Abstract
Six clinical CTX-M-producing isolates of the family Enterobacteriaceae were detected between 1999 and 2000 in different French hospitals. Two strains produced CTX-M-1 and CTX-M-3 and four strains produced CTX-M-14, a mutant Ala-231→Val of CTX-M-9. A putative transposable element, ISEcp-1, was located 43 bp upstream of all the blaCTX-M genes. Two CTX-M-14-encoding plasmids exhibited similar restriction patterns. The CTX-M-1- and CTX-M-3-encoding plasmids were related to the CTX-M-1- and CTX-M-3-encoding plasmids previously reported in 1990 in France and in 1998 in Poland, respectively.
The early extended-spectrum β-lactamases (ESBLs) arose as the result of a few amino acid substitutions from the common plasmid-mediated TEM and SHV-1 β-lactamases. At the beginning of the 1990s, a new class A type of ESBL was characterized in the first reports of the CTX-M-1 (MEN-1) enzyme (2, 3). These CTX-M-type enzymes are typical ESBLs, characterized by much greater hydrolytic activity against cefotaxime than against ceftazidime. Thus, they confer a high level of resistance to cefotaxime and have only marginal effects on the MIC of ceftazidime.
The family of CTX-M-type ESBLs comprises at least 12 members belonging to four major phylogenetic branches on the basis of their amino acid sequence similarities (8): the CTX-M-1 (MEN-1) branch (CTX-M-1, CTX-M-3, and CTX-M-10), the CTX-M-2 branch (CTX-M-2, Toho-1, and CTX-M-4 to CTX-M-6), the CTX-M-9 branch (CTX-M-9 and Toho-2), and the CTX-M-8 branch. These enzymes have been reported in several species of the family Enterobacteriaceae and in Vibrio cholerae serovar El Tor and have been isolated in three main geographical areas: South America (7, 8; M. Galas, F. Pasteran, R. Melano, A. Petroni, G. Lopez, A. Corso, A. Rossi, and Whonet Collaborative Group, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-109, p. 201, 1998), Eastern Europe (11, 12, 13, 20), and Japan (14, 19).
We describe six clinical strains of the family Enterobacteriaceae (Table 1) which were collected in different French hospitals on the basis of their positive synergy test results (16) and their higher levels of resistance to cefotaxime than ceftazidime (MICs, 16 to 128 and 2 to 8 μg/ml, respectively). Escherichia coli CF-1 and Enterobacter cloacae CF-2 were isolated at the teaching hospital of Clermont-Ferrand, France, from the urine of a patient hospitalized in 1999 and from a pulmonary sample of a patient admitted in 2000, respectively. E. coli Mnt-1 and Klebsiella pneumoniae Mnt-2 were isolated from blood and stool samples, respectively, of a Vietnamese child admitted to Montpellier hospital, Montpellier, France, in 1999. E. coli Roa-1 was isolated in Roannes hospital, Roannes, France, in 1999 from blood, and E. cloacae Ver-1 was isolated in Versailles hospital, Versailles, France, in 1999 from a urine sample (10). CTX-M-1-producing strain MEN (2) and CTX-M-3-encoding plasmid A1 (20) were used as references.
TABLE 1.
Clinical strains and plasmids used in the study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Clinical strains | ||
| E. coli CF-1 | Clinical strain harboring natural plasmid pCF-1 (Clermont-Ferrand, France, 1999) | This study |
| E. cloacae CF-2 | Clinical strain harboring natural plasmid pCF-2 (Clermont-Ferrand, France, 2000) | This study |
| E. coli Roa-1 | Clinical strain harboring natural plasmid pRoa-1 (Roanne, France, 1999) | This study |
| E. coli Mnt-1 | Clinical strain harboring natural plasmid pMnt-1 (Montpellier, France, 1999) | This study |
| K. pneumoniae Mnt-2 | Clinical strain producing CTX-M-14 (Montpellier, France, 1999) | This study |
| E. cloacae Ver-1 | Clinical strain harboring natural plasmid pVer-1 (Versailles, France, 1999) | 10 |
| E. coli MEN | Clinical strain harboring natural plasmid pMEN (Paris, France, 1999) | 2 |
| Plasmids | ||
| pCF-1 | 150-kb natural plasmid from E. coli strain CF-1 containing the blaCTX-M-14 gene | This study |
| pCF-2 | 55-kb natural plasmid from E. cloacae strain CF-2 containing the blaCTX-M-1 gene | This study |
| pMnt-1 | 110-kb natural plasmid from E. coli strain Mnt-1 containing the blaCTX-M-14 gene | This study |
| pRoa-1 | 120-kb natural plasmid from E. coli strain Roa-1 containing the blaCTX-M-14 gene | This study |
| pVer-1 | 180-kb natural plasmid from E. cloacae strain Ver-1 containing the blaCTX-M-3 gene | 10 |
| pMEN | 40-kb natural plasmid from E. coli strain MEN containing the blaCTX-M-1 gene | 2 |
| A1 | 110-kb natural A1 plasmid containing the blaCTX-M-3 gene | 20 |
| pClCF-1 | 13-kb recombinant plasmid of pBK-CMV containing the blaCTX-M-14 gene | This study |
| pBK-CMV | Phagemid vector; kanamycin resistance phenotype | Stratagene, La Jolla, Calif. |
Analytical isoelectric focusing was performed as described previously (7). The following β-lactamases with known pIs were used as standards: TEM-1 (pI 5.4), SHV-1 (pI 7.6), and CTX-M-1 (pI 8.4). All strains tested produced an enzyme of pI 5.4, associated with a second β-lactamase of alkaline pI: pI 8.4 for strains Ver-1 and CF-2 and pI 7.9 for strains CF-1, Mnt-1, Mnt-2, and Roa-1.
PCR TEM and direct sequencing of the PCR product (23) identified the β-lactamase of pI 5.4 as being the TEM-1 penicillinase. No PCR products were obtained with primers specific for blaSHV genes. Positive amplifications were obtained with primers CTX-MA (5′-CGCTTTGCGATGTGCAG-3′) and CTX-MB (5′-ACCGCGATATCGTTGGT-3′), which amplified 550-bp internal fragments of the blaCTX-M genes (8). From the results obtained for the sequence of this 550-bp fragment, the complete blaCTX-M open reading frames (ORFs) of strains Ver-1 and CF-2 were amplified and sequenced with primers specific for the blaCTX-M-1 and blaCTX-M-3 genes (primers CTX-M1A [5′-CTTCCAGAATAAGGAATC-3′] and CTX-M-1B [5′-CCGTTTCCGCTATTACAA-3′]; temperature of annealing, 52°C). The sequence obtained had 100% identity with those of blaCTX-M-3 (13) and blaCTX-M-1 (4) for strains Ver-1 and CF-2, respectively. The blaCTX-M gene of strain CF-1 was cloned in plasmid vector pBK-CMV (Stratagene, La Jolla, Calif.) by partial digestion of its plasmid content with Sau3A, as reported previously (7, 22). A 13-kb recombinant plasmid, designated pClCF-1, was sequenced with primers CTX-M-A and CTX-M-B, which are specific for the internal blaCTX-M sequence. The sequence of the blaCTX-M gene was identical to that of blaCTX-M-14 sequenced by another group (GenBank accession number AF252622). Strains Mnt-1, Mnt-2, and Roa-1 harbored the same blaCTX-M-14 gene.
The deduced amino acid sequence of the CTX-M-14 β-lactamase differed from that of CTX-M-9 (21) by the amino acid substitution Ala-231→Val (numbering of Ambler et al. [1]). Position 231 is located at the beginning of CTX-M beta sheet β3 (15), which contains at position 234 the KTG conserved element of class A β-lactamases. Val is usually encountered at positions 231 of CTX-M enzymes.
The CTX-M-14 enzyme was extracted from recombinant E. coli DH5α(pClCF-1) by sonification and was purified to homogeneity as described previously (7). The purities of the CTX-M extracts (≥97%) were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described previously (7).
The kinetic constants of CTX-M-14 were obtained by a computerized microacidimetric method described elsewhere (18) and were compared with those of CTX-M-9 (Table 2). The concentrations of the inhibitors (clavulanate and tazobactam) required to inhibit enzyme activity by 50% (IC50s) were determined with penicillin G as described previously (7). IC50s and Ki values were monitored with penicillin G (200 mM) as the reporter substrate. CTX-M-14 and CTX-M-9 had similar kinetic constants (Table 2). High catalytic efficiencies (kcat,/km values) were observed against penicillin G (11.8 to 14.5 μM−1 · s−1), amoxicillin (4.5 to 5 μM−1 · s−1), piperacillin (4.2 to 5.5 μM−1 · s−1), cephalothin (15.4 to 20 μM−1 · s−1), and cefuroxime (7 to 8 μM−1 · s−1). As reported previously for the other CTX-M enzymes, CTX-M-14 and CTX-M9 had better catalytic activities against methoximino cephems such as cefuroxime, cefotaxime, and cefpirome (kcat values, 320 to 950 s−1) than against carboxylic propyloximino β-lactams such as ceftazidime and aztreonam (kcat values, 2 to 10 s−1). CTX-M-14 and CTX-M-9 were susceptible to the β-lactam inhibitors clavulanate (IC50s, 0.033 and 0.036 μM, respectively), tazobactam (IC50s, 0.008 and 0.007 μM, respectively), and, to a lesser extent, sulbactam (IC50s, 3.4 and 3.0 μM, respectively).
TABLE 2.
Substrate profile of CTX-M-14 and CTX-M-9 β-lactamases
| Substrate | CTX-M-14
|
CTX-M-9
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | |
| Penicillin G | 290 | 20 | 14.5 | 295 | 25 | 11.8 |
| Amoxicillin | 100 | 20 | 5.0 | 90 | 20 | 4.5 |
| Ticarcillin | 45 | 24 | 1.9 | 60 | 35 | 1.7 |
| Piperacillin | 200 | 48 | 4.2 | 110 | 20 | 5.5 |
| Cephalothin | 2,700 | 175 | 15.4 | 3,000 | 150 | 20.0 |
| Cefuroxime | 320 | 40 | 8.0 | 350 | 50 | 7.0 |
| Cefotaxime | 415 | 130 | 3.2 | 450 | 120 | 3.7 |
| Cefpirome | 940 | 1,000 | 0.9 | 950 | 800 | 1.2 |
| Aztreonam | 10 | 200 | 0.05 | 10 | 220 | 0.04 |
| Ceftazidime | 3 | 610a | 0.005 | 2 | 600a | 0.003 |
Km values were determined as the Ki values by substrate competition with penicillin G.
Five E. coli transconjugants of the six clinical strains were selected on agar containing cefotaxime (2 μg/ml) and rifampin (30 μg/ml). All these transconjugants produced the CTX-M enzyme associated with the TEM-1 penicillinase.
Table 3 lists the MICs of the β-lactams alone and in combination with β-lactamase inhibitors for the CTX-M-producing transconjugants. They were determined by a dilution method on Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes la Coquette, France) with an inoculum of 104 CFU per spot. The antibiotics were provided as powders by SmithKline Beecham Pharmaceuticals (amoxicillin, ticarcillin, and clavulanate); Lederle Laboratories (piperacillin and tazobactam); Eli Lilly, Paris, France (cephalothin); Roussel-Uclaf (cefotaxime and cefpirome); Glaxo Wellcome Research and Development (ceftazidime); and Bristol-Myers Squibb (cefepime). The E. coli transconjugants exhibited a high level of resistance to amoxicillin, ticarcillin, cephalothin, and cefuroxime (MICs, >1,024 μg/ml). The MICs of cefotaxime (16 to 32 μg/ml) were 8- to 32-fold higher than those of ceftazidime (1 to 2 μg/ml) and 2- to 8-fold higher than those of aztreonam (4 to 8 μg/ml) and cefpirome (2 to 16 μg/ml). Clavulanate restored partially or totally the activities of the β-lactams. All strains were susceptible to associations of clavulanate and broad-spectrum cephalosporins (MICs, 0.06 to 0.12 μg/ml).
TABLE 3.
Comparison of β-lactam MICs for CTX-M-producing E. coli transconjugants
| MIC (μg/ml) for E. coli C600 with plasmid (pI):
|
|||||
|---|---|---|---|---|---|
| pCF-2a (5.4, 8.4) | pVer-1b (5.4, 8.4) | pCF-1c (5.4, 7.9) | pMnt-1d (5.4, 7.9) | pRoa-1e (5.4, 7.9) | |
| Amoxicillin | 2,048 | >2,048 | >2,048 | >2,048 | >2,048 |
| Amoxicillin + CLAf | 8 | 16 | 16 | 32 | 32 |
| Ticarcillin | 2,048 | >2,048 | >2,048 | >2,048 | >2,048 |
| Ticarcillin + CLA | 8 | 64 | 32 | 32 | 64 |
| Piperacillin | 128 | 512 | 256 | 1,024 | 1,024 |
| Piperacillin + TZBg | 0.5 | 1 | 1 | 1 | 1 |
| Cephalothin | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 |
| Cephalothin + CLA | 8 | 8 | 8 | 8 | 8 |
| Cefuroxime | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 |
| Cefuroxime + CLA | 4 | 8 | 4 | 4 | 8 |
| Cefotaxime | 16 | 16 | 16 | 32 | 32 |
| Cefotaxime + CLA | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 |
| Cefpirome | 2 | 8 | 8 | 8 | 16 |
| Cefpirome + CLA | 0.03 | 0.06 | 0.06 | 0.03 | 0.03 |
| Ceftazidime | 1 | 2 | 1 | 1 | 2 |
| Ceftazidime + CLA | 0.12 | 0.25 | 0.12 | 0.12 | 0.12 |
| Aztreonam | 4 | 8 | 4 | 4 | 8 |
| Aztreonam + CLA | 0.06 | 0.12 | 0.06 | 0.06 | 0.06 |
Natural CTX-M-1-encoding plasmid of strain CF-2.
Natural CTX-M-3-encoding plasmid of strain Ver-1.
Natural CTX-M-14-encoding plasmid of strain CF-1.
Natural CTX-M-14-encoding plasmid of strain Mnt-1.
Natural CTX-M-14-encoding plasmid of strain Roa-1.
CLA, clavulanate at a fixed concentration of 2 μg/ml.
TZB, tazobactam at a fixed concentration of 4 μg/ml.
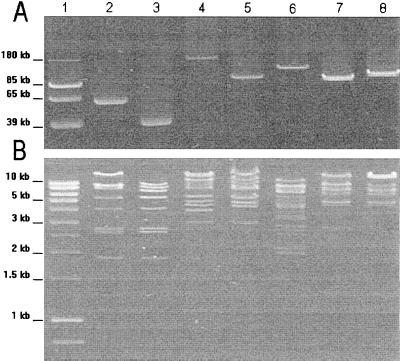
The plasmid contents of the transconjugants after extraction by the method of Kado and Liu (17) are shown in Fig. 1A. Plasmid sizes were determined by comparison with 39.5-, 65-, 85-, and 180-kb reference plasmids (6). The CTX-M-encoding plasmids were 55-kb plasmid pCF-2 for strain CF-2 (CTX-M-1), 180-kb plasmid pVer-1 for strain Ver-1 (CTX-M-3), 150-kb plasmid pCF-1 for strain CF-1 (CTX-M-14), 110-kb plasmid pMnt-1 for strain Mnt-1 (CTX-M-14), and 120-kb plasmid pRoa-1 for strain Roa-1 (CTX-M-14) (Table 1).
FIG. 1.
(A) Agarose (0.7%) electrophoresis of plasmid DNA from CTX-M-producing E. coli transconjugants. (B) Agarose (0.7%) electrophoresis of HpaI-restricted plasmid DNA from CTX-M-producing E. coli transconjugants. Lanes: 1, molecular size marker; 2, CTX-M-1-encoding plasmid pCF-2; 3, CTX-M-1-encoding plasmid pMEN (2); 4, CTX-M-3-encoding plasmid pVer-1; 5, CTX-M-3-encoding plasmid A1 (20); 6, CTX-M-14-encoding plasmid pCF-1; 7, CTX-M-14-encoding plasmid pMnt-1; 8, CTX-M-14-encoding plasmid pRoa-1.
These CTX-M-encoding plasmids were extracted by the method of Birnboim and Doly (5) and were digested with restriction endonucleases HpaI, EcoRI, and SalI (Boehringer Mannheim), according to the manufacturer’s recommendations. The restriction patterns are shown in Fig. 1B. The CTX-M-1-, CTX-M-3-, and CTX-M-9-type enzymes were encoded by plasmids with distinct restriction patterns. Of the three CTX-M-14-encoding plasmids, pCF-1 presented a distinct restriction pattern, whereas plasmids pRoa-1 and pMnt-1 exhibited related restriction patterns. CTX-M-1-encoding plasmid pCF-2 had a restriction pattern related to that of blaCTX-M-1-harboring plasmid pMEN of strain MEN, which was isolated in Paris, France, from an Italian patient in 1990 (2). Likewise, CTX-M-3-encoding plasmid pVer-1 was related to blaCTX-M-3-harboring plasmid A1, which was characterized in 1997 in Poland (20). Closely related plasmids encoding the same CTX-M ESBL were thus observed in different geographical areas (central France and the south of France for CTX-M-14; France and Poland for CTX-M-3) and at different times (in 1999 and 1987 for CTX-M-1). These findings suggest the establishment and diffusion of the CTX-M-encoding plasmids in Europe.
Genes encoding an identical CTX-M enzyme could be harbored by different plasmids, as observed for blaCTX-M-3 (20), blaCTX-M-8 (8), and blaCTX-M-14 (this work). The CTX-M-5 enzyme is encoded by genes located on plasmids in different isolates of the family Enterobacteriaceae (9) and in the chromosome of Kluyvera ascorbata. In addition, the putative insert sequence ISEcp-1 initially reported upstream of the plasmid-encoded CMY-type cephalosporinases (24) was observed upstream of the gene of the CTX-M-5 enzyme in the chromosome (C. Humeniuk, G. Arlet, R. Labia, P. Grimont, and A. Philippon, 20th Réunion Interdisciplinaire Chimiothér. Anti-Infectieuse, abstr. 20/C4, 2000). We used PCR and hybridization of the restricted plasmids to detect ISEcp-1 in our CTX-M-encoding plasmids: the ISEcp-1 probe and the amplifications were performed at an annealing temperature of 50°C with primers ISEcp1A (5′-AATCTAACATCAAATGCAGG-3′) and ISEcp-1B (5′-TTTTGCTGCAAGAAATACATA-3′), whose sequences are located in the transposase gene of ISEcp-1 (24). The specificities of the primers were confirmed by sequencing of the PCR products. ISEcp-1 was observed upstream of the CTX-M-encoding ORF in plasmids pCF1, pCF-2, pRoa-1, pVer-1, pMnt-1, and A1; but only one copy was detected. These results confirm the close association of ISEcp-1 with the blaCTX-M genes and suggest that the mobility of the blaCTX-M genes involves the ISEcp-1 element. If ISEcp-1 is really involved in the transfer of blaCTX-M genes, this element should be able to mobilize adjacent DNA sequences alone. Another possibility is that ISEcp-1 is part of an altered composite transposon from which the second ISEcp-1, initially located downstream of the blaCTX-M gene, was deleted, perhaps by a transposition event. Besides, the location of ISEcp1 only 43 bp upstream of blaCTX-M genes may contribute to blaCTX-M gene expression.
In this work, we have reported on six clinical strains of the family Enterobacteriaceae which produced CTX-M-1, CTX-M-3, and CTX-M-14 ESBLs. The study of their genetic support suggests that the CTX-M-encoding plasmids have become established in Europe and are spreading. In addition, the blaCTX-M genes were associated with ISEcp-1, which could be implicated in their spread and/or expression.
Acknowledgments
We thank Rolande Perroux, Marlène Jan, and Dominique Rubio for technical assistance. We are also grateful to M. Gniadkowski, Sera and Vaccines Central Research Laboratory, Warsaw, Poland, for the type A blaCTX-M-3-encoding plasmid and Jeffrey Watts for reading the English manuscript.
This work was supported in part by a grant from Ministère de l’Education Nationale de la Recherche et de la Technologie.
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J.-M. Frére, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Lévesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthélémy, M., J. Péduzzi, H. Bernard, C. Tancrede, and R. Labia. 1992. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15–22. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilhelm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158–163. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R., C. De Champs, D. Sirot, C. Chanal, R. Labia, and J. Sirot. 1999. Diversity of TEM mutants in Proteus mirabilis. Antimicrob. Agents Chemother. 43:2671–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, R., J. L. Sampaio, C. Chanal, D. Sirot, C. De Champs, J. L. Viallard, R. Labia, and J. Sirot. 2000. A novel class A extended-spectrum β-lactamase (BES-1) in Serratia marcescens isolated in Brazil. Antimicrob. Agents Chemother. 44:3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet, R., J. L. M. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucet-Populaire, F., R. Bonnet, J. C. Ghnassia, and J. Sirot. 2000. First isolation of a CTX-M-3-producing Enterobacter cloacae in France. Antimicrob. Agents Chemother. 44:3239–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gazouli, M., E. Tzelepi, A. Markogiannakis, N. J. Legakis, and L. S. Tzouvelekis. 1998. Two novel plasmid-mediated cefotaxime-hydrolyzing β-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol. Lett. 165:289–293. [DOI] [PubMed] [Google Scholar]
- 12.Gazouli, M., E. Tzelepi, S. V. Sidorenko, and L. S. Tzouvelekis. 1998. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob. Agents Chemother. 42:1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibuka, A., A. Taguchi, M. Ishiguro, S. Fushinobu, Y. Ishii, S. Kamitori, K. Okuyama, K. Yamaguchi, M. Konno, and H. Matsuzawa. 1999. Crystal structure of the E166A mutant of extended-spectrum β-lactamase Toho-1 at 1.8 Å resolution. J. Mol. Biol. 285:2079–2087. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867–878. [DOI] [PubMed] [Google Scholar]
- 17.Kado, C. I., and L. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labia, R., J. Andrillon, and F. Le Goffic. 1973. Computerized microacidimetric determination of β-lactamase Michaelis-Menten constants. FEBS Lett. 33:42–44. [DOI] [PubMed] [Google Scholar]
- 19.Ma, L., Y. Ishii, M. Ishiguro, H. Matsuzawa, and K. Yamaguchi. 1998. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob. Agents Chemother. 42:1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palucha, A., B. Mikiewicz, W. Hryniewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44:489–499. [DOI] [PubMed] [Google Scholar]
- 21.Sadate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolysing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]