 5′-hydroxymethyl moiety, not just the 5′-hydroxy moiety, demonstrate that the moiety in this position is not critical for the antiviral activity of the molecule.
Acknowledgments
We thank Donald McKenzie for expert technical performance of certain antiviral assays with HCMV.
This study was supported by research grants U01-AI31718 and U19-AI31718 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by research agreement DRDA-942941 with Glaxo Wellcome Co. P.M.K. was supported in part by NIH training grant GM07767.
REFERENCES
- 1.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377–2384. [DOI] [PubMed] [Google Scholar]
- 2.Alford, C. A., and W. J. Britt. 1993. Cytomegalovirus, p.227–255. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpesviruses. Raven Press, New York, N.Y.
- 3.Balfour, H. H., B. A. Chace, J. T. Stapleton, R. L. Simmons, and D. S. Fryd. 1989. A randomized, placebo-controlled trial of oral acyclovir for the prevention of cytomegalovirus disease in recipients of renal allographs. N. Engl. J. Med. 320:1381–1387. [DOI] [PubMed] [Google Scholar]
- 4.Biron, K. K., and G. B. Elion. 1980. In vitro susceptibility of varicella-zoster virus to acyclovir. Antimicrob. Agents Chemother. 18:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biron, K. K., S. C. Stanat, J. B. Sorrell, J. A. Fyfe, P. M. Keller, C. U. Lambe, and D. J. Nelson. 1985. Metabolic activation of the nucleoside 9-{[2-hydroxy-1(hydroxymethyl)ethoxy]methyl}guanine in human diploid fibroblasts infected with human cytomegalovirus. Proc. Natl. Acad. Sci. USA 82:2473–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the Pac motif and has specific nuclease activity. J. Virol. 72:2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chrisp, P., and S. P. Clissold. 1991. Foscarnet: a review of its antiviral activity, pharmacokinetic properties, and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs 41:104–129. [DOI] [PubMed] [Google Scholar]
- 8.Crumpacker, C. S. 1996. Ganciclovir. Drug Ther. 335:721–729. [DOI] [PubMed] [Google Scholar]
- 9.Davidson, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9–14. [DOI] [PubMed] [Google Scholar]
- 10.Egyhazi, E., M. Holst, and U. Tayip. 1983. Unmetabolized 5,6-dichlororibofuranosylbenzimidazole rather than its monophosphate metabolites is probably the active transcription inhibitor. Eur. J. Biochem. 130:223–226. [DOI] [PubMed] [Google Scholar]
- 11.Egyhazi, E., A. Ossoinak, M. Holst, K. Rosendahl, and U. Tayip. 1980. Kinetic analysis of uptake and phosphorylation of 5,6-dichlororibofuranosylbenzimidazole (DRB) by salivary gland cells of Chironomus tentans. J. Biol. Chem. 255:7807–7812. [PubMed] [Google Scholar]
- 12.Egyhazi, E., A. Ossoinak, U. Tayip, Z. Kazimierczuk, and D. Shugar. 1982. Specific inhibition of hnRNA synthesis by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole: requirement of a free 3′-hydroxyl group but not 2′- or 5′-hydroxyls. Biochim. Biophys. Acta 697:213–220. [DOI] [PubMed] [Google Scholar]
- 13.Egyhazi, E., and D. Shugar. 1979. 5,6-Dichlororibofuranosylbenzimidazole (DRB) is phosphorylated in salivary gland cells of Chironomus tentans. FEBS Lett. 107:431–435. [DOI] [PubMed] [Google Scholar]
- 14.Erriksson, B., B. Öberg, and B. Wahren. 1982. Pyrophosphate analogs as inhibitors of DNA polymerases of cytomegalovirus, herpes simplex virus and cellular origin. Biochim. Biophys. Acta 696:115–123. [DOI] [PubMed] [Google Scholar]
- 15.Furman, P. A., P. de Miranda, M. H. St. Clair, and G. B. Elion. 1981. Metabolism of acyclovir in virus-infected and uninfected cells. Antimicrob. Agents Chemother. 20:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fyfe, J. A., P. M. Keller, P. A. Furman, R. L. Miller, and G. B. Elion. 1978. Thymidine kinase from herpes simplex virus phosphorylates the new antiviral compound, 9-(2-hydroxyethoxymethyl)guanine. J. Biol. Chem. 253:8721–8727. [PubMed] [Google Scholar]
- 17.Good, S. S., B. S. Owens, L. B. Townsend, and J. C. Drach. 1994. The disposition in rats and monkeys of 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole (BDCRB) and its 2,5,6-trichloro congener (TCRB). Antivir. Res. 23:103. [Google Scholar]
- 18.Grattan, M. T., C. E. Moreno-Cabral, V. A. Starnes, E. B. Stinson, and N. E. Shumway. 1989. Cytomegalovirus infection is associated with cardiac allograph rejection and atherosclerosis. JAMA 261:3561–3566. [PubMed] [Google Scholar]
- 19.Gudmundsson, K. S., J. C. Drach, L. L. Wotring, and L. B. Townsend. 1997. Synthesis and antiviral activity of certain 5′-modified analogs of 2,5,6-trichloro-1-(β-d-ribofuranosyl)benzimidazole. J. Med. Chem. 40:785–793. [DOI] [PubMed] [Google Scholar]
- 20.Gudmundsson, K. S., J. Tidwell, N. Lippa, G. W. Koszalka, N. van Draanen, R. Ptak, J. C. Drach, and L. B. Townsend. 2000. Synthesis and antiviral evaluation of halogenated β-d- and -l-erythrofuranosylbenzimidazoles. J. Med. Chem. 43:2464–2472. [DOI] [PubMed] [Google Scholar]
- 21.He, Z., Y.-S. He, Y. Kim, L. Chu, C. Ohmstede, K. K. Biron, and D. M. Coen. 1997. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J. Virol. 71:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitchcock, M. J. M., H. S. Jaffe, J. C. Martin, and R. J. Stagg. 1996. Cidofovir, a new agent with potent anti-herpesvirus activity. Antimicrob. Agents Chemother. 7:115–127. [Google Scholar]
- 23.Ho, H.-T., K. L. Woods, J. J. Bronson, H. DeBoeck, J. C. Martin, and M. J. M. Hitchcock. 1992. Intracellular metabolism of the antiherpes agent (S)-1-[3-hydroxy-2-(phophonylmethoxy)propyl]cytosine. Mol. Pharmacol. 41:197–202. [PubMed] [Google Scholar]
- 24.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118–133. [DOI] [PubMed] [Google Scholar]
- 25.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W.-H. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole nucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mar, E., J. Chiou, Y. Cheng, and E. Huang. 1985. Inhibition of cellular DNA polymerase α and human cytomegalovirus-induced DNA polymerase by the triphosphates of 9-(2-hydroxyethoxymethyl)guanine and 9-(1,3-dihydroxy-2-propoxymethyl)guanine. J. Virol. 53:776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marx, J. 1994. CMV-p53 interaction may help explain clogged arteries. Science 265:320. [DOI] [PubMed] [Google Scholar]
- 28.McKenzie, R., M. W. D. Travis, S. A. Dolan, S. Pittaluga, I. M. Feuerstein, J. Shelhamer, R. Yarchoan, and H. Masur. 1991. The cause of death in patients with human immunodeficiency virus infection: a clinical and pathological study with emphasis on the role of pulmonary disease. Medicine 70:326–343. [DOI] [PubMed] [Google Scholar]
- 29.Mocarski, E. S. J. 1996. Cytomegaloviruses and their replication, p.2447–2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Field’s virology, vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 30.Mulamba, G. B., A. Hu, R. F. Azad, K. P. Anderson, and D. M. Coen. 1998. Human cytomegalovirus mutant with sequence-dependent resistance to the phosphorothioate oligonucleotide fomivirsen (ISIS 2922). Antimicrob. Agents Chemother. 42:971–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nassiri, M. R., S. G. Emerson, R. V. Devivar, L. B. Townsend, J. C. Drach, and R. S. Taichman. 1996. Comparison of benzimidazole nucleosides and ganciclovir on the in vitro proliferation and colony formation of human bone marrow progenitor cells. Br. J. Haematol. 93:273–279. [DOI] [PubMed] [Google Scholar]
- 32.Pelling, J. C., J. C. Drach, and C. Shipman, Jr. 1981. Internucleotide incorporation of arabinosyladenine into herpes simplex virus and mammalian cell DNA. Virology 109:323–335. [DOI] [PubMed] [Google Scholar]
- 33.Perry, C. M., and J. A. Balfour. 1999. Fomivirsen. Drugs 57:375–380. [DOI] [PubMed] [Google Scholar]
- 34.Plosker, G. L., and S. Noble. 1999. Cidofovir: a review of its use in cytomegalovirus retinitis in patients with AIDS. Drugs 58:325–345. [DOI] [PubMed] [Google Scholar]
- 35.Pogolotti, A. L. J., and D. V. Santi. 1982. High-pressure liquid chromotography—ultraviolet analysis of intracellular nucleotides. Anal. Biochem. 126:335–345. [DOI] [PubMed] [Google Scholar]
- 36.Prichard, M. N., S. R. Turk, L. A. Coleman, S. L. Englehardt, C. J. Shipman, and J. C. Drach. 1990. A microtiter virus yield reduction assay for the evaluation of antiviral compounds against human cytomegalovirus and herpes simplex virus. J. Virol. Methods 28:101–106. [DOI] [PubMed] [Google Scholar]
- 37.Puck, T. T., S. J. Cieciura, and A. Robinson. 1958. Genetics of somatic mammalian cells. III. Long-term cultivation of euploid cells from human and animal subjects. J. Exp. Med. 108:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, V. B., and L. W. Black. 1988. Cloning, overexpression, and purification of the terminase proteins gp16 and gp17 of bacteriophage T4. Construction of a defined in vitro DNA packaging system using purified terminase proteins. J. Mol. Biol. 200:475–488. [DOI] [PubMed] [Google Scholar]
- 39.Shipman, C., Jr. 1969. Evaluation of 4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid (HEPES) as a tissue culture buffer. Proc. Soc. Exp. Biol. 130:305–310. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan, V., C. L. Talarico, S. C. Stanat, M. Davis, D. M. Coen, and K. K. Biron. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 358:162–164. [DOI] [PubMed] [Google Scholar]
- 41.Talarico, C. L., T. C. Burnette, W. H. Miller, S. L. Smith, M. G. Davis, S. C. Stanat, T. I. Ng, Z. He, D. M. Coen, B. Roizman, and K. K. Biron. 1999. Acyclovir is phosphorylated by the human cytomegalovirus UL97 protein. Antimicrob. Agents Chemother. 43:1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation requires the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townsend, L. B., R. V. Devivar, S. R. Turk, M. R. Nassiri, and J. C. Drach. 1995. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(β-d-ribofuranosyl)benzimidazoles. J. Med. Chem. 38:4098–4105. [DOI] [PubMed] [Google Scholar]
- 44.Townsend, L. B., and J. C. Drach. November 1994. Polysubstituted benzimidazoles as antiviral agents. U.S. patent 5,360,795.
- 45.Turk, S. R., C. Shipman, R. Nassiri, G. Genzlinger, S. H. Krawczyk, L. B. Townsend, and J. C. Drach. 1987. Pyrrolo[2,3-d]pyrimidine nucleosides as inhibitors of human cytomegalovirus. Antimicrob. Agents Chemother. 31:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]