Abstract
Previous studies have indicated that human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PIs) are less active at blocking viral replication in HIV-1 infected peripheral blood monocytes/macrophages (M/M) than in HIV-1-infected T cells. We explored the hypothesis that oxidative modification and/or metabolism of the PIs in M/M might account for this reduced potency. We first tested the susceptibility of several PIs (kynostatin-272 [KNI-272], saquinavir, indinavir, ritonavir, or JE-2147) to oxidation after exposure to hydrogen peroxide (H2O2): only KNI-272 was highly susceptible to oxidation. Treatment of KNI-272 with low millimolar concentrations of H2O2 resulted in mono-oxidation of the sulfur in the S-methyl cysteine (methioalanine) moiety, as determined by reversed-phase high-performance liquid chromatography and mass spectrometry (RP-HPLC/MS). Higher concentrations of H2O2 led to an additional oxidation of the sulfur in the thioproline moiety of KNI-272. None of the PIs were metabolized or oxidized when added to T cells and cultured for up to 12 days. However, when KNI-272 was added to M/M, the concentration of the original KNI-272 steadily decreased with a corresponding increase in the production of three KNI-272 metabolites as identified by RP-HPLC/MS. The structures of these metabolites were different from those produced by H2O2 treatment. The two major products of M/M metabolism of KNI-272 were identified as isomeric forms of KNI-272 oxidized solely on the thioproline ring. Both metabolites had reduced capacities to inhibit HIV-1 protease activity when tested in a standard HIV-1 protease assay. These studies demonstrate that antiviral compounds can be susceptible to oxidative modification in M/M and that this can affect their antiviral potency.
Inhibitors of the human immunodeficiency virus type 1 (HIV-1) protease prevent the production of infectious virus by blocking the function of virus-encoded aspartyl protease (28). Inhibition of the protease leads to the production of noninfectious immature virions containing uncleaved Gag and Gag-Pol polyproteins (11, 19, 31). Several HIV-1 protease inhibitors (PIs) are now in widespread clinical use and are highly potent when used in combination with other anti-HIV agents (21, 36). While highly active antiretroviral therapy can lead to undetectable plasma viral loads, there is increasing evidence that cells of the monocyte/macrophage (M/M) lineage act as a host reservoir for HIV (1, 5, 20, 34). Moreover, there is evidence that PIs may have reduced activity in M/M and that smoldering infection in such cells can act as viral reservoirs that lead to the selection of resistant virus (28). It has been recognized that there is a need to achieve more complete inhibition of viral replication in potential viral reservoirs, and there is interest in developing more effective methods to block HIV-1 replication in M/M.
In a previous study, we found that certain HIV-1 PIs were less potent (higher 50% effective concentration) at blocking HIV-1 production in M/M than in T cells (28). We hypothesized that increased PI metabolism may occur, in part, as a result of increased oxidative metabolism of the inhibitors. In response to a variety of stimuli, M/M produce superoxide and hydrogen peroxide (H2O2) by using the NADPH oxidase system (25). In addition, M/M have an increased mitochondrial oxidative metabolism compared to other cells (17). Therefore, modification of the inhibitors could occur via reactive oxygen intermediates (ROIs) generated by M/M or through oxidative metabolism (28). Many HIV-1 PIs are metabolized through oxidative pathways in vivo (8, 15, 18, 36, 37), and it seemed possible that the PIs might also be oxidized by certain target cells in HIV-infected patients.
Increased oxidative metabolism and the production of ROIs by M/M occur after HIV-1 infection, possibly due to increased exposure to cytokines and HIV-1-related antigens (2, 25, 26, 29). These ROIs have been reported to play an important role in HIV-1 replication in M/M (26) and in enhancing the cell-to-cell spread of virus (14). In addition a number of clinical studies reveal that the overall redox state of HIV-1-infected patients is altered to a more prooxidant state (2, 35). Together, these studies indicate the presence of altered oxidative metabolism in HIV-1-infected M/M and in HIV-1-infected patients, and we have hypothesized that this could have an impact on the drugs that target HIV-1 replication, especially in M/M. An examination of the structures of several PIs showed that certain of these agents have functional groups that could be susceptible to oxidation. To explore this possibility, we examined the susceptibility of several PIs to oxidative modification and, when such changes were observed, studied the metabolism of the drugs in HIV-1-susceptible cells.
MATERIALS AND METHODS
Chemicals.
The PIs JE-2147 (22, 38), saquinavir (4), indinavir (3), and ritonavir (21) were obtained as gifts from Hiroaki Mitsuya (National Cancer Institute, National Institutes of Health, Bethesda, Md.). Kynostatin-272 (KNI-272) (13) was obtained from the Japan Energy Corporation. All inhibitors were stored at −20°C as 10 to 40 mM stock solutions in 100% dimethyl sulfoxide (DMSO). Granulocyte-macrophage colony-stimulating factor (GM-CSF) and tissue necrosis factor alpha (TNF-α) were obtained from R&D (Minneapolis, Minn.). Lipopolysaccharide (LPS) was obtained from Invitrogen (Gaithersburg, Md.).
Cells and culture conditions.
Primary M/M were prepared and purified as described previously (10). Briefly, peripheral blood mononuclear cells were obtained from healthy HIV-1-negative donors through the Department of Transfusion Medicine, Warren G. Magnuson Clinical Center (Bethesda, Md.). Cells were separated over a Ficoll-Hypaque gradient and seeded in 25-cm2 tissue culture flasks at a density of 3 × 107 cells/flask in complete medium consisting of RPMI 1640 with 10% AB+ heat-inactivated human serum; 20% heat-inactivated, low-endotoxin, mycoplasma-free fetal bovine serum; 4 mM l-glutamine; 50 U of penicillin/ml; and 50 mg of streptomycin/ml. Cultures were maintained in 5% CO2 in air at 37°C. After 4 to 5 days in culture, cell monolayers were gently washed three times with phosphate-buffered saline (calcium- and magnesium-free) to remove nonadherent cells. Monolayers of M/M were then maintained on the complete medium described above without human serum. The nonadherent cell population removed during washing of the M/M monolayers was resuspended at 5 × 106 cells/ml and stimulated with phytohemagglutinin (PHA) for 2 days and then with interleukin-2 (IL-2) (10 ng/ml; R&D) in complete RPMI 1640 medium. Five days after culture of M/M or 3 days after PHA and IL-2 stimulation of the peripheral blood T-cell population, the PIs were added into the cell culture medium to assess metabolism of the inhibitors by these cells. In some experiments, HIV-infected M/M were used. M/M were infected with HIV-1BaL (Advanced Biotechnology, Inc., Columbia, Md.) at 50 ng of p24/ml, and peripheral blood T cells were infected with 50 ng of p24/ml of the HIV-1 HTLV-IIIB isolate (30). Assays for p24 (RIA; Perkin-Elmer Life Sciences, Boston, Mass.) released into the culture medium were done to verify infection before the addition of the test compounds. At different time points an aliquot (250 μl) of the medium was removed to analyze the PI concentration and evidence of any metabolites. The 250-μl aliquot was added to 750 μl of 8 M guanidine-HCl to give a final concentration of 6 M guanidine-HCl. The guanidine-HCl was used to inactivate any enzymes present in the media that could lead to further metabolism and to denature proteins and release any protein-bound inhibitor prior to reversed-phase high-pressure liquid chromatography (RP-HPLC) analysis. These samples were stored at −20°C until further analysis by RP-HPLC.
H9, H9/IIIB, and Jurkat T cells were used in some experiments to assess the metabolism of PIs. These cell lines were maintained in complete RPMI 1640 medium, with 15% fetal bovine serum as previously described (30). Cells were plated at 105 cells per ml, and test compounds were then added to give a final concentration of 5 or 10 μM. On different days as indicated in the results, a 250-μl sample of the medium was removed and added to 750 μl of 8 M guanidine-HCl. These samples were stored at −20°C until further analysis by RP-HPLC.
Oxidation of PIs with hydrogen peroxide.
Stock solutions (40 to 100 mM) of PIs in DMSO were diluted into phosphate-buffered saline at concentrations of 1 to 200 μM. The samples were then incubated for 15 to 30 min at 42°C in the presence of different concentrations (1 to 200 mM) of H2O2. Samples were then analyzed directly by RP-HPLC as described below.
RP-HPLC analysis.
RP-HPLC analysis was used to assess the susceptibility of the PIs to oxidation by H2O2 and to assess metabolism of the PIs by M/M, peripheral blood T cells, and the other cell lines. The samples (obtained from PI-treated cells) in guanidine-HCl were first passed through a Microcon-10 filtration device (Amicon, Beverly, Mass.) to remove proteinaceous and cellular debris. The flowthrough containing the PI was then analyzed by RP-HPLC on a Vydac (Resolution Systems, Holland, Mich.) C18 column (0.21 by 5 cm). Samples were eluted from the column over a period of 25 min at a flow rate of 0.3 ml/min with a linear gradient of 2.5 to 52.5% acetonitrile in 0.05% trifluoroacetic acid (TFA) and water. The eluate was monitored with a multiwavelength diode array detector with specific monitoring at 205, 276, and 350 nm. The inhibitor concentrations were calculated by using a standard curve obtained with different concentrations of inhibitor in the tested range of 0.5 to 10 μM diluted from stock solutions into 6 M guanidine-HCl. Metabolites were purified by collecting the eluting peak from RP-HPLC, lyophilized, resuspended in 10% DMSO, and then analyzed by liquid chromatography-mass spectrometry (LC/MS). Medium extracts were subjected to LC/MS directly to identify the metabolites.
Methionine sulfoxide reductase treatment of oxidized KNI-272.
KNI-272 oxidized at the S-methyl cysteine residue was purified by RP-HPLC as described above and used in assays with purified recombinant yeast methionine sulfoxide reductase EC 1.8.4.6 (MsrA) to determine whether the oxidized form could be reduced by this enzyme. Oxidized KNI-272 dissolved in 3% DMSO was incubated at a final concentration of 5 μM in 150 mM sodium phosphate buffer (pH 8.0), 20 mM dithiothreitol (DTT), and 0.5 mM EDTA with or without MsrA at 170 μg/ml isolated as described previously (23). The samples were then incubated for 30 min at 37°C, at which time they were stopped by acidification with 5 μl of 10% trifluoroacetic acid. Prior to RP-HPLC analysis, the sample was diluted with a solution of 8 M guanidine to bring the sample to a final concentration of 6 M guanidine, which was necessary for good recovery of the oxidized and reduced forms of KNI-272.
Enzymatic assays.
HIV-1 protease activity was determined as described previously (6). Briefly, HIV-1HXB2 protease at a final dimeric concentration of 200 nM was incubated in 40 μl of protease assay buffer (150 mM sodium acetate, pH 5.7; 10% glycerol; 5% ethylene glycol; 1 mM EDTA) in the presence of the test compounds dissolved in DMSO. After a 5-min preincubation, a 9-amino-acid peptide substrate (dissolved in distilled water) spanning the matrix capsid cleavage site in gag was added at a final concentration of 3 mM and incubated for 5 to 10 min. The assay was stopped with an aliquot of 10% TFA, and the cleavage products were quantified by RP-HPLC as described previously (6).
MS.
Capillary HPLC was performed with a HP1100 binary gradient pump (Agilent Technologies, Palo Alto, Calif.), operating at 100 μl/min. An Accurate AC-100-VAR flow splitter (LC Packings, San Francisco, Calif.), fitted with a CAL-100-0.3 calibrator, reduced the flow being delivered to a C18 capillary column (100 by 0.3 mm, 5 μm; Keystone Scientific, Bellefonte, Pa.) to ca. 5 μl/min. Solvent A was a 95:5:0.1 mixture of water, acetonitrile, and formic acid, and solvent B was a 95:5:0.1 mixture of acetonitrile, isopropanol, and formic acid. Next, 10 μl of each sample (5 μg) was desalted with C18 Ziptips (Millipore, Bedford, Mass.), according to the manufacturer’s instructions. A 2-μl sample was injected and eluted by the following mixed-gradient method: 100% solvent A held for 5 min, switched to 20% solvent B, followed by a gradient from 20 to 30% solvent B for 20 min, followed by 30% solvent B held for 5 min, and then a gradient from 30 to 95% solvent B for 5 min. The eluent flowed directly into a Finnigan LCQ (ThermoQuest, San Jose, Calif.) for mass analysis. No sheath or auxiliary gases were used, and the voltage was applied directly to the eluent. Centroid LC/MS data were collected with the triple-play method (full-scan, high-resolution zoom scan; mass spectrometry-mass spectrometry [MS/MS]) by using the Xcalibur Software package.
RESULTS
Susceptibility of PIs to oxidation by hydrogen peroxide.
To assess the potential for different PIs (saquinavir, indinavir, ritonoavir, JE-2147, and KNI-272) to be modified by oxidation, we tested their susceptibility to modification by hydrogen peroxide (H2O2). Saquinavir, ritonoavir, and KNI-272 were chosen since they had been shown previously by our group to be less effective as inhibitors in M/M than in infected T cells (28). The activity of these drugs in chronically infected M/M was 2- to 10-fold lower than for infected T-cell lines, and it was suggested that this may be due to increased oxidative modification by M/M. JE-2147 was chosen because it is an allophyenylnorstatin-containing inhibitor similar to KNI-272 and was recently developed to inhibit highly resistant HIV-1 isolates (38), although the comparative activity of JE-2147 in infected M/M cultures and infected T cells is unknown (38). Treatment with high concentrations of H2O2 (100 mM) resulted in evidence of modification of saquinavir, JE-2147, and KNI-272, as indicated by RP-HPLC analysis (data not shown). The mass of JE-2147 increased by 32 after treatment with high concentrations (100 mM) of H2O2, and this indicated dioxidation of the compound (data not shown). Although we did not further characterize the nature of this oxidation, it is likely that oxidation occurred on the sulfur of the thioproline moiety.
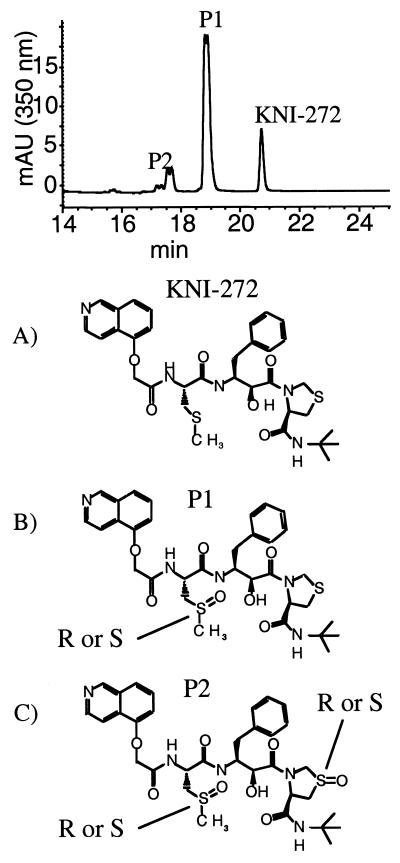
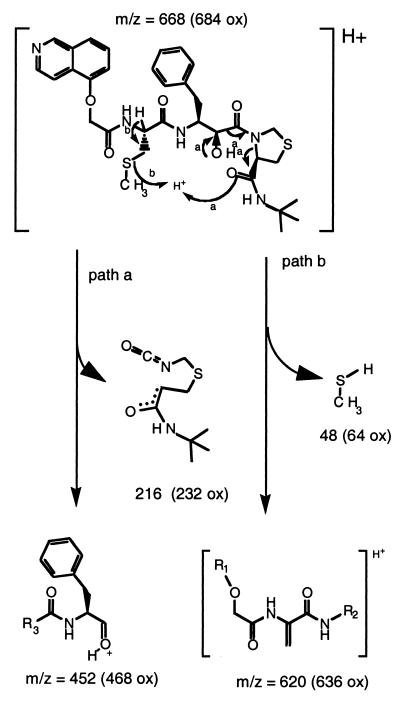
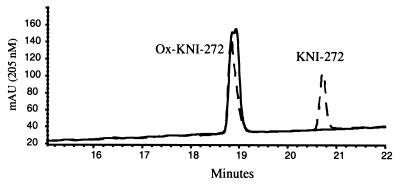
KNI-272 was clearly more sensitive to modification by H2O2 than the other PIs, as indicated by RP-HPLC analysis. After treatment of KNI-272 with H2O2 there were several new peaks detected that eluted earlier than untreated KNI-272 (Fig. 1, top). The majority of KNI-272 was converted to a doublet peak eluting at 18.9 and 19.1 min designated P1 (Fig. 1, top). In addition, there were four closely eluting peaks between 17 and 18 min designated P2 (Fig. 1, top). KNI-272 (Fig. 1A) would be predicted to be most susceptible to oxidation at the sulfur in the thioproline ring and/or the sulfur in the S-methyl cysteine moiety yielding the sulfoxides. To determine the nature of the oxidations, the new peaks generated after H2O2 treatment were subjected to MS/MS analysis and compared to untreated KNI-272. After MS/MS of untreated KNI-272 (M+H = 668) we detect an m/z fragment ion of 452 that shows a loss of 216, representing the thioproline ring-containing fragment (Fig. 2). If the thioproline sulfur becomes oxidized, then we would detect an m/z ion of 452 as with native KNI-272 but with a loss of 232 (216 + 16) from the original mass of 684 (668 + 16) representing the thioproline oxidized fragment (Fig. 2, path a). Alternatively, if the methioalanine sulfur becomes oxidized after treatment, one would predict an unfragmented m/z of 684 and an m/z fragment ion of 468 (452 + 16) and a loss of 216 via path a (Fig. 2). These fragments would also show a loss of 64 (48 + 16) via path b (Fig. 2), representing the oxidized methyl sulfur fragment. Indeed, these were the fragments detected after MS/MS analysis of P1 eluting at 18.9 and 19.1 min on RP-HPLC, demonstrating that P1 represented KNI-272 oxidized only at the S-methyl cysteine sulfur (Fig. 1B). Since the nonspecific oxidation of the S-methyl cysteine sulfur with hydrogen peroxide would be expected to yield both the R and S configurations of the sulfoxide, it is likely that the doublet peak represents these two epimers. To explore this possibility further, we purified the material eluting as a doublet and treated it with MsrA. It has been reported that MsrA only reduces the S forms of methionine sulfoxide (24). RP-HPLC analysis of the P1 doublet after treatment with MsrA demonstrated that only the later eluting form of the doublet peak could be reduced back to native KNI-272 (Fig. 3). This indicates that the doublet peak represents two epimeric forms of KNI-272 (see Fig. 1B), oxidized at the S-methyl cysteine residue (either R or S) that are generated after treatment of KNI-272 with H2O2.
FIG. 1.
RP-HPLC analysis and chemical structures for KNI-272 and its oxidized forms generated by treatment with hydrogen peroxide. (Top) RP-HPLC tracing of KNI-272 after treatment with 10 mM H2O2. KNI-272 and the oxidized products, P1 and P2, are indicated in the RP-HPLC tracing. Structures: A, KNI-272; B, KNI-272 oxidized at the sulfur in the S-methyl cysteine moiety; C, KNI-272 oxidized on the sulfur of the S-methyl cysteine residue and on the sulfur in the thioproline ring.
FIG. 2.
Schematic diagram indicating the important fragmentation paths for fragmentation of KNI-272 after MS/MS analysis that allowed us to assign the sites of oxidation. Path a provides information for the determination of the oxidation state of the thioproline. Path b provides information for the determination of the oxidation state of the S-methyl cysteine. Numbers in parentheses represent the expected molecular weights if the sulfur in the fragment is oxidized.
FIG. 3.
RP-HPLC analysis of purified oxidized KNI-272 (S-methyl cysteine sulfoxide) before and after treatment with MsrA. Oxidized KNI-272 was purified by RP-HPLC, incubated in the presence of 20 mM DTT, and then treated with or without MsrA as described in Materials and Methods. After treatment, the sample was analyzed by RP-HPLC. Curves: ——, without MsrA; - - -, with MsrA. The locations of oxidized and unoxidized KNI-272 are indicated.
The other oxidation products of KNI-272 after treatment with H2O2 and represented by the four closely eluting peaks on RP-HPLC were also subjected to MS/MS analysis. Each of these peaks was 32 Da greater than KNI-272 (M+H = 700). The fragmentation results from MS/MS analysis were consistent with these four peaks representing KNI-272 oxidized at both the S-methyl cysteine residue and the thioproline ring. There are four possible forms of P2 which likely represent the four peaks: the R and S sulfoxides on S-methyl cysteine and the R and S sulfoxides on the thioproline, thus generating the RR, RS, SS, and SR sulfoxide forms of P2 (Fig. 1C). Interestingly, hydrogen peroxide treatment of KNI-272 did not generate KNI-272 oxidized solely on the thioproline ring.
KNI-272 is metabolized by M/M but not by T cells.
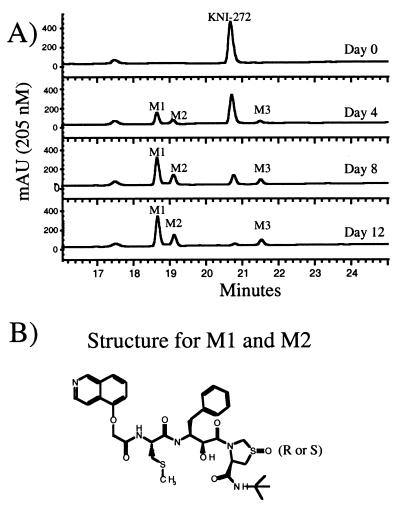
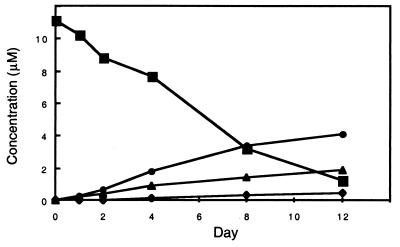
In the presence of M/M, the concentration of KNI-272, but not those of the other inhibitors, decreased steadily over the 12-day period tested. During this time, there was a steady increase in the concentration of three metabolites (Fig. 4). Data from several experiments in which ca. 5 million M/M (the adherent cells) were cultured in 8 ml of medium indicated that the concentration of the parent compound, KNI-272, decreased 30 to 50% by day 4 from the initial concentration of 10 μM and was barely detectable after 12 days (Fig. 4). Considering the relatively small volume occupied by the macrophages, relative to the total volume of the cultures, this rate of metabolism of KNI-272 by the cells appears to be substantial. We hypothesized that because of this intracellular metabolism of KNI-272, the concentration of unmetabolized drug in the cells would be less than in the media during the culture period. However, we were unable to successfully measure the intracellular concentrations of drug due to the large number of M/M required for the analysis.
FIG. 4.
RP-HPLC analysis of medium extracts from KNI-272-treated M/M at different time points. (A) The peaks corresponding to KNI-272 and metabolites M1, M2, and M3 are indicated. (B) Structure of M1 and M2 as determined by MS. M1 and M2 were produced after addition of KNI-272 to macrophages. KNI-272 is oxidized at the thioproline ring in both M1 and M2 but in different epimeric configurations.
As determined by RP-HPLC analysis, the initial decrease in KNI-272 corresponded with the appearance of two new metabolites (designated here as M1 and M2), with both eluting earlier than native KNI-272 (Fig. 4). In addition, M1 and M2 retained the characteristic UV spectrum of KNI-272 with absorbance maxima at 220, 260, and 340 nm showing that there were no modifications near the aromatic rings. M1 had a retention time of 18.6 min and a mass of 683, which is equivalent to the mass of KNI-272 containing an additional oxygen atom. LC/MS/MS analysis of M1 identified this peak as corresponding to KNI-272 oxidized only at the sulfur residue of the thioproline ring because of its loss of 232 Da via path a and its loss of 48 Da via path b (see Fig. 2). M2, which had a retention time of 19.1 min, also had a mass of 683 and had the same fragmentation pattern as M1, again indicating it to be KNI-272 oxidized solely at the sulfur residue in the thioproline ring. This indicates that both the R and S configurations at the thioproline sulfoxide epimeric center were produced when KNI-272 was exposed to M/M cultures. KNI-272 oxidized at the S-methyl cysteine residue (P1) was detected at low levels by LC/MS/MS and eluted between the two thioproline oxidized products on RP-HPLC. Surprisingly, the S-methyl cysteine oxidized form did not represent a major metabolite of KNI-272, even though this was the predominant form generated after hydrogen peroxide treatment. A third major metabolite (designated M3), which eluted later than KNI-272 (Fig. 4) with a retention time of 21.5 min, also had a mass of 683, again indicating an oxidized form of KNI-272. LC/MS/MS analysis showed a loss of 216 Da via path a (Fig. 2), indicating that oxidation did not occur on the thioproline. While we were not able to ascertain by MS/MS the site of oxidation in M3, the later retention time compared to the native KNI-272 is indicative of the ring being oxidized adjacent to the nitrogen generating a neutral, less-polar form of KNI-272. In addition, the loss of 64 Da via path b, which is characteristic of S-methyl cysteine oxidation, was absent. M3 had an altered UV spectrum compared to KNI-272 with a shift in absorbance maxima in the UV from 245 nm to 270 nm. This shift in the absorbance maxima would be consistent with KNI-272 oxidized on the isoquinol moiety. To determine whether the PIs were oxidized in cells that are susceptible to HIV infection, we treated peripheral blood T cells, H9, H9/HTLVIIIB, and Jurkat T cells or human M/M with the different PIs and then analyzed the media for the production of metabolites. The cells were incubated in the presence of each protease inhibitor, and aliquots of the cell culture media were taken at different time points and analyzed by RP-HPLC. We did not detect metabolites of any of the inhibitors after more than 10 days of incubation with peripheral blood T cells, H9, H9/HTLVIIIB, or Jurkat cells (data not shown). In addition, there was no measurable decrease in the concentration of each tested inhibitor, including KNI-272, after a 10-day incubation, suggesting that these compounds were highly stable in these cell types.
Time course of M1, M2, and M3 production by M/M after treatment with KNI-272.
In the presence of M/M, the metabolites of KNI-272, M1 and M2, were detected within 2 days and steadily increased up to day 12 (Fig. 5). M1 was the major metabolite, followed by M2 and M3. M3 was first detected on day 4 and steadily increased over 12 days. By day 12 >90% of the KNI-272 was metabolized to other products by the M/M. By day 12, the metabolites M1, M2, and M3 accounted for a little more than 50% of the original KNI-272. This suggests that further metabolism of the oxidized forms of KNI-272 likely occurs in M/M.
FIG. 5.
Time course of KNI-272 metabolism in media by M/M. Approximately 5 million M/M (the adherent cells from 30 million plated cells) were cultured in 8 ml of medium, to which was added KNI-272 at a final concentration of 10 μM. Small aliquots of the media were periodically removed and analyzed for KNI-272 and metabolites M1, M2, and M3 by RP-HPLC as described in Materials and Methods. Symbols: ▪, KNI-272; •, M1; ▴, M2; and ⧫, M3.
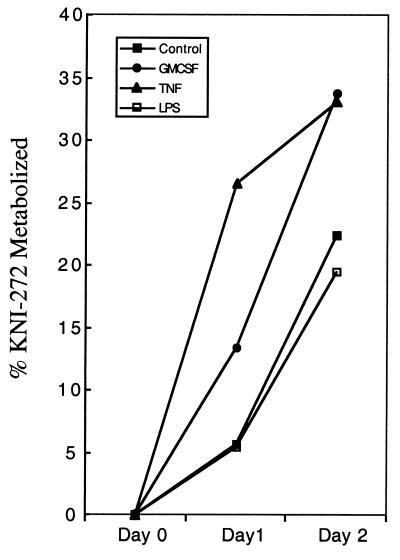
We tested a number of conditions reported to activate the oxidative burst in cells to determine whether the level of oxidative stress might influence the extent at which M/M metabolized KNI-272. HIV infection is thought to increase oxidative stress (27), so we first compared the ability of uninfected and HIV-infected M/M to metabolize KNI-272 to M1, M2, and M3. However, there was little difference in the rate or extent of KNI-272 metabolism between the two conditions tested (data not shown). We also sought to determine whether GM-CSF, TNF-α, and LPS affected the extent of KNI-272 metabolism since these agents have been shown to elicit oxidative stress in cells (9, 32, 33). After 1 day, untreated macrophages metabolized ca. 6% of the KNI-272 added (Fig. 6). However, the percentage of KNI-272 metabolized increased to 13 and 26% 1 day after addition of KNI-272 to the cells treated with 10 ng of GM-CSF or 1 ng of TNF-α/ml, respectively (Fig. 6). LPS at 25 ng/ml or higher, however, did not increase the percentage of KNI-272 metabolized and was found to decrease the rate of metabolism two or more days after treatment (Fig. 6 and data not shown).
FIG. 6.
Effect of GM-CSF, TNF-α, or LPS on KNI-272 metabolism in macrophages. Cultures of M/M in medium to which KNI-272 had been added at a final concentration of 10 μM were cultured as as in Fig. 5. Aliquots (250 ml) were withdrawn at days 0, 1, and 2, and the concentration of the remaining KNI-272 was determined as described in Materials and Methods.
Comparison of the inhibition of HIV-1 protease activity by KNI-272 and its metabolites derived from M/M.
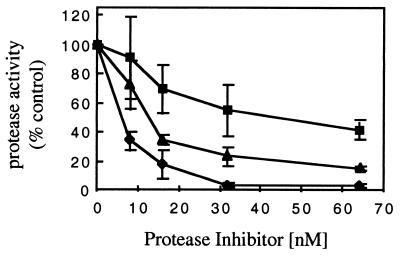
To determine whether the oxidation of KNI-272 by M/M affected its potency as an HIV-1 protease inhibitor, we purified native KNI-272 and the major oxidized forms (M1 and M2) by RP-HPLC from medium extracts of treated M/M. The identity of the compounds was first verified by MS analysis, the concentration was determined by RP-HPLC analysis, and then the purified compounds were tested in a standard HIV-1 protease assay. As shown in Fig. 7, the inhibitory activity of M1 and M2 was impaired compared to that of native KNI-272. The calculated 50% inhibitory concentrations for KNI-272, M1, and M2, were 8, 45, and 16 nM, respectively. KNI-272 at 30 nM decreased protease activity by >90% compared to decreases of 50 and 75% with 30 nM M1 and M2, respectively (Fig. 7).
FIG. 7.
HIV-1 protease activity in the presence of increasing concentrations of KNI-272 and M1 and M2 (KNI-272 metabolites). Each compound was separated and purified by RP-HPLC. The compounds were dissolved in DMSO and then tested for inhibitory activity toward the HIV-1 protease as described in Materials and Methods. Each point represents the mean of three independent measurements ± the standard deviation. Symbols: ⧫, KNI-272; ▪, M1; and ▴, M2.
DISCUSSION
The ability of various HIV-1 PIs to block HIV-1 replication in infected M/M is decreased compared to that for infected T cells (28). In this study, we investigated the degree to which PIs were metabolized by peripheral blood T cells, T-cell lines, and M/M to determine whether this might account for the observed differences in antiviral activity. We were particularly interested in the susceptibility of PIs to oxidative metabolism since M/M are known to generate a number of ROIs that could modify the inhibitors. Although some of the inhibitors (saquinavir, JE-2147, and KNI-272) contained functional groups that were susceptible to oxidation in vitro with H2O2, only one of these, KNI-272, was substantially affected when treated with H2O2 or added to M/M. KNI-272 was readily oxidized at the S-methyl cysteine moiety (R and S configurations of S-methyl cysteine sulfoxide) after hydrogen peroxide treatment, while oxidation of KNI-272 in the presence of M/M occurred predominantly at the thioproline ring, once again without stereospecificity. In addition, the primary macrophage-derived metabolites of KNI-272, M1 and M2, were less active as inhibitors of the HIV-1 protease compared to KNI-272. In contrast, KNI-272 and the other inhibitors were not metabolized when they were incubated with peripheral blood T cells or various T-cell lines for up to 12 days. Therefore, differences in oxidative metabolism by M/M compared to T cells could explain the reduced potency of KNI-272 in M/M, although there are apparently other explanations for the decrease in potency of the other PIs when tested in M/M.
The mechanism by which KNI-272 is oxidized and/or metabolized by M/M was not directly addressed in this study. Kiriyama et al. found three major metabolites when they measured the metabolism of KNI-272 in rat liver microsomes exposed to KNI-272 (15). The production of these three metabolites was NADPH dependent and was inhibited in the presence of the P450 monooxygenase inhibitor, ketoconazole. These metabolites which were not identified were presumably oxidized forms of KNI-272 and may be similar if not identical to the ones that we have identified here by LC/MS analysis. M/M may metabolize KNI-272 through a P450 monooxygenase pathway or by an NADPH oxidase system. Interestingly, the percentage of KNI-272 metabolized after just 1 day was increased from two- to sixfold when the cells were treated with GM-CSF or TNF-α. This suggests that activating oxidative stress with various agents may further increase the extent of metabolism of protease inhibitors. These agents are known to activate NADPH oxidase and this would suggest that superoxide produced from this system may be involved in the metabolism of KNI-272.
The S-methyl cysteine moiety of KNI-272 was found to be more sensitive to hydrogen peroxide oxidation than the thioproline moiety. Indeed, we even detected low levels of oxidation of S-methyl cysteine in stored samples of KNI-272 (data not shown). However, the S-methyl cysteine oxidized form of KNI-272 did not accumulate to any great degree when KNI-272 was added to M/M. In contrast, the thioproline group was substantially oxidized. Interestingly, thioproline itself is a known antioxidant shown to improve macrophage function. Therefore, the thioproline within KNI-272 probably reacts with ROIs in a similar way.
We found that M/M oxidatively modified KNI-272 while other cell types did not. This is consistent with other studies indicating that the oxidative environment of M/M is different from that of T cells and other cell types (16). Such differences in the oxidative environment can also affect HIV-1 replication. We previously reported that the activity of HIV-1 protease can be affected by oxidative conditions, and we suggested (28) that the oxidative environment of M/M may favor the production of a more active glutathionylated form of the HIV-1 protease (6, 7). These effects of oxidative conditions on HIV-1 protease may provide a reason why HIV-1-infected M/M require higher concentrations of PIs to reach similar levels of inhibition compared to HIV-1-infected T cells. In addition, the present observation that M/M and T cells can differentially metabolize a potent PI may provide another possible mechanism for decreased PI activity in M/M. It is unclear whether the metabolism of KNI-272 under oxidative conditions contributed to the relatively poor activity of this compound in clinical testing (12). While the decline in KNI-272 concentration in the media in cultures of M/M (ca. 5 million cells in 8 ml of medium) was relatively slow, it should be remembered that the total intracellular volume of the M/M in the cultures was only a small fraction of the total volume of media. We were unable to assay the intracellular concentration of KNI-272 in the M/M but would expect these levels to be substantially lower than that in the medium. Thus, metabolism of susceptible protease inhibitors is likely to result in lower intracellular levels in M/M than in serum, and the results of the present study suggest that when developing novel antiviral therapies one should consider the possibility that metabolism by target cells under oxidative conditions may lead to altered activity. In particular, it will be of importance to assess the fate of potentially useful antiviral compounds in M/M and M/M exposed to various immune activating agents.
Acknowledgments
We thank Hiroaki Mitsuya and the Japan Energy Corporation for providing the PIs used in this study. We thank Hiroaki Mitsuya for helpful discussions.
Funding for this study was provided in part by a Cooperative Research and Development Agreement between the National Cancer Institute and the Japan Energy Corporation.
REFERENCES
- 1.Aquaro, S., E. Balestra, A. Cenci, M. Francesconi, R. Calio, and C. F. Perno. 1997. HIV infection in macrophage: role of long-lived cells and related therapeutical strategies. J. Biol. Regul. Homeostatic Agents 11:69–73. [PubMed] [Google Scholar]
- 2.Baier-Bitterlich, G., D. Fuchs, and H. Wachter. 1997. Chronic immune stimulation, oxidative stress, and apoptosis in HIV infection. Biochem. Pharmacol. 53:755–763. [DOI] [PubMed] [Google Scholar]
- 3.Balani, S. K., B. H. Arison, L. Mathai, L. R. Kauffman, R. R. Miller, R. A. Stearns, I. W. Chen, and J. H. Lin. 1995. Metabolites of L-735,524, a potent HIV-1 protease inhibitor, in human urine. Drug Metab. Dispos. 23:266–270. [PubMed] [Google Scholar]
- 4.Bragman, K. 1996. Saquinavir: an HIV proteinase inhibitor. Adv. Exp. Med. Biol. 394:305–317. [DOI] [PubMed] [Google Scholar]
- 5.Crowe, S. M., and S. Sonza. 2000. HIV-1 can be recovered from a variety of cells including peripheral blood monocytes of patients receiving highly active antiretroviral therapy: a further obstacle to eradication. J. Leukoc. Biol. 68:345–350. [PubMed] [Google Scholar]
- 6.Davis, D. A., K. Dorsey, P. T. Wingfield, S. J. Stahl, J. Kaufman, H. M. Fales, and R. L. Levine. 1996. Regulation of HIV-1 protease activity through cysteine modification. Biochemistry 35:2482–2488. [DOI] [PubMed] [Google Scholar]
- 7.Davis, D. A., F. M. Newcomb, D. W. Starke, D. E. Ott, J. J. Mieyal, and R. Yarchoan. 1997. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. J. Biol. Chem. 272:25935–25940. [DOI] [PubMed] [Google Scholar]
- 8.Denissen, J. F., B. A. Grabowski, M. K. Johnson, A. M. Buko, D. J. Kempf, S. B. Thomas, and B. W. Surber. 1997. Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug Metab. Dispos. 25:489–501. [PubMed] [Google Scholar]
- 9.Dusi, S., M. Donini, D. Lissandrini, P. Mazzi, V. D. Bianca, and F. Rossi. 2001. Mechanisms of expression of NADPH oxidase components in human cultured monocytes: role of cytokines and transcriptional regulators involved. Eur. J. Immunol. 31:929–938. [DOI] [PubMed] [Google Scholar]
- 10.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219. [DOI] [PubMed] [Google Scholar]
- 11.Humphrey, R. W., A. Ohagen, D. A. Davis, T. Fukazawa, H. Hayashi, S. Höglund, H. Mitsuya, and R. Yarchoan. 1997. Removal of human immunodeficiency virus type 1 (HIV-1) protease inhibitors from preparations of immature HIV-1 virions does not result in an increase in infectivity or the appearance of mature morphology. Antimicrob. Agents Chemother. 41:1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphrey, R. W., K. M. Wyvill, B. Y. Nguyen, L. E. Shay, D. R. Kohler, S. M. Steinberg, T. Ueno, T. Fukasawa, M. Shintani, H. Hayashi, H. Mitsuya, and R. Yarchoan. 1999. A phase I trial of the pharmacokinetics, toxicity, and activity of KNI- 272, an inhibitor of HIV-1 protease, in patients with AIDS or symptomatic HIV infection. Antivir. Res. 41:21–33. [DOI] [PubMed] [Google Scholar]
- 13.Kageyama, S., T. Mimoto, Y. Murakawa, M. Nomizu, H. Ford, Jr., T. Shirasaka, S. Gulnik, J. Erickson, K. Takada, H. Hayashi, S. Broder, Y. Kiso, and H. Mitsuya. 1993. In vitro anti-human immunodeficiency virus (HIV) activities of transition state mimetic HIV protease inhibitors containing allophenylnorstatine. Antimicrob. Agents Chemother. 37:810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kameoka, M., T. Kimura, and K. Ikuta. 1993. Superoxide enhances the spread of HIV-1 infection by cell-to-cell transmission. FEBS Lett. 331:182–186. [DOI] [PubMed] [Google Scholar]
- 15.Kiriyama, A., T. Nishiura, H. Yamaji, and K. Takada. 1999. Metabolic characterization of a tripeptide human immunodeficiency virus type 1 protease inhibitor, KNI-272, in rat liver microsomes. Antimicrob. Agents Chemother. 43:549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight, J. A. 2000. Review: free radicals, antioxidants, and the immune system. Ann. Clin. Lab. Sci. 30:145–158. [PubMed] [Google Scholar]
- 17.Kobzik, L., J. J. Godleski, and J. D. Brain. 1990. Oxidative metabolism in the alveolar macrophage: analysis by flow cytometry. J. Leukoc. Biol. 47:295–303. [PubMed] [Google Scholar]
- 18.Kumar, G. N., V. Jayanti, R. D. Lee, D. N. Whittern, J. Uchic, S. Thomas, P. Johnson, B. Grabowski, H. Sham, D. Betebenner, D. J. Kempf, and J. F. Denissen. 1999. In vitro metabolism of the HIV-1 protease inhibitor ABT-378: species comparison and metabolite identification. Drug Metab. Dispos. 27:86–91. [PubMed] [Google Scholar]
- 19.Lambert, D. M., S. R. Petteway, Jr., C. E. McDanal, T. K. Hart, J. J. Leary, G. B. Dreyer, T. D. Meek, P. J. Bugelski, D. P. Bolognesi, B. W. Metcalf, and T. J. Matthews. 1992. Human immunodeficiency virus type 1 protease inhibitors irreversibly block infectivity of purified virions from chronically infected cells. Antimicrob. Agents Chemother. 36:982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambotte, O., Y. Taoufik, M. G. de Goer, C. Wallon, C. Goujard, and J. F. Delfraissy. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 23:114–119. [DOI] [PubMed] [Google Scholar]
- 21.Lea, A. P., and D. Faulds. 1996. Ritonavir. Drugs 52:541–548. [DOI] [PubMed] [Google Scholar]
- 22.Mimoto, T., R. Kato, H. Takaku, S. Nojima, K. Terashima, S. Misawa, T. Fukazawa, T. Ueno, H. Sato, M. Shintani, Y. Kiso, and H. Hayashi. 1999. Structure-activity relationship of small-sized HIV protease inhibitors containing allophenylnorstatine. J. Med. Chem. 42:1789–1802. [DOI] [PubMed] [Google Scholar]
- 23.Moskovitz, J., B. S. Berlett, J. M. Poston, and E. R. Stadtman. 1997. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc. Natl. Acad. Sci. USA 94:9585–9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskovitz, J., J. M. Poston, B. S. Berlett, N. J. Nosworthy, R. Szczepanowski, and E. R. Stadtman. 2000. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 275:14167–14172. [DOI] [PubMed] [Google Scholar]
- 25.Nathan, C. F., and S. Tsunawaki. 1986. Secretion of toxic oxygen products by macrophages: regulatory cytokines and their effects on the oxidase. Ciba Found. Symp. 118:211–230. [DOI] [PubMed] [Google Scholar]
- 26.Nottet, H. S., B. S. van Asbeck, L. de Graaf, N. M. de Vos, M. R. Visser, and J. Verhoef. 1994. Role for oxygen radicals in self-sustained HIV-1 replication in monocyte-derived macrophages: enhanced HIV-1 replication by N-acetyl-l-cysteine. J. Leukoc. Biol. 56:702–707. [DOI] [PubMed] [Google Scholar]
- 27.Pace, G. W., and C. D. Leaf. 1995. The role of oxidative stress in HIV disease. Free Radic. Biol. Med. 19:523–528. [DOI] [PubMed] [Google Scholar]
- 28.Perno, C. F., F. M. Newcomb., D. A. Davis, S. Aquaro, R. W. Humphrey, R. Calio, and R. Yarchoan. 1998. Relative antiviral efficacy of protease inhibitors in monocytes/macrophages acutely and chronically infected by human immunodeficiency virus. J. Infect. Dis. 178:413–422. [DOI] [PubMed] [Google Scholar]
- 29.Pietraforte, D., E. Tritarelli, U. Testa, and M. Minetti. 1994. gp120 HIV envelope glycoprotein increases the production of nitric oxide in human monocyte-derived macrophages. J. Leukoc. Biol. 55:175–182. [DOI] [PubMed] [Google Scholar]
- 30.Popovic, M., M. G. Sarngadharan, E. Read, and R. C. Gallo. 1984. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224:497–500. [DOI] [PubMed] [Google Scholar]
- 31.Rayner, M. M., B. C. Cordova, R. P. Meade, P. E. Aldrich, P. K. Jadhav, Y. Ru, and P. Y. Lam. 1994. DMP 323, a nonpeptide cyclic urea inhibitor of human immunodeficiency virus (HIV) protease, specifically and persistently blocks intracellular processing of HIV Gag polyprotein. Antimicrob. Agents Chemother. 38:1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roilides, E., C. A. Lyman, S. D. Mertins, D. J. Cole, D. Venzon, P. A. Pizzo, S. J. Chanock, and T. J. Walsh. 1996. Ex vivo effects of macrophage colony-stimulating factor on human monocyte activity against fungal and bacterial pathogens. Cytokine 8:42–48. [DOI] [PubMed] [Google Scholar]
- 33.Sasada, M., M. J. Pabst, and R. B. Johnston, Jr. 1983. Activation of mouse peritoneal macrophages by lipopolysaccharide alters the kinetic parameters of the superoxide-producing NADPH oxidase. J. Biol. Chem. 258:9631–9635. [PubMed] [Google Scholar]
- 34.Schrager, L. K., and M. P. D’Souza. 1998. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA 280:67–71. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz, K. B. 1996. Oxidative stress during viral infection: a review. Free Radic. Biol. Med. 21:641–649. [DOI] [PubMed] [Google Scholar]
- 36.Vella, S., and M. Floridia. 1998. Saquinavir. Clinical pharmacology and efficacy. Clin. Pharmacokinet. 34:189–201. [DOI] [PubMed] [Google Scholar]
- 37.Yeh, K. C., P. J. Deutsch, H. Haddix, M. Hesney, V. Hoagland, W. D. Ju, S. J. Justice, B. Osborne, A. T. Sterrett, J. A. Stone, E. Woolf, and S. Waldman. 1998. Single-dose pharmacokinetics of indinavir and the effect of food. Antimicrob. Agents Chemother. 42:332–338. (Erratum, 42: 1308.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimura, K., R. Kato, K. Yusa, M. F. Kavlick, V. Maroun, A. Nguyen, T. Mimoto, T. Ueno, M. Shintani, J. Falloon, H. Masur, H. Hayashi, J. Erickson, and H. Mitsuya. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. USA 96:8675–8680. [DOI] [PMC free article] [PubMed] [Google Scholar]