The story of antituberculosis chemotherapy is a miniature of the history of anti-infective chemotherapy. In the first half of the 20th century the problem of tuberculosis appeared insoluble: the lipid-rich cell wall was believed to make chemotherapy impossible (21). This gloomy view seemed to be confirmed when the first antibiotics developed, sulfonamides and penicillin, had no useful activity against Mycobacterium tuberculosis. With this in mind it is easy to understand the early euphoria surrounding Albert Schatz and Selman Waksman’s discovery of streptomycin while working at Rutgers University in New Jersey (70) and Harold Lehmann’s discovery of para-aminosalicylic acid (PAS) shortly afterwards (47).
The clinical trials that followed the description of streptomycin rapidly dispersed the first hopes of a conquest of tuberculosis. Although patients improved compared with those patients not on therapy (the British Medical Research Council [BMRC] trial is widely considered to have been the first randomized controlled clinical trial), relapse occurred in many patients and the organisms were found to be resistant to streptomycin. Combined streptomycin and PAS trials proved that combination therapy prevented the emergence of resistance (14). The subsequent descriptions of isoniazid (19), pyrazinamide (52), rifampin (34), ethambutol (30), and other drugs gave the medical community the basic tools for tuberculosis control. The subsequent series of trials conducted under the auspices of the U.S. Public Health Service, the BMRC, and others produced data indicating that cure rates of over 95% with minimal relapse rates were possible in as little as 6 months, a reduction from the first regimens, which required treatment for 2 years (13, 22, 32, 33). Using these tools many countries have seen the virtual eradication of tuberculosis (82) and others, including some of the poorest, have seen a steady decline in the disease until the human immunodeficiency virus (HIV) epidemic caused the number of cases to spiral out of control (74). The tragedy of tuberculosis treatment is that, 50 years after the introduction of effective specific chemotherapy, the number of cases is higher worldwide and, more threateningly, there is an increasing number of cases of infections with organisms resistant to the major antituberculosis agents (25, 26, 62).
The circumstances in which drug resistance emerges are well known and have been so since shortly after the first clinical trials became available and their lessons were digested (51). In recent years the molecular basis for the mechanism of action of antituberculosis agents and the way in which the organisms become resistant have begun to be unraveled. In this review the clinical circumstances of resistance are described. The molecular mechanisms whereby resistance emerges are also outlined together with the insights that this brings to controlling the threat of an epidemic of multiple-drug resistance.
CLINICAL CIRCUMSTANCE FOR RESISTANCE DEVELOPMENT
The approach to chemotherapy for tuberculosis is very different from that for other bacterial infections. The organism has a long generation time and a capacity for dormancy, when its low metabolic activity makes it a difficult therapeutic target (53, 61, 83). In addition, M. tuberculosis may be located in pulmonary cavities, empyema pus, or solid caseous material, where penetration of antibiotics is difficult or the pH is sufficiently low to inhibit the activity of most antibiotics (29, 43). A series of animal and human clinical trials has led to the concept that there are different populations of bacteria present within the host. (8–10, 44, 57). Organisms in pulmonary cavities are thought to be multiplying in an aerobic environment and consequently behave in a way that can be mimicked by in vitro tests. Organisms located within caseous foci are in a milieu where the low pH is likely to inhibit the activity of agents such as aminoglycosides but to provide the conditions necessary for pyrazinamide activity. Bacteria found within macrophages probably only exhibit occasional spurts of metabolism and may be in relatively microaerophilic conditions, where mycobacterial dormancy can be induced (83).
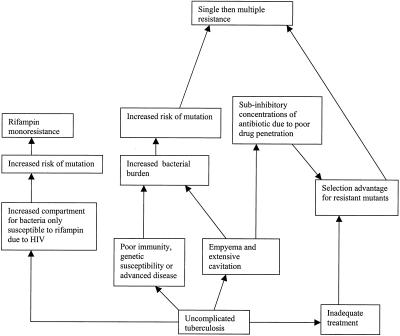
Each of the antituberculosis drugs has a major role in dealing with one of these populations. For example, isoniazid is critical early in therapy; its bactericidal activity rapidly reduces the sputum viable count because it is active mainly against the organisms growing aerobically in pulmonary cavities (23, 40). Pyrazinamide is only active at low pH, making it ideally suitable for killing the organisms inside caseous necrotic foci. This explains the finding that pyrazinamide appears to have no benefit after the second month of therapy (27). Rifampin is important in killing organisms that are metabolizing slowly, killing the persisters, and sterilizing the patient’s sputum, as demonstrated by animal studies (35) and clinical trials (27). Mathematical models suggest that increases in the size of the starting bacterial population are associated with the emergence of resistance. Poor adherence, effectively exposing organisms to monotherapy, is predicted to be very important in determining a resistance outcome (49) (Fig. 1).
FIG. 1.
Diagrammatic representation of the link between mutation rate, population size, and clinical complication in the emergence of resistance in M. tuberculosis infection. This demonstrates that individuals with tuberculosis in which the bacterial population increases or drug penetration is compromised by empyema or poor adherence are more likely to produce resistant mutants.
COMPARTMENTALIZATION
Compartmentalization of infection makes it more likely that bacteria will be exposed to monotherapy, especially when the patient receives inadequate therapy. This may arise due to an inadequate dosage because of inadequate prescription by the physician or nonadherence by the patient.
The presence of lung cavities that permit bacteria to grow in sites that are protected from the penetration of antituberculosis agents in adequate concentrations (29, 43 and, in empyema pus, may be compounded by low pH, which may reduce drug activity. There is also a strong association between HIV infection and multiple drug resistance although the reasons for this are not known (67). Partly this is due to circulation of multidrug-resistant tuberculosis strains in HIV-seropositive communities (1), but this association may occur because the contribution of the immune system in eradicating bacteria during chemotherapy is crucial in preventing the emergence of resistance. This may happen because the patient with HIV immunosuppression is unable to contain the size of individual lesions, thus increasing the number of organisms available for mutation (see below). The reason is likely to be more complex than this since, if enlarged bacterial population size were the only reason for resistance, then isoniazid monodrug resistance would be more likely. Patients with tuberculosis and HIV may be unable to absorb their drugs due to concomitant gastrointestinal disease, exposing the organisms to subtherapeutic concentrations. Extrapulmonary disease is more common in cases of HIV infection, and this may provide the opportunity for the growth of organisms in protected compartments (Fig. 1).
MOLECULAR MECHANISMS FOR THE EMERGENCE OF DRUG RESISTANCE
Within the last 10 years, the mechanism of action of most of the antituberculosis agents has been described, and we are beginning to understand some of the molecular mechanisms whereby M. tuberculosis becomes resistant (66).
M. tuberculosis is often acquired early in life with acute infection and with developing immunity, granuloma formation, and calcification. This is followed by a long latent period, which continues until reactivation occurs in a proportion of the individuals. This means that individual strains of M. tuberculosis have little opportunity to interact and exchange genetic information with other strains compared with, for example, organisms that colonize the nasopharynx or the gastrointestinal tract. In these locations, other bacteria may transmit antibiotic resistance determinants through transmissible genetic elements, transposons, integrons, and plasmids, by transduction or transformation. This option is not available for M. tuberculosis, so resistance can only occur through chromosomal mutation although rarely movement of mobile genetic elements, such as the insertion sequence IS6110, has been associated with new resistance emerging through the inactivation of critical genes (15, 48).
MUTATION
In any prokaryotic genome mutations are constantly occurring due to base changes caused by exogenous agents, DNA polymerase errors, deletions, insertions, and duplications. For prokaryotes there is a constant rate of spontaneous mutation of 0.0033 mutations/DNA replication that is uniform for a diverse spectrum of organisms (23). The mutation rate for individual genes varies significantly between and within genes. The reasons for these variations are uncertain but are thought to be under the influence of the local DNA sequence. For example significant differences between the evolutionary rates of heat shock protein genes within the Mycobacterium genus have been detected. The nonsynonymous sites of the GroEL gene have evolved twice as fast as those of the HSP65 (42) gene. The antibiotic resistance genes encoding fundamental replication functions of the organism such as rpoB and gyrA are typically highly conserved (24, 80).
GENETIC BASIS OF RESISTANCE
Telenti and colleagues were the first to determine the site of mutation that resulted in rifampin resistance in M. tuberculosis. They used the evidence that Escherichia coli became resistant to rifampin through mutation in the beta subunit of the rpoB gene and sequenced this gene from a series of epidemiologically unrelated strains (80). They showed that almost all rifampin-resistant isolates had mutations in a small region of rpoB. Subsequently, further clinical studies indicated that mutations are found in this region in up to 95% of resistant isolates (66). A similar approach has been adopted to detect mutations conferring resistance to other antibiotics. Since then the molecular mechanism of resistance to all of the main antituberculosis drugs, including isoniazid, pyrazinamide, streptomycin, ethambutol, and fluoroquinolones, has been described (7, 38, 39, 48, 75, 77). The different genes that have been associated with resistance to antituberculosis agents are summarized in Table 1.
TABLE 1.
Summary of the molecular mechanisms of antituberculosis drug resistance
| Drug | Associated mutated gene or mutation |
|---|---|
| Rifampin | rpoB |
| Isoniazid | katC, inhA, oxyR, ahpC, furA |
| Streptomycin | rrs, rpsL |
| Pyrazinamide | pncA, IS6110 insertion |
| Ethambutol | embB |
| Fluoroquinolones | gyrA, gyrB |
The genetic basis of resistance for some antituberculosis agents is not fully known. For example, streptomycin resistance emerges through mutations in rrs and rpsL that produce an alteration in the streptomycin binding site, but these changes are identified in just over one-half of the strains studied to date (41, 75). Thus there is a considerable amount of research into the mechanisms of resistance that is still required. It should be noted that in many cases mutations found in association with drug-resistant organisms may cause different levels of resistance and also may not be directly related to the mechanism of resistance. Isoniazid-resistance is a case in point. Modification of KatG, partial or total deletions, point mutations, or insertions, leads to the abolition or diminution of catalase activity and high-level resistance to isoniazid (37, 85). Catalase activity is essential in activating isoniazid to the active hydrazine derivative. A deficiency in enzyme activity produces high-level resistance and is found in more than 80% of isoniazid-resistant strains (66). Alternatively, low-level resistance can be caused by point mutations in the regulatory region of inhA operon, resulting in overexpression of inhA. Strains with this mutation have normal mycolic acid synthesis but low-level resistance to isoniazid. Point mutations in the regulatory region of ahpC have also been demonstrated; these are a compensation for the effects of absent or reduced catalase (KatG) function and do not directly result in resistance (38, 76). Most pyrazinamide-resistant organisms have mutations in the pyrazinamidase gene, although the gene may also be inactivated through the insertion of IS6110 (48). Pyrazinamide is essential in producing the active pyrazinoic acid derivative, and mutants are unable to produce an active drug. In addition to this, some resistant strains have no defined mutation (18). For additional details and a review of other mechanisms Ramaswamy and Musser’s review is recommended (66).
MUTATION RATE
The rate at which resistance emerges differs for all of the antituberculosis agents, being highest for ethambutol and lowest for rifampin and quinolones. The risks of mutation for most of the antibiotics used in tuberculosis treatment have been defined previously (16); for rifampin, isoniazid, streptomycin, and ethambutol, they are 3.32 × 10−9, 2.56 × 10−8, 2.29 × 10−8, and 1.0 × 10−7 mutations per bacterium per cell division, respectively. The mutation rate, rather than the mutation frequency, is the most reliable measure, as it records the risk of mutation per cell division rather than the proportion of mutant cells. Mutation frequency is significantly affected by “jackpot” mutation occurring early in the culture. There are several different calculation methods used to determine the mutation rate accurately, and the mathematics behind the calculations is beyond the scope of this review. Readers seeking further information can find a detailed description of these methods in one of several review articles (45, 78). These methods have been applied to antituberculosis drugs to calculate the estimated rates of mutation to resistance to the major antituberculosis drugs (16).
It has been assumed that the risk that an organism will develop resistance to two agents is the product of the risks of developing resistance to each separately. For example the risk of resistance for a combination of rifampin, streptomycin, and isoniazid is10−25/bacterium/generation. The risk of mutants emerging in a patient depends partly on this and the size of bacterial populations within compartments. Therefore, risk of resistance may be more accurately calculated using the formula P = 1 − (1 − r)n where P is the probability of drug resistance emerging, r is the mutation rate, and n is the number of bacilli in a lesion, usually calculated to be 108 per lesion (72). If single-drug therapy with a risk of mutation of 10−6 is used, the risk of resistance emerging is 100%. If two drugs with a combined mutation rate of 10−12 are used, then the risk is 0.01%; however, if the bacterial population in a lesion is 1010 and the mutation rate is 10−12, then there is a 1% risk of resistance emerging.
If mycobacteria are found in different compartments or in different physiological states, it is likely that this equation is an oversimplification. It is likely that, even if a patient is receiving optimum chemotherapy, there are populations of mycobacterial cells that are effectively receiving monotherapy or dual therapy. This means that the above equation is an optimistic estimate of the risk of resistance. This is in accord with clinical experience, which suggests that a relatively small deviation from the standard regimen may lead to the emergence of resistance.
HETERORESISTANCE AND PREEXISTENT MUTANTS
The considerations of the preceding paragraph imply that a patient with a large population of M. tuberculosis organisms may already have preexistent resistant organisms. This phenomenon was recognized soon after the introduction of chemotherapy, when the nature of resistance was first investigated. Naturally occurring streptomycin-resistant organisms were found in large broth cultures of H37Rv (81) (the standard laboratory strain). Streptomycin-resistant mutants derived from two patients and H37Rv were shown, in vitro, to segregate into three populations, susceptible populations and those with low- and high-level resistance (56). An early clinical study with patients treated with streptomycin monotherapy provides a fascinating insight into the emergence of resistance. Sputum was inoculated from patients before and during streptomycin monotherapy onto plates containing different concentrations of streptomycin. It was noted that, in seven out of eight cases before treatment, a small number of colonies grew resistant to streptomycin at 5 μg/liter and fewer grew resistant to it at 10 μg/liter but none grew resistant to it at 25 μg/liter. After 4 or 5 months of monotherapy predominantly susceptible strains were replaced by strains resistant to the drug at more than 1,000 μg/liter in four cases. The proportion of relatively resistant organisms was calculated as therapy progressed. Before chemotherapy 1 in 88,750 bacilli was resistant to the drug at 10 μg/liter, after 2 weeks 1 in 13,174 was resistant, and after 4 weeks 1 in 588 was resistant (64).
Shortly after the introduction of isoniazid into clinical practice it was shown that resistance to this antibiotic could arise by a single step in Mycobacterium bovis BCG and Mycobacterium ranae (63, 79) and the rate of mutation to isoniazid resistance was calculated as between 1 × 10−6 and 3 × 10−6/cell division. Resistant colonies were also cultured from the sputum of patients before they had been given any chemotherapy (40). There are some more-recent studies that throw light on the molecular basis of this phenomenon. In one study some samples from which isoniazid-susceptible M. tuberculosis was grown were examined by PCR-restriction fragment length polymorphism for mutation of katG. The cloning and sequencing of the PCR products demonstrated that the original specimen contained M. tuberculosis with two different katG alleles, a phenomenon that the authors described as heteroresistance (68). Similar heteroresistance was found for ethambutol (69). Heteroresistance may represent natural variation in the population of M. tuberculosis cells and may be an important mechanism for the emergence of resistance. What is not yet certain is whether every patient has a heteroresistant population or whether this is only found in strains with an increased likelihood of developing drug resistance.
MUTATION TYPE
The type of mutation that emerges depends on the selecting-antibiotic concentration. It has been shown that a different spectrum of mutants is selected at different concentrations of antibiotics. Most quinolone-resistant organisms, of whatever species, have mutations in a small region of the DNA gyrase genes (or topoisomerase IV genes if they possess them, which M. tuberculosis does not) known as the quinolone resistance-determining region (QRDR). Zhou and colleagues used Mycobacterium smegmatis and M. tuberculosis as a model system, growing bacteria in liquid culture and then plating out onto different concentrations of fluoroquinolone. At low concentrations colonies growing on concentrations close to the original MIC did not have evidence of mutation in the QRDR of gyrA (86). In this study no mutation events were detected in association with these small reductions in susceptibility. In contrast colonies selected on plates containing a higher concentration of fluoroquinolone had mutations mainly in the gyrA gene. Another way of thinking about this is that the non-gyrA mutations brought only a modest rise in the MIC. Only mutation in gyrA brought about an increase in resistance sufficient to be detected on plates containing a higher concentration of antibiotic. It must be remembered that there are a large number of mutants that are not detected in such a system, such as those that do not encode any increase in the MIC. Similarly, mutations that significantly interfere with the function of gyrA may cause the death of the mutated cell. These lethal mutations represent another extreme case that will not be detected in this system. In between these two extremes strains that encode resistance may do so at a physiological cost. Such mutants possess enzymes that do not perform their allotted task as efficiently as the wild-type enzymes, imposing a metabolic burden on the organism. This may be detected as reduced growth rates measured in in vitro systems or in vivo in animal models. They will be detected in this system only if their growth rate is sufficient for them to be detected within the experimental parameters. This is discussed in more detail below. Alternatively, if the concentration of antibiotic exceeds the maximum resistance level of mutant cells, no resistant organisms will emerge. This concept, the mutant prevention concentration, can be used to compare the activities of antibiotics in suppressing the emergence of resistance (20, 73).
MUTANT ANTAGONISM
It has long been recognized clinically that patients infected with an organism that has developed resistance to one agent appear more likely to develop resistance to another antibiotic. This clinical impression lacks hard data, and it is particularly difficult to obtain it as resistance emerges within patients where nonadherence is usually thought to be the main underlying reason. Once one resistance determinant has developed a second is even more likely to develop, as the patient may continue not to adhere to therapy and there are fewer active drugs available to suppress the emergence of resistant mutants. Some outbreaks have been associated with the sequential development of resistance, for example, the New York strain W epidemic (59). Recent studies have shown that there may be a molecular explanation for this phenomenon. Early studies of rifampin and streptomycin in E. coli showed that there was antagonism between rpoB and rpsL mutations, making double resistance more likely (11, 12). Paired streptomycin- and rifampin-resistant E. coli mutants have a temperature-sensitive phenotype, suggesting that a mutant with combined resistance produced a ribosome and RNA polymerase that were unable to function as effectively as the wild type. A study investigating this phenomenon in M. smegmatis showed that a similar antagonism existed in a mycobacterial system with an opposite effect (46). When streptomycin-resistant mutants were plated on a medium containing rifampin, double mutants arose at a lower frequency. However, mutation frequencies were enhanced up to fourfold during the stationary phase of growth, making resistance more likely. The presence of such a hypermutable state, if it were to occur in M. tuberculosis, would be of considerable importance in understanding the development of multiple drug resistance (46). M. tuberculosis grows under conditions of stress within cells, and these conditions may provide a hypermutable background, making resistance more likely.
STREPTOMYCIN RESISTANCE AND FITNESS
Early observations suggested that streptomycin-resistant strains grew more slowly than their wild-type parents (64). Mutations in rpsL that result in a high level of resistance to streptomycin fall into two categories, restrictive and nonrestrictive (54). Restrictive mutations are associated with an attenuation of virulence, whereas nonrestrictive mutations are not. Resistance to streptomycin was selected in vitro by plating M. smegmatis and M. tuberculosis on medium containing the drug. The rpsL gene of bacteria isolated on drug-containing medium was sequenced. This showed that for M. smegmatis resistant mutants were equally divided between restrictive and nonrestrictive genotypes. For M. tuberculosis only two out of six mutants had nonrestrictive mutations. A survey of resistant M. tuberculosis strains isolated from clinical cases showed a different picture, with all but 1 of 90 isolates having nonrestrictive genotypes (5). This suggests that the strains isolated in clinical practice are more likely to have normal virulence than would be expected from the results of in vitro studies.
RIFAMPIN
Resistance to rifampin arises due to mutations in the beta subunit of RNA polymerase encoded by the gene rpoB. Almost all of the corresponding mutations in rpoB occur in a small region of less than 100 bp, with less than 5% occurring outside of this region (36). This includes point mutations, deletions, and insertions (66). Despite the large number of different possible mutations three are found in more than 70% of clinical isolates. An experimental investigation of this phenomenon used in vitro resistance induction to investigate this. A limited repertoire of mutations was detected. When the growth rate of these organisms was compared with that of their parents in competition, a range of fitness was detected. There was considerable variation in the fitness of the rifampin-resistant strains, with some showing a severe physiological burden with a relative fitness (rf) of as little as 0.21, whereas other strains had a fitness similar to that of the susceptible parent (rf = 1.05; in these experiments relative fitness was defined as the ratio of the growth velocity of the mutant strain to that of the susceptible parent). The relative frequency of clinical isolation correlated significantly with the relative fitness of each mutation. These data suggest that many mutant strains may arise in a patient being treated with rifampin but that the strain most likely to survive and dominate the clinical culture is determined by the physiological deficit imposed on the strain by the mutation.
However, a recent study has suggested that differences in the mutation rate may contribute to this correlation. The mutants growing on rifampin-containing media were characterized by sequencing the rifampin resistance-determining region by a classical Luria-Delbruck methodology (50). A large number of different mutations were identified from the 60 cultures examined. In this experimental system a rapid growth rate would not have been an advantage as most of the broths contained only a single mutant cell. The Ser351-to-Leu mutation was shown to occur in 60.9% of cultures, suggesting that there is a higher mutation rate at this position (58).
The idea that changes in “fitness” occur on acquisition of resistance is supported by animal studies. An early study of resistant M. tuberculosis using a guinea pig infection model showed that some isoniazid-resistant strains caused much less severe disease than susceptible strain H37Rv while some resistant strains were fully virulent. The spectrum of virulence and resistance detected led the authors to conclude that the degree of isoniazid resistance was related to the virulence of the strains: strains with a greater degree of resistance were less virulent (2). A panel of strains resistant to one or more antimycobacterial drugs were tested in a mouse model of infection and demonstrated a range of virulence (60). The problem with both of these studies is that the genetic backgrounds of the organisms under test are unknown. They may have been markedly different, and thus differences in the virulence demonstrated could be explained by differences in genes other than those for antibiotic resistance. In vivo studies of defined isoniazid-resistant mutants have indicated a link between resistance and virulence. Experimental studies using the model of guinea pigs infected with M. tuberculosis in which the katG gene was inactivated showed that the virulence of these strains was significantly reduced compared with that of the parent strain and was restored when a functional katG was reintegrated into the genome (84). For isoniazid resistance, the resistance gene is important for survival of the organism inside macrophages.
The contention that isoniazid-resistant organisms might be less virulent than their susceptible parents is based of these and previous animal studies (51, 55). But clinical studies suggested that patients treated with isoniazid alone were likely to suffer a poor outcome as organisms continue to cause progressive disease (65). The authors did note that organisms with low catalase activity were more likely to be bacteriologically quiescent than resistant organisms with normal catalase activity although this result did not achieve statistical significance. This emphasizes that attenuation of virulence, if it occurs in isoniazid-resistant organisms, only develops when catalase in inactivated.
ADAPTATION
There is evidence from several experimental systems that the initial fitness deficit associated with development of resistance disappears with repeated multiplication. In general it is known that organisms in artificial culture will readily adapt to the in vitro conditions. This can be measured in a number of ways; for example, the cell volume of cultured cells increases with the number of generations in culture or the speed of growth. Adaptation to the physiological cost of resistance has been shown to occur for strains that have acquired resistance by acquisition of a plasmid or by mutation in chromosomal genes. Using streptomycin-resistant mutant E. coli as a model system Schrag and colleagues demonstrated that initial rpsL mutants had a 14 to 19% selective disadvantage per generation as measured by the chain elongation rate (71). After serial passage in the absence of antibiotic selection, adaptation took place without any evidence of reversion to susceptibility, indicating the degree to which the fitness deficit had been eliminated (71). If a susceptible genotype was reinserted into the adapted resistant strain, the new mutant paradoxically showed a fitness deficit relative to the adapted resistant strain. These data have been reproduced in other bacterial species and suggest that M. tuberculosis is likely to behave in a similar way (3, 4). Adaptation experiments with rpoB mutant strains of M. tuberculosis show that, after passage for 88 generations, mutants initially less fit than the sensitive parent improve their fitness value to match and even exceed the parent (O. J. Billington, personal communication).
HUMAN STUDIES OF RESISTANCE EMERGENCE
Evidence to support the idea that variation in the biological fitness influences the outcome of therapy is beginning to emerge. There is a report of a pair of cases of tuberculosis, a brother and sister, in which both suffered multiple relapses due to nonadherence; the organism in one case exhibited multiple drug resistance, and that in the other was consistently susceptible. Both strains had a lower in vitro fitness than the laboratory control, H37Rv, and the resistant strain had a lower fitness than the susceptible but otherwise indistinguishable strain (17). This suggested that the fitness deficit was directly related to the difference in susceptibility. However, strains from an outbreak of multiple-drug-resistant M. tuberculosis with identical susceptibilities had significantly different in vitro fitnesses. The index case was that of an immunocompetent female (strain rf, 0.8) whose disease followed a progressive course and who died. The strain from a second patient, who was HIV positive, had an rf of 1.0, and his disease progressed rapidly to death. The strain isolated from a third patient, who had acquired infection later in the outbreak, had an rf of 0.5, and this patient remained alive and on treatment 2 years after diagnosis. It is impossible to dissect the contribution of host resistance from these results, but the individual with the index case, who was immunocompetent, died, and the HIV-seropositive individual with the least-fit strain continues to survive. These data suggest that subtle changes in the strains occur on passage between human hosts. Since we know that initially resistant strains have a fitness deficit, transmission within a group of immunocompromised individuals may allow the organisms to multiply and be transmitted while adapting. This conjecture is in accord with the history of recent outbreaks of multiple-drug-resistant tuberculosis that have arisen initially among patients who are severely immunocompromised (28, 31). Transmission within this group may have produced organisms fully capable of producing disease in immunocompetent individuals (6).
SUMMARY
Drug-resistant tuberculosis poses a significant threat to human health, and it is important to understand how the resistance emerges if we are to reverse the upward trend. Treatment with internationally approved regimens results in a very high cure rate with few relapses and without the emergence of resistance. These regimens are effective in preventing the emergence of resistance because combination chemotherapy makes it highly unlikely that there will be a spontaneous mutant resistant to all of the components of chemotherapy. Patients with uncomplicated tuberculosis who receive inadequate treatment provide a selection advantage for resistant mutants because bacteria may be exposed to monotherapy, permitting the emergence of resistance to single agents and then to multiple agents as the protection of combination chemotherapy is eroded. That M. tuberculosis cells within the body are susceptible to different components of antituberculosis chemotherapy means that the risk of resistant mutants emerging is higher than would be expected if the whole population of bacterial cells could be counted together. Clinical complications such as empyema and extensive cavitation permit a large population to develop in a compartment into which drugs may not penetrate. This large bacterial pool increases the population for mutation, and with poor penetration there is an increased likelihood of resistance emerging. A similar situation may develop in patients with extensive disease or poor immunity (Fig. 1).
We have learned that some physiological conditions may induce a hypermutable state, making multiple resistance more likely. The assumption that resistant organisms are less fit than wild-type strains may not be correct, as the initial fitness deficits may be attenuated by adaptation by multiple passage. Instances of isoniazid resistance, where attenuated virulence is common, may occur because the molecular mechanism of resistance directly affects a system required by the organism for intracellular survival.
The important lesson these clinical and molecular studies teach us is that resistant organisms over time will be fully virulent and that if we are to prevent an epidemic of multiple-drug-resistant tuberculosis we must take steps to ensure that all patients are diagnosed and effectively treated so that resistant strains are not created and transmitted in the community.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710–1716. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, M., S. R. M. Bushby, and D. A. Mitchison. 1953. Tubercle bacilli resistant to isoniazid: virulence and response to treatment with isoniazid in guinea pigs and mice. Br. J. Exp. Pathol. 34:568–581. [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479–1482. [DOI] [PubMed] [Google Scholar]
- 5.Bottger, E. C., B. Springer, M. Pletschette, and P. Sander. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4:1343–1344. [DOI] [PubMed] [Google Scholar]
- 6.Breathnach, A. S., A. de Ruiter, G. M. Holdsworth, N. T. Bateman, D. G. O’Sullivan, P. J. Rees, D. Snashall, H. J. Milburn, B. S. Peters, J. Watson, F. A. Drobniewski, and G. L. French. 1998. An outbreak of multi-drug-resistant tuberculosis in a London teaching hospital. J. Hosp. Infect. 39:111–117. [DOI] [PubMed] [Google Scholar]
- 7.Cambau, E., W. Sougakoff, M. Besson, C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1994. Selection of a gyrA mutant of Mycobacterium tuberculosis resistant to fluoroquinolones during treatment with ofloxacin. J. Infect. Dis. 170:479–483. [DOI] [PubMed] [Google Scholar]
- 8.Canetti, G. 1955. The tubercle bacillus in the pulmonary lesion of man. Springer Publishing Company, New York, N.Y.
- 9.Canetti, G. 1966. Direct bacteriologic study of treated tuberculomas. Rev. Tuberc. Pneumol. 30:912–915. (In French.) [PubMed] [Google Scholar]
- 10.Canetti, G., M. Le Lirzin, G. Porven, N. Rist, and F. Grumbach. 1968. Some comparative aspects of rifampicin and isoniazid. Tubercle 49:367–376. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti, S. L., and L. Gorini. 1975. A link between streptomycin and rifampicin mutation. Proc. Natl. Acad. Sci. USA 72:2084–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti, S. L., and L. Gorini. 1977. Interaction between mutations of ribosomes and RNA polymerase: a pair of strA and rif mutants individually temperature-insensitive but temperature-sensitive in combination. Proc. Natl. Acad. Sci. USA 74:1157–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Combs, D. L., R. J. O’Brien, and L. J. Geiter. 1990. USPHS tuberculosis short-course chemotherapy trial 21: effectiveness, toxicity, and acceptability. The report of final results. Ann. Intern. Med. 112:397–406. [DOI] [PubMed] [Google Scholar]
- 14.Crofton, J., and D. A. Mitchison. 1948. Streptomycin resistance in pulmonary tuberculosis. Br. Med. J. 2:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale, J. W. 1995. Mobile genetic elements in mycobacteria. Eur. Respir. J. Suppl. 20:633s–648s. [PubMed] [Google Scholar]
- 16.David, H. L. 1970. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 20:810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, A. P., O. J. Billington, B. A. Bannister, W. R. Weir, T. D. McHugh, and S. H. Gillespie. 2000. Comparison of fitness of two isolates of Mycobacterium tuberculosis, one of which had developed multi-drug resistance during the course of treatment. J. Infect. 41:184–187. [DOI] [PubMed] [Google Scholar]
- 18.Davies, A. P., O. J. Billington, T. D. McHugh, D. A. Mitchison, and S. H. Gillespie. 2000. Comparison of phenotypic and genotypic methods for pyrazinamide susceptibility testing with Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3686–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domagk, G., and P. Klee. 1952. Die behandlung der tuberkulose mit neoteben (isonikotinsaurehydrazid). Dtsch. Med. Wochenschr. 77:578–581. [DOI] [PubMed] [Google Scholar]
- 20.Dong, Y., X. Zhao, B. N. Kreiswirth, and K. Drlica. 2000. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2581–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dormandy, T. 1999. The white death: a history of tuberculosis. Hambledon Press, London, United Kingdom.
- 22.Doster, B., F. J. Murray, R. Newman, and S. F. Woolpert. 1973. Ethambutol in the initial treatment of pulmonary tuberculosis. U.S. Public Health Service tuberculosis therapy trials. Am. Rev. Respir. Dis. 107:177–190. [DOI] [PubMed] [Google Scholar]
- 23.Drake, J. W. 1999. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann. N. Y. Acad. Sci. 870:100–107. [DOI] [PubMed] [Google Scholar]
- 24.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dye, C. 2000. Tuberculosis 2000–2010: control, but not elimination. Int. J. Tuberc. Lung Dis. 4:S146–S152. [PubMed] [Google Scholar]
- 26.Dye, C., and M. A. Espinal. 2000. Will tuberculosis become resistant to all antibiotics? Proc. R. Soc. Lond. B Biol. Sci. 267:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.East African/British Medical Research Councils. 1981. Controlled trial of five short course regimens of chemotherapy regimens for pulmonary tuberculosis. Am. Rev. Respir. Dis. 123:165–170. [DOI] [PubMed] [Google Scholar]
- 28.Edlin, B. R., J. I. Tokars, M. H. Grieco, J. T. Crawford, J. Williams, E. M. Sordillo, K. R. Ong, J. O. Kilburn, S. W. Dooley, and K. G. Castro. 1992. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 326:1514–1521. [DOI] [PubMed] [Google Scholar]
- 29.Elliott, A. M., S. E. Berning, M. D. Iseman, and C. A. Peloquin. 1995. Failure of drug penetration and acquisition of drug resistance in chronic tuberculous empyema. Tuber. Lung Dis. 76:463–467. [DOI] [PubMed] [Google Scholar]
- 30.Ferebee, S. H., B. E. Doster, and F. J. Murray. 1966. Ethambutol: a substitute for para-aminosalicylic acid in regimens for pulmonary tuberculosis. Ann. N. Y. Acad. Sci. 135:910–920. [DOI] [PubMed] [Google Scholar]
- 31.Fischl, M. A., R. B. Uttamchandani, G. L. Daikos, R. B. Poblete, J. N. Moreno, R. R. Reyes, A. M. Boota, L. M. Thompson, T. J. Cleary, and S. Lai. 1992. An outbreak of tuberculosis caused by multiple-drug-resistant tubercle bacilli among patients with HIV infection. Ann. Intern. Med. 117:177–183. [DOI] [PubMed] [Google Scholar]
- 32.Fox, W. 1978. The current status of short course chemotherapy. Bull. Int. Union Tuberc. 53:268–280. [Google Scholar]
- 33.Fox, W., G. A. Ellard, and D. A. Mitchison. 1999. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946–1986, with relevant subsequent publications. Int. J. Tuberc. Lung Dis. 3:S231–S279. [PubMed] [Google Scholar]
- 34.Furesz, S., and M. T. Timball. 1963. The antibacterial activity of rifamycins. Chemotherapia 7:200. [DOI] [PubMed] [Google Scholar]
- 35.Grumbach, F., G. Canetti, and M. Le Lirzin. 1970. Durable character of the sterilization of experimental tuberculosis in mice by rifampicin-isoniazid association: cortisone test. Rev. Tuberc. Pneumol. 34:312–319. (In French.) [PubMed] [Google Scholar]
- 36.Heep, M., U. Rieger, D. Beck, and N. Lehn. 2000. Mutations in the beginning of the rpoB gene can induce resistance to rifamycins in both Helicobacter pylori and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:1075–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heym, B., P. M. Alzari, N. Honore, and S. T. Cole. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15:235–245. [DOI] [PubMed] [Google Scholar]
- 38.Heym, B., B. Saint-Joanis, and S. T. Cole. 1999. The molecular basis of isoniazid resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:267–271. [DOI] [PubMed] [Google Scholar]
- 39.Hirano, K., M. Takahashi, Y. Kazumi, Y. Fukasawa, and C. Abe. 1997. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 78:117–122. [DOI] [PubMed] [Google Scholar]
- 40.Hobby, G. L., and T. F. Lenert. 1952. Resistance to isonicotinic acid. Am. Rev. Tuberculosis 65:771. [PubMed] [Google Scholar]
- 41.Honore, N., and S. T. Cole. 1994. Streptomycin resistance in mycobacteria. Antimicrob. Agents Chemother. 38:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes, A. L. 1993. Contrasting evolutionary rates in the duplicate chaparonin genes of Mycobacterium tuberculosis and M. leprae. Mol. Biol. Evol. 10:1343–1359. [DOI] [PubMed] [Google Scholar]
- 43.Iseman, M. D., and L. A. Madsen. 1991. Chronic tuberculous empyema with bronchopleural fistula resulting in treatment failure and progressive drug resistance. Chest 100:124–127. [DOI] [PubMed] [Google Scholar]
- 44.Jindani, A., V. R. Aber, E. A. Edwards, and D. A. Mitchison. 1980. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am. Rev. Respir. Dis. 121:939–949. [DOI] [PubMed] [Google Scholar]
- 45.Jones, M. E., S. M. Thomas, and A. Rogers. 1994. Luria-Delbruck fluctuation experiments: design and analysis. Genetics 136:1209–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karunakaran, P., and J. Davies. 2000. Genetic antagonism and hypermutability in Mycobacterium smegmatis. J. Bacteriol. 182:3331–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehman, J. 1946. Para-aminosalacylic acid in the treatment of tuberculosis. Lancet i:15. [DOI] [PubMed] [Google Scholar]
- 48.Lemaitre, N., W. Sougakoff, C. Truffot-Pernot, and V. Jarlier. 1999. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob. Agents Chemother. 43:1761–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipsitch, M., and B. R. Levin. 1998. Population dynamics of tuberculosis treatment: mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Int. J. Tuberc. Lung Dis. 2:187–199. [PubMed] [Google Scholar]
- 50.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDermott, W. 1960. Antimicrobial therapy of pulmonary tuberculosis. Bull. W. H. O. 23:427–461. [PMC free article] [PubMed] [Google Scholar]
- 52.McKenzie, D., L. Malone, S. Kushner, J. J. Olesson, and Y. Subbarow. 1948. The effect of nicotinic acid amide on experimental tuberculosis of white mice. J. Lab. Clin. Med. 33:1249–1253. [PubMed] [Google Scholar]
- 53.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. [DOI] [PubMed] [Google Scholar]
- 54.Meier, A., P. Sander, K. J. Schaper, M. Scholz, and E. C. Bottger. 1996. Correlation of molecular resistance mechanisms and phenotypic resistance levels in streptomycin-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:2452–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Middlebrook, G., and S. H. Dressler. 1954. Clinical evaluation of isoniazid. Am. Rev. Tuberc. 70:1102–1103. [DOI] [PubMed] [Google Scholar]
- 56.Mitchison, D. 1951. The segregation of streptomycin resistant variants of Mycobacterium tuberculosis into groups with characteristic resistance. J. Gen. Microbiol. 5:596–604. [DOI] [PubMed] [Google Scholar]
- 57.Mitchison, D. A. 1985. The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66:219–225. [DOI] [PubMed] [Google Scholar]
- 58.Morlock, G. P., B. B. Plikaytis, and J. T. Crawford. 2000. Characterization of spontaneous, in vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moss, A. R., D. Alland, E. Telzak, D. Hewlett, Jr., V. Sharp, P. Chiliade, V. LaBombardi, D. Kabus, B. Hanna, L. Palumbo, K. Brudney, A. Weltman, K. Stoeckle, K. Chirgwin, M. Simberkoff, S. Moghazeh, W. Eisner, M. Lutfey, and B. Kreiswirth. 1997. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int. J. Tuberc. Lung Dis. 1:115–121. [PubMed] [Google Scholar]
- 60.Ordway, D. J., M. G. Sonnenberg, and S. A. Donahue. 1995. Drug-resistant strains of Mycobacterium tuberculosis exhibit a range of virulence for mice. Infect. Immun. 63:741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pablos-Mendez, A. 2000. Working alliance for TB drug development, Cape Town, South Africa, February 8th, 2000. Int. J. Tuberc. Lung Dis. 4:489–490. [PubMed] [Google Scholar]
- 62.Pablos-Mendez, A., M. C. Raviglione, A. Laszlo, N. Binkin, H. L. Rieder, F. Bustreo, D. L. Cohn, C. S. Lambregts-van Weezenbeek, S. J. Kim, P. Chaulet, and P. Nunn. 1998. Global surveillance for antituberculosis-drug resistance, 1994–1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 338:1641–1649. [DOI] [PubMed] [Google Scholar]
- 63.Pansy, F., H. Stander, and R. Donovick. 1952. In vitro studies on isonicotinic acid hydrazine. Am. Rev. Tuberc. 65:761–764. [PubMed] [Google Scholar]
- 64.Pyle, M. M. 1947. Relative numbers of resistant tubercle bacilli in sputa of patients before and during treatment with streptomycin. Proc. Staff Meet. Mayo Clin. 22:465–473. [PubMed] [Google Scholar]
- 65.Ramakrishnan, C. V., A. L. Bhatia, A. Devadatta, W. Fox, A. S. L. Narayana, J. B. Selkon, and S. Velu. 1962. The course of pulmonary tuberculosis in patients excreting organisms which have acquired resistance to isoniazid. Bull. W. H. O. 26:1–18. [PMC free article] [PubMed] [Google Scholar]
- 66.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3–29. [DOI] [PubMed] [Google Scholar]
- 67.Ridzon, R., C. G. Whitney, M. T. McKenna, J. P. Taylor, S. H. Ashkar, A. T. Nitta, S. M. Harvey, S. Valway, C. Woodley, R. Cooksey, and I. M. Onorato. 1998. Risk factors for rifampin mono-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 157:1881–1884. [DOI] [PubMed] [Google Scholar]
- 68.Rinder, H., K. Feldmann, E. Tortoli, J. Grosset, M. Casal, E. Richter, M. Rifai, V. Jarlier, M. Vaquero, S. Rusch-Gerdes, E. Cambau, J. Gutierrez, and T. Loscher. 1999. Culture-independent prediction of isoniazid resistance in Mycobacterium tuberculosis by katG gene analysis directly from sputum samples. Mol. Diagn. 4:145–152. [DOI] [PubMed] [Google Scholar]
- 69.Rinder, H., K. T. Mieskes, E. Tortoli, E. Richter, M. Casal, M. Vaquero, E. Cambau, K. Feldmann, and T. Loscher. 2001. Detection of embB codon 306 mutations in ethambutol resistant Mycobacterium tuberculosis directly from sputum samples: a low-cost, rapid approach. Mol. Cell Probes 15:37–42. [DOI] [PubMed] [Google Scholar]
- 70.Schatz, A., and S. A. Waksman. 1944. Effect of streptomycin and other antibioic substances upon Mycobacterium tuberculosis and related organisms. Proc. Soc. Exp. Biol. Med. 57:244–248. [Google Scholar]
- 71.Schrag, S. J., V. Perrot, and B. R. Levin. 1997. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. Lond. B Biol. Sci. 264:1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimao, T. 1987. Drug resistance in tuberculosis control. Tubercle 68:5–18. [DOI] [PubMed] [Google Scholar]
- 73.Sindelar, G., X. Zhao, A. Liew, Y. Dong, T. Lu, J. Zhou, J. Domagala, and K. Drlica. 2000. Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria. Antimicrob. Agents Chemother. 44:3337–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snider, D. E., Jr., and J. R. La Montagne. 1994. The neglected global tuberculosis problem: a report of the 1992 World Congress on Tuberculosis. J. Infect. Dis. 169:1189–1196. [DOI] [PubMed] [Google Scholar]
- 75.Sreevatsan, S., X. Pan, K. E. Stockbauer, D. L. Williams, B. N. Kreiswirth, and J. M. Musser. 1996. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents Chemother. 40:1024–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sreevatsan, S., X. Pan, Y. Zhang, V. Deretic, and J. M. Musser. 1997. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob. Agents Chemother. 41:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41:1677–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stewart, F. M., D. M. Gordon, and B. R. Levin. 1990. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics 124:175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szyblaski, W., and V. Bryson. 1952. Bacterial resistance studies with derivatives of isonicotinic acid. Am. Rev. Tuberc. 65:770. [PubMed] [Google Scholar]
- 80.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650. [DOI] [PubMed] [Google Scholar]
- 81.Vennesland, K., R. H. Ebert, and R. G. Bloch. 1947. The demonstration of naturally occurring streptomycin resistant variants in the human strain of tubercule bacillus H-37RV. Science 106:476–477. [DOI] [PubMed] [Google Scholar]
- 82.Vynnycky, E., and P. E. Fine. 1997. The annual risk of infection with Mycobacterium tuberculosis in England and Wales since 1901. Int. J. Tuberc. Lung Dis. 1:389–396. [PubMed] [Google Scholar]
- 83.Wayne, L. G. 1994. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur. J. Clin. Microbiol. Infect. Dis. 13:908–914. [DOI] [PubMed] [Google Scholar]
- 84.Wilson, T. M., G. W. De Lisle, and D. M. Collins. 1995. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol. Microbiol. 15:1009–1015. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, Y., T. Garbe, and D. Young. 1993. Transformation with katG restores isoniazid-sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol. Microbiol. 8:521–524. [DOI] [PubMed] [Google Scholar]
- 86.Zhou, J., Y. Dong, X. Zhao, S. Lee, A. Amin, S. Ramaswamy, J. Domagala, J. M. Musser, and K. Drlica. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517–525. [DOI] [PubMed] [Google Scholar]