Abstract
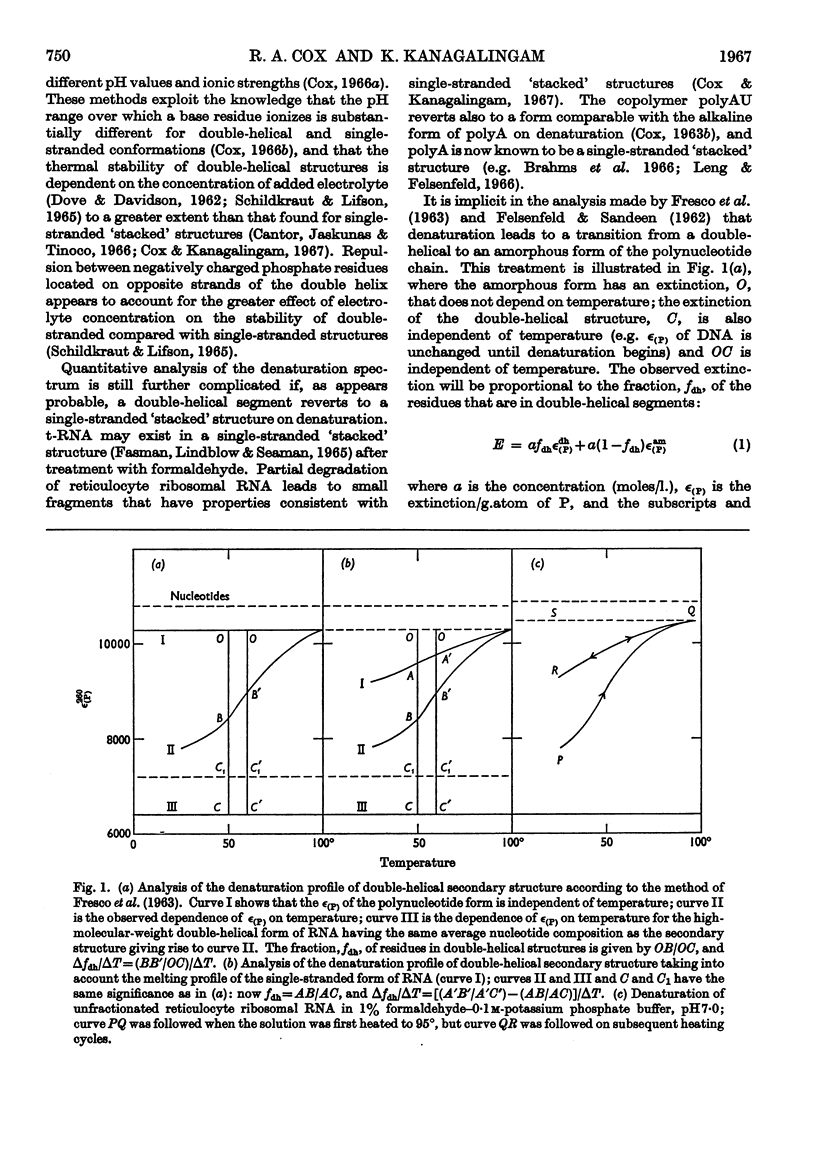
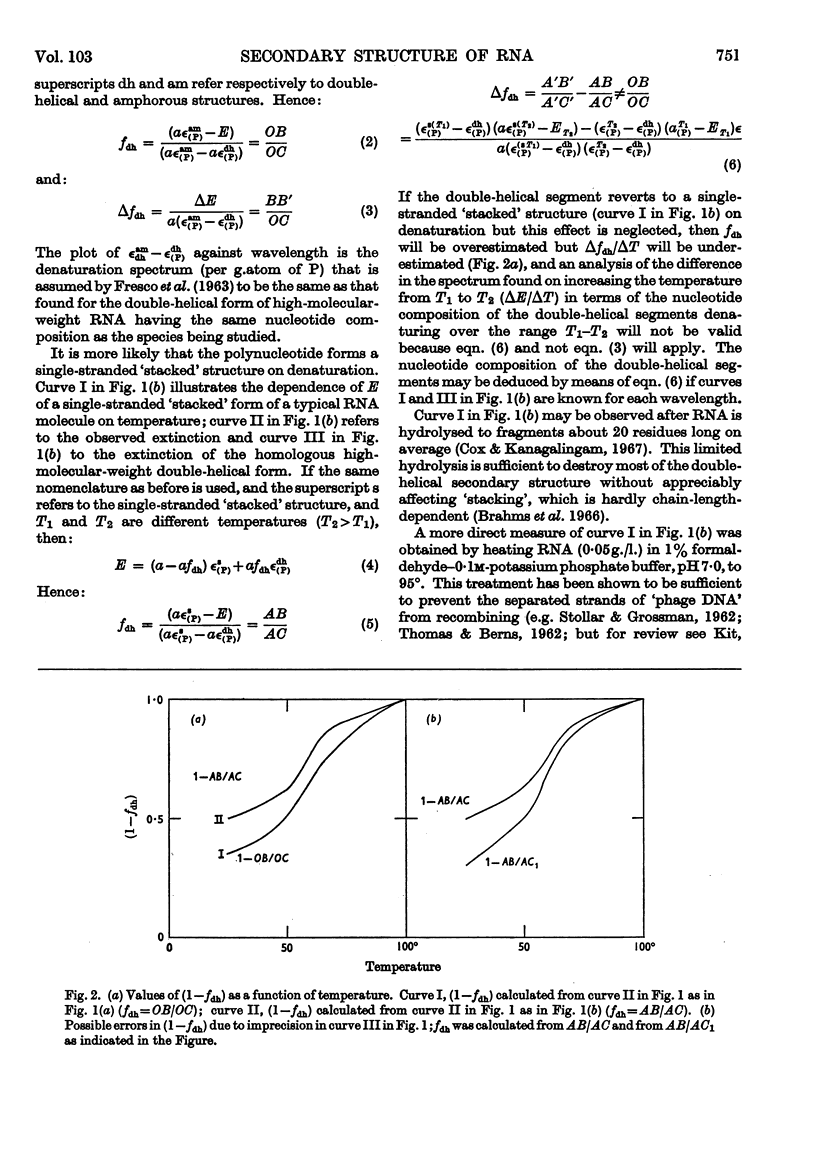
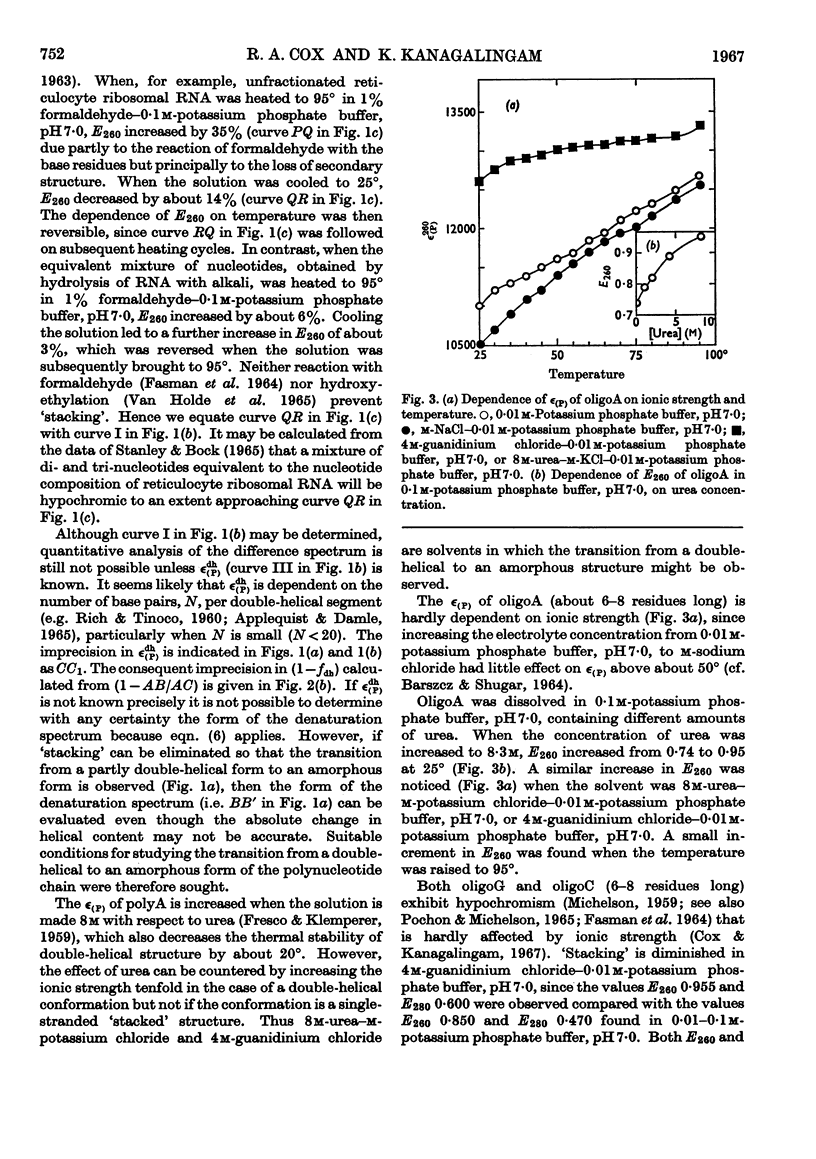
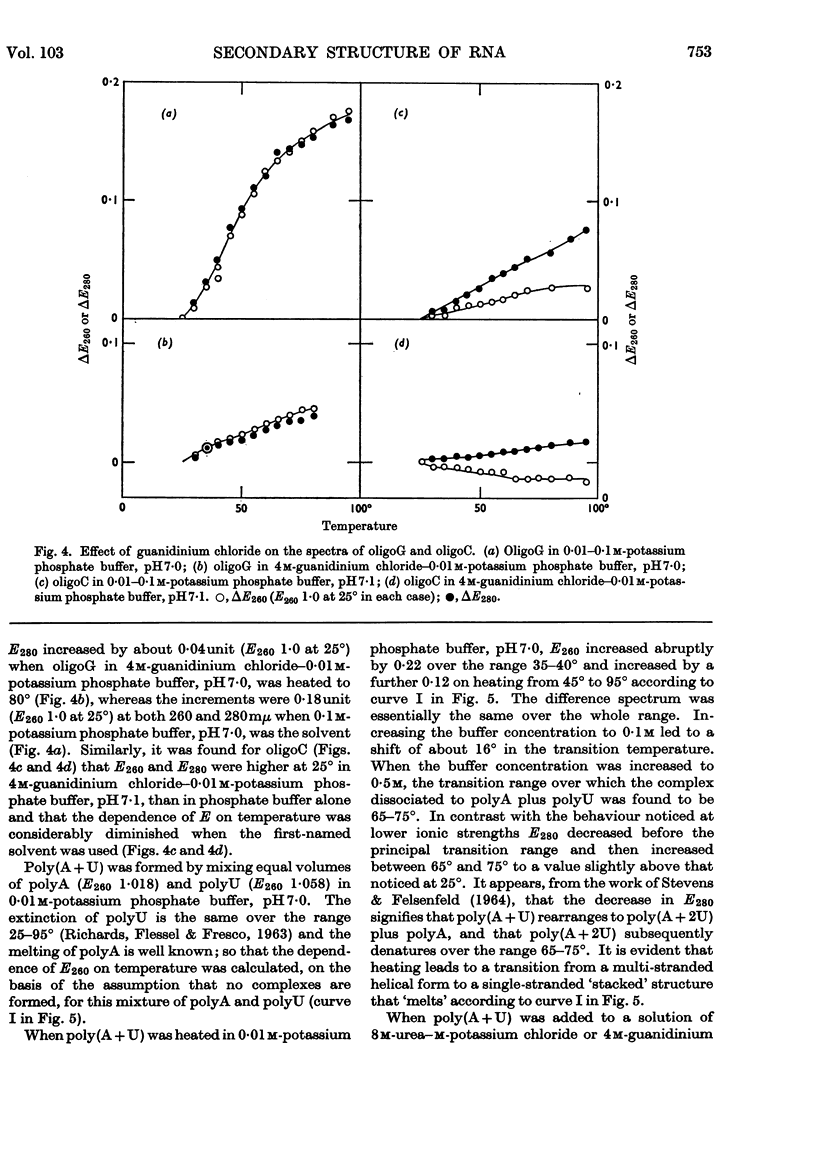
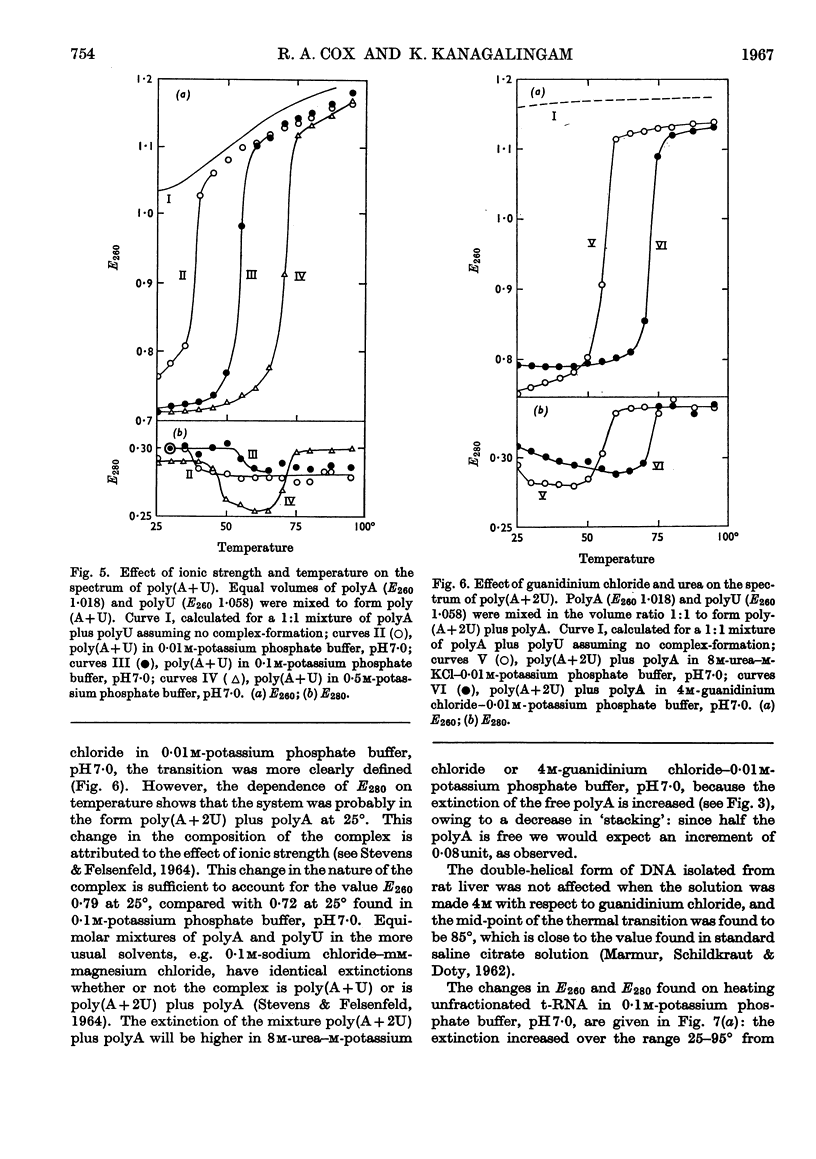
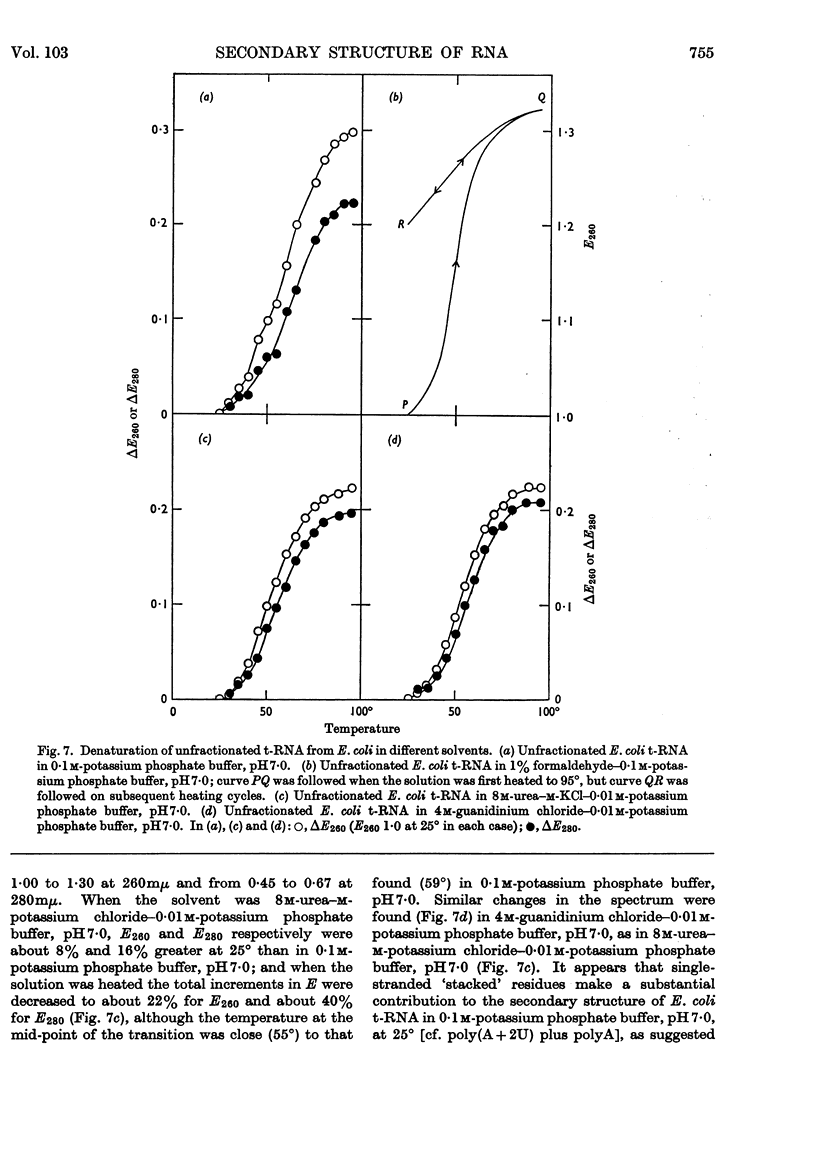
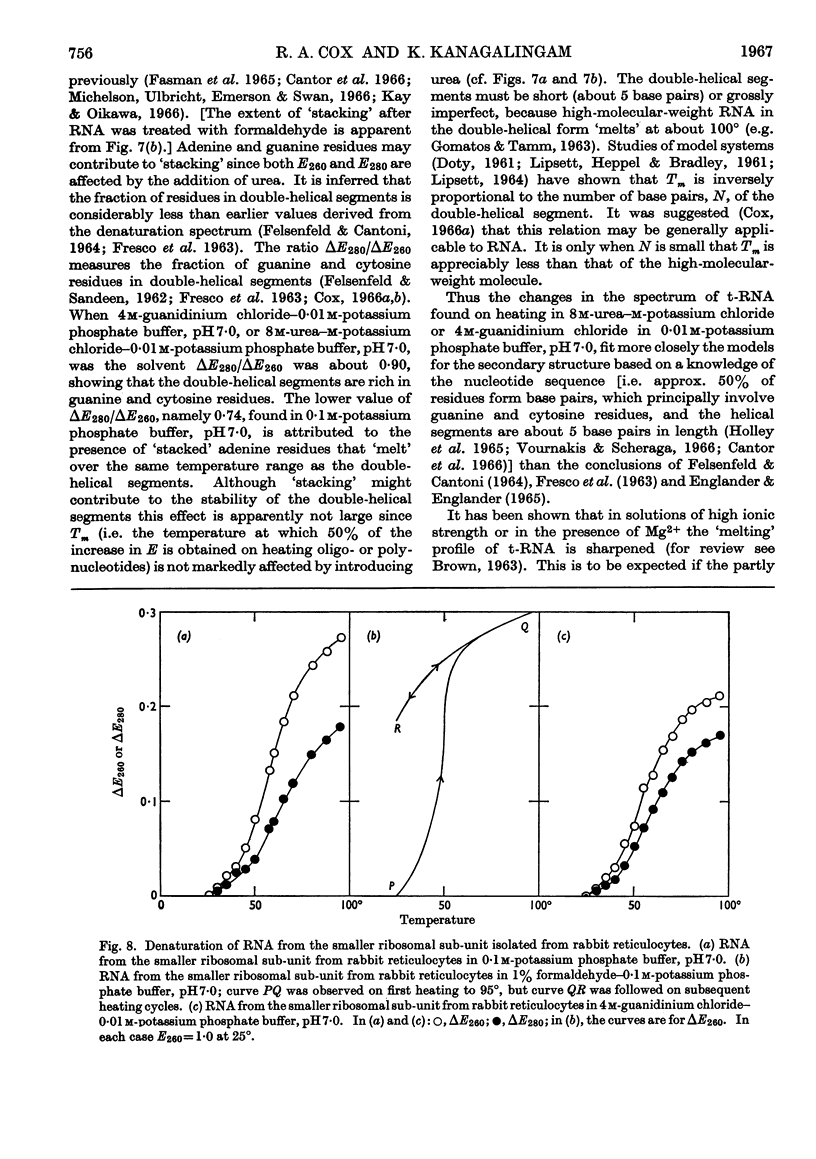
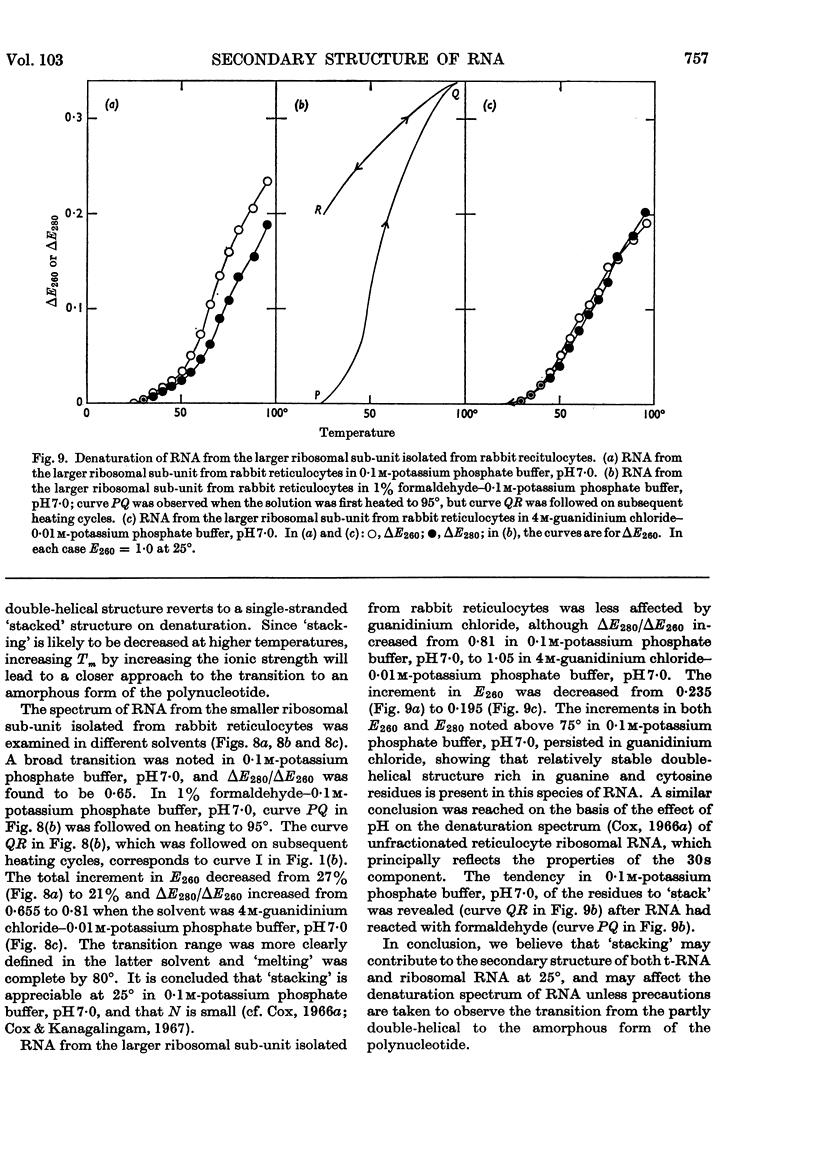
1. On the basis of studies with model compounds it was concluded that in 8m-urea–m-potassium chloride (or 4m-guanidinium chloride) in 0·01m-potassium phosphate buffer, pH7·0, multi-helical structures have about the same stability as in 0·1m-potassium phosphate buffer, pH7·0, whereas the tendency of base residues to `stack' along a single polynucleotide chain is much decreased. 2. Base-pairing was eliminated whereas base-`stacking' persisted after RNA in 1% formaldehyde–0·1m-potassium phosphate buffer, pH7·0, was heated to 95°. 3. From a study of the thermal denaturation of unfractionated transfer RNA from Escherichia coli and of RNA from the fractionated sub-units of rabbit reticulocyte ribosomes in 8m-urea–m-potassium chloride (or 4m-guanidinium chloride) in 0·01m-potassium phosphate buffer, pH7·0, it was inferred that `stacked' residues may account for up to 25% of the increase in E260 found on heating RNA in solvents such as 0·1m-potassium phosphate buffer, pH7·0. 4. Changes in the spectrum with temperature were analysed on the basis of the assumptions that (a) the polynucleotide chain is amorphous on denaturation (which is probable in 8m-urea–m-potassium chloride–0·01m-potassium phosphate buffer, pH7·0) and that (b) the polynucleotide chain adopts a single-stranded `stacked' conformation on denaturation (which is probable when ordinary solvents such as 0·1m-potassium phosphate buffer, pH7·0, are used).
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brahms J., Michelson A. M., Van Holde K. E. Adenylate oligomers in single- and double-strand conformation. J Mol Biol. 1966 Feb;15(2):467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- COX R. A. Dissociation properties of ribonucleic acid. I. Titration of rat-liver RNA and model polynucleotides. Biochim Biophys Acta. 1963 Mar 26;68:401–410. doi: 10.1016/0006-3002(63)90161-6. [DOI] [PubMed] [Google Scholar]
- COX R. A. The acid-base properties of ribonucleic acid. II. Spectrophotometric titration studies of poly-AU. Biochim Biophys Acta. 1963 Jun 25;72:203–208. doi: 10.1016/0006-3002(63)90235-x. [DOI] [PubMed] [Google Scholar]
- Cox R. A. A possible method for characterizing the secondary structure of ribonucleic acids. Biochem J. 1966 Jul;100(1):146–168. doi: 10.1042/bj1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Kanagalingam K. A study of the hydrolysis of unfractionated reticulocyte ribosomal ribonucleic acid by pancreatic ribonuclease and its relevance to secondary structure. Biochem J. 1967 May;103(2):431–452. doi: 10.1042/bj1030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A. The secondary structure of ribosomal ribonucleic acid in solution. Biochem J. 1966 Mar;98(3):841–857. doi: 10.1042/bj0980841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander S. W., Englander J. J. HYDROGEN EXCHANGE STUDIES OF sRNA. Proc Natl Acad Sci U S A. 1965 Feb;53(2):370–378. doi: 10.1073/pnas.53.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FASMAN G. D., LINDBLOW C., GROSSMAN L. THE HELICAL CONFORMATIONS OF POLYCYTIDYLIC ACID: STUDIES ON THE FORCES INVOLVED. Biochemistry. 1964 Aug;3:1015–1021. doi: 10.1021/bi00896a002. [DOI] [PubMed] [Google Scholar]
- FELSENFELD G., CANTONI G. L. USE OF THERMAL DENATURATION STUDIES TO INVESTIGATE THE BASE SEQUENCE OF YEAST SERINE SRNA. Proc Natl Acad Sci U S A. 1964 May;51:818–826. doi: 10.1073/pnas.51.5.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FELSENFELD G., SANDEEN G. The dispersion of the hyperchromic effect in thermally induced transitions of nucleic acids. J Mol Biol. 1962 Dec;5:587–610. doi: 10.1016/s0022-2836(62)80088-6. [DOI] [PubMed] [Google Scholar]
- FRESCO J. R., KLEMPERER E. Polyriboadenylic acid, a molecular analogue of ribonucleic acid and desoxyribonucleic acid. Ann N Y Acad Sci. 1959 Sep 4;81:730–741. doi: 10.1111/j.1749-6632.1959.tb49354.x. [DOI] [PubMed] [Google Scholar]
- Fasman G. D., Lindblow C., Seaman E. Optical rotatory dispersion studies on the conformational stabilization forces of yeast soluble ribonucleic acid. J Mol Biol. 1965 Jul;12(3):630–640. doi: 10.1016/s0022-2836(65)80317-5. [DOI] [PubMed] [Google Scholar]
- Gomatos P. J., Tamm I. THE SECONDARY STRUCTURE OF REOVIRUS RNA. Proc Natl Acad Sci U S A. 1963 May;49(5):707–714. doi: 10.1073/pnas.49.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- KIT S. DEOXYRIBONUCLEIC ACIDS. Annu Rev Biochem. 1963;32:43–82. doi: 10.1146/annurev.bi.32.070163.000355. [DOI] [PubMed] [Google Scholar]
- Kay C. M., Oikawa K. Hydrodynamic and optical rotatory dispersion studies on wheat germ soluble ribonucleic acid. Biochemistry. 1966 Jan;5(1):213–223. doi: 10.1021/bi00865a028. [DOI] [PubMed] [Google Scholar]
- Klucis E. S., Gould H. J. Zonal ultracentrifuge for the separation of ribosomal subunits. Science. 1966 Apr 15;152(3720):378–378. doi: 10.1126/science.152.3720.378. [DOI] [PubMed] [Google Scholar]
- LIPSETT M. N. COMPLEX FORMATION BETWEEN POLYCYTIDYLIC ACID AND GUANINE OLIGONUCLEOTIDES. J Biol Chem. 1964 Apr;239:1256–1260. [PubMed] [Google Scholar]
- LIPSETT M. N., HEPPEP L. A., BRADLEY D. F. Complex formation between oligonucleotides and polymers. J Biol Chem. 1961 Mar;236:857–863. [PubMed] [Google Scholar]
- Leng M., Felsenfeld G. A study of polyadenylic acid at neutral pH. J Mol Biol. 1966 Feb;15(2):455–466. doi: 10.1016/s0022-2836(66)80121-3. [DOI] [PubMed] [Google Scholar]
- MARTIN E. M., YEGIAN C., STENT G. S. SPECIFICITY OF THE AMINO ACID TRANSFER REACTION IN E. COLI STRAINS CARRYING DIFFERENT ALLELES OF THE RNA CONTROL (RC) GENE. Z Vererbungsl. 1963 Nov 21;94:303–315. doi: 10.1007/BF00894774. [DOI] [PubMed] [Google Scholar]
- Michelson A. M., Ulbricht T. L., Emerson T. R., Swan R. J. Optical rotatory dispersion of oligoadenylic acids and a consideration of the factors stabilizing helical polynucleotides. Nature. 1966 Feb 26;209(5026):873–874. doi: 10.1038/209873a0. [DOI] [PubMed] [Google Scholar]
- Poland D., Vournakis J. N., Scheraga H. A. Cooperative interactions in single-strand oligomers of adenylic acid. Biopolymers. 1966;4(2):223–235. doi: 10.1002/bip.1966.360040209. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Van Holde K. E., Brahms J., Michelson A. M. Base interactions of nucleotide polymers in aqueous solution. J Mol Biol. 1965 Jul;12(3):726–739. doi: 10.1016/s0022-2836(65)80323-0. [DOI] [PubMed] [Google Scholar]
- Vournakis J. N., Scheraga H. A. Optical rotatory dispersion studies of yeast alanine and tyrosine transfer ribonucleic acids. Evidence for intramolecular hydrogen bonding and discussion of conformational aspects. Biochemistry. 1966 Sep;5(9):2997–3006. doi: 10.1021/bi00873a032. [DOI] [PubMed] [Google Scholar]
- Zachau H. G., Dütting D., Feldmann H. Nucleotide sequences of two serine-specific transfer ribonucleic acids. Angew Chem Int Ed Engl. 1966 Apr;5(4):422–422. doi: 10.1002/anie.196604221. [DOI] [PubMed] [Google Scholar]


