Abstract
We have found a novel phospholipid antibiotic (named bacilysocin) which accumulates within (or associates with) the cells of Bacillus subtilis 168 and determined the structure by nuclear magnetic resonance and mass spectrometry analyses. The structure of bacilysocin elucidated was 1-(12-methyltetradecanoyl)-3-phosphoglyceroglycerol. Bacilysocin demonstrated antimicrobial activity, especially against certain fungi. Production of bacilysocin commenced immediately after growth ceased and before the formation of heat-resistant spores. The production of bacilysocin was completely blocked when the ytpA gene, which encodes a protein homologous to lysophospholipase, was disrupted, but blockage of the ytpA gene did not significantly affect growth. Sporulation was also impaired, with a 10-fold reduction in heat-resistant spore titers being detected. Since the ytpA disruptant actually lacked phospholipase activity, we propose that the YtpA protein functions as an enzyme for the biosynthesis of bacilysocin.
The gram-positive bacterium Bacillus subtilis produces a large number of antibiotics, which are classified as ribosomal or nonribosomal. Nonribosomally synthesized circular oligopeptides that contain a fatty acid chain exhibit potent antibacterial or antifungal activity, as represented by surfactin, the iturinic group, and fengycin (16). B. subtilis 168 is the best-studied strain in the genus Bacillus, the genome of which was completely sequenced in 1997. Strain 168 is known to produce three ribosomal antibiotics, TasA (12), subtilosin (1), sublancin (10), and two nonribosomal antibiotics, surfactin (14) and bacilysin (9). The production of other antibiotics by strain 168 has also been predicted on the basis of genome sequence analysis, as exemplified by plipastatin (13). The ribosomal peptide antibiotics are synthesized during active growth, while nonribosomal ones are synthesized after growth has ceased. The role of antibiotic production for the producing organism is still under speculation. The best-accepted theory is that nonribosomal antibiotics may play a role in competition with other microorganisms during spore germination (for a review, see references 5 and 16). The detection of novel antibiotics produced by B. subtilis 168 would therefore be helpful in providing an understanding of the intrinsic (if any) role of antibiotics in the life cycle of this organism. In the present paper, we describe the isolation and identification of a new antibiotic (named bacilysocin) produced by B. subtilis 168.
MATERIALS AND METHODS
Strain, media, and culture conditions.
B. subtilis strain 168 (trpC2) was precultured at 30°C for 24 h in NG medium (2) supplemented with 50 μg of tryptophan per ml, and then 0.1 ml of the resulting culture was inoculated into 10 ml of fresh NG medium, followed by incubation under shaking at 30°C. The titers of the heat-resistant spores were determined by heating the cultures for 10 min at 80°C and then plating them onto an NG agar plate.
Detection and bioassay of bacilysocin.
Bacilysocin was extracted with butanol from cells grown for 24 h in NG medium. A butanol (BuOH) extract was then subjected to thin-layer chromatography (TLC) with CHCl3-methanol (MeOH)-H2O (60/25/4) as a development solvent. The bioassay was performed with Staphylococcus aureus 209P as the test organism. TLC plates were placed in bioassay plates and overlaid with Muller-Hinton agar (0.5% [wt/vol]; Difco) which had been seeded with S. aureus 209P. The inhibitory zones were visible after 12 h of incubation at 37°C, followed by staining with GelCodeR Blue Stain reagent (Pierce).
NMR and MS analyses.
Three milligrams of the sample was dissolved in CD3OD. Analyses were performed within 3 days, since the degraded signals were detected after the sample had been dissolved for a few days. The 1H nuclear magnetic resonance (NMR) spectra were obtained at 500 MHz, and the 13C NMR spectra were obtained at 125.8 MHz (JEOL JNM-A500 spectrometer). Mass spectrometric (MS) analysis was done with a JEOL JMS-SX102 spectrometer.
DNA manipulations and strain construction.
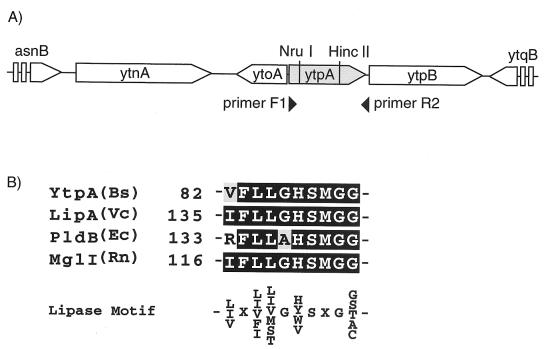
The methods described by Sambrook et al. (11) were used for the construction, isolation, and identification of recombinant DNA. Construction of a strain harboring a disrupted ytpA allele was carried out with plasmid pAE41 (15). The ytpA fragment was amplified by PCR with primers ytpA-F1 (5′-ATGTGGACCTGGAAAGCAG-3′) and ytpA-R2 (5′-CGGTACTGTCAACACATGC-3′). An 812-bp NruI-HincII fragment whose sequence was specific for a sequence internal to ytpA was excised from the PCR product and ligated with pAE41 treated with SmaI to create a gene disruption plasmid, pLE41 (see Fig. 5A). Strain 168 was transformed with pLE41, which selects for the vector-encoded erythromycin marker. Correct integration (via Campbell-like recombination of pLE41) was confirmed by PCR. A representative strain designated JJ25 was used for further experiments.
FIG. 5.
(A) Gene organization of ytpA locus for B. subtilis. The open reading frames are shown by horizontal arrows that designate the direction of transcription. NruI and HincII indicate the reaction positions for these restriction enzymes. (B) Alignment of amino acid sequences of the segments with a motif conserved in the lipase. Bs, Bacillus subtilis; Vc, Vibrio comma; Ec, Escherichia coli, Rn; Rattus norvegicus.
Phospholipase assay.
A crude cell extract was prepared as described by Hrafnsdottir and Menon (3), and the phospholipase reaction was done by the method of Nishijima et al. (6), with slight modifications, as follows. Cells from a 10-ml culture were suspended in 1 ml of 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-2 mM CaCl2-1% Triton X-100 (buffer A). The cells were disrupted with a French press at a pressure of 18,000 kPa. After removal of the cell debris by centrifugation (8,000 × g, 20 min), the supernatant thus obtained was used as a crude extract. The protein concentration was adjusted with buffer A to give a concentration of 5 mg/ml. Each reaction mixture contained 5 μg of dl-α-phosphatidylcholine dimyristoyl (Sigma) and 500 μg of protein in 130 μl of buffer A. After the reaction was allowed to proceed for 2 h at 37°C, the reaction mixture was subjected to extraction with 400 μl of CHCl3-MeOH (2/1). The extract was concentrated, redissolved in 10 μl of MeOH, and applied to a silica gel TLC plate, followed by development with CHCl3-MeOH-water (65/25/4, by volume). After the development of the plate, the TLC plates were stained with 0.1% difluorocein, and the reaction products were detected under UV light.
RESULTS
Production and isolation of an antibiotic.
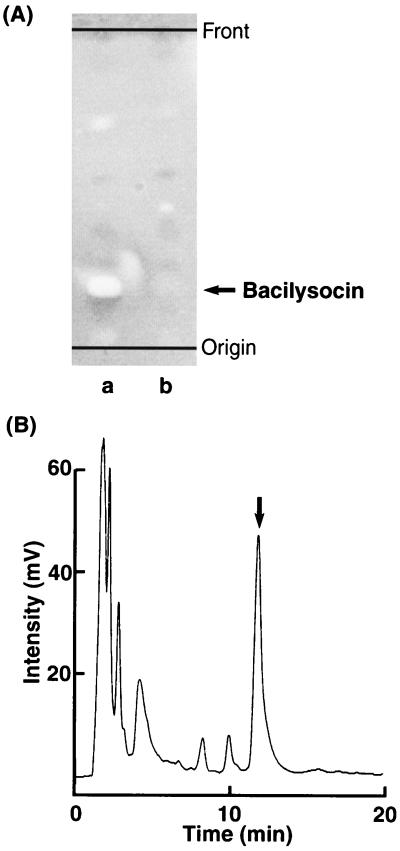
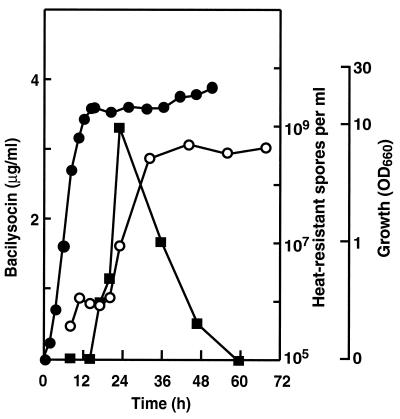
We found that B. subtilis 168 produces an antimicrobial substance (later named bacilysocin; see below) intracellularly when it is grown in NG medium. The activity of the compound was detected as a superimposed spot after TLC-bioautography (Fig. 1A). Its production commenced just after the cells entered the stationary phase but before the formation of heat-resistant spores (Fig. 2). Later, the activity of the antibiotic decreased rapidly.
FIG. 1.
Detection and isolation of the antimicrobial substance. (A) TLC-bioautography of antimicrobial substance. A BuOH extract from cells of strain 168 (lane a) or strain JJ25 (lane b) was subjected to TLC. The arrow indicates the bioactive position. (B) HPLC profile of the crude antimicrobial substance. The arrow indicates the bioactive fraction. Details of the procedures for TLC bioautography and HPLC are described in Materials and Methods.
FIG. 2.
Antibiotic production and sporulation during growth of B. subtilis 168. Symbols: •, growth; ▪, bacilysocin; ○, heat-resistant spores. OD660, optical density at 660 nm.
We next attempted to isolate the antimicrobial substance. Cultured cells (cultured in 40 liters) were collected, and the cellular contents were extracted three times with 4 liters of 50% n-butanol. The organic layer was collected and concentrated in vacuo. The resulting crude extract (12.9 g) was suspended in 1.6 liters of 10% MeOH, adjusted to pH 7 with NaOH, and extracted three times with an equal volume of ethyl acetate. The aqueous layer was then adjusted to pH 2 and was again extracted three times with ethyl acetate. The solvent of the acidic organic layer was evaporated, resulting in 4.78 g of crude extract. Silica gel (400 ml) was equilibrated with the development solution (CHCl3-MeOH-H2O [20/5/1]), and the extract, dissolved in 20 ml of the same solution, was applied to a chromatography column. Chromatography was carried out with 1.2 liters of the development solution. The active fraction was collected and dried in vacuo, and then the resulting crude sample was redissolved in 4 ml of CHCl3-MeOH (4/1). This mixture was applied to a DEAE Sepharose column (200 ml; Pharmacia) equilibrated with CHCl3-MeOH (4/1). After the column was washed, development was performed with 600 ml of CHCl3-MeOH (4/1) containing 2% aqueous NH3OH and 0.02 M ammonium acetate. The active fraction was collected and vacuum evaporated, resulting in a crude substance that weighed 0.25 g. Finally, this crude fraction was purified by high-pressure liquid chromatography (HPLC) with a Sephasil peptide C18 column (Pharmacia) with 62% MeOH in 1 mM phosphate buffer (pH 7.5) as an elution solvent (Fig. 1B). Eventually, 170 mg of pure antimicrobial substance was obtained.
Determination of structure.
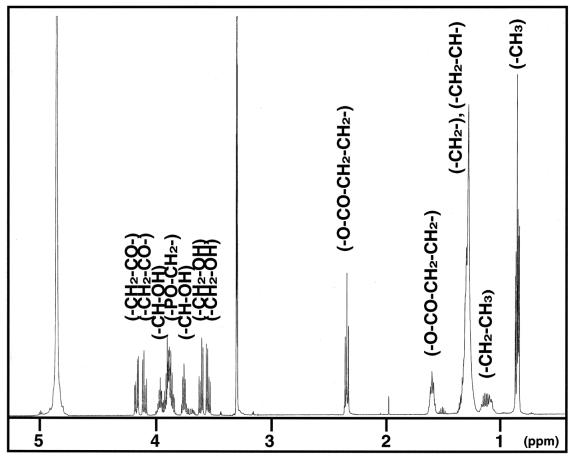
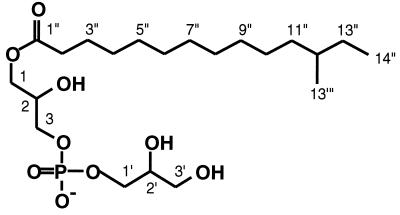
The 1H NMR spectrum of the purified substance (Fig. 3) displayed signals typical for polar glycerolipids (signals from the acyl groups at δ 1.60 to 2.35 ppm and from protons adjacent to oxygen at δ 3.55 to 4.17 ppm). The presence of a long aliphatic hydrocarbon chain is indicated by a large band of methylene protons at δ 1.2 to 1.4 ppm without any detectable olefinic proton or carbon signals. The 13C NMR spectrum (data not shown) displayed the presence of an acyl moiety (δ 25.99 to 34.93 ppm), one carbonyl moiety (δ 175.40 ppm), and aliphatic moieties (δ 28.2 to 37.8 ppm). The 31P NMR signal (data not shown) showed a connection between the carbon signals at δ 67.58 and 67.61 ppm and the methylene protons at δ 3.89 ppm, which indicates the existence of a phosphatidylglycerol structure. The length of the aliphatic chain predicted by the 1H NMR signals was confirmed by fast atom bombardment (FAB)-MS analysis. By FAB-MS analysis the parent peak was detected as m/z 515.32 (M + 2Na)+ (data not shown). Consequently, we determined the structure of the substance, as shown in Fig. 4, and named this novel antibiotic bacilysocin. All proton and carbon signals were successfully assigned by two-dimensional NMR analysis. A detailed assignment of the NMR spectra is summarized in Table 1. There are three chiral centers in this compound (at positions C-2, C-2′, and C-12). The stereochemistries of the hydroxyl groups at positions C-2 and C-2′ and the methyl group at position C-12 have yet to be determined.
FIG. 3.
1H NMR spectrum of the antimicrobial substance.
FIG. 4.
Structure of the antimicrobial substance, bacilysocin.
TABLE 1.
NMR chemical shifts for bacilysocin in MeOH-d4
| Moiety | Carbon no. | δH | δC (ppm) |
|---|---|---|---|
| Acyl | 13′″ | 0.85 (3H,d) | 19.63 |
| 14″ | 0.87 (3H, t) | 11.74 | |
| 13″ | 1.15, 1.3 (2H, m) | 30.72 | |
| 12″ | 1.3 (H, m) | 35.70 | |
| 11″ | 1.1, 1.3 (2H, m) | 37.80 | |
| 4″–10″ | 1.2–1.4 (14H, m) | 28.20, 30.23, 30.43, 30.59, 30.62, 30.78, 31.11 | |
| 3″ | 1.60 (2H, m) | 25.99 | |
| 2″ | 2.35 (2H, dt) | 34.93 | |
| 1″ | 175.40 | ||
| Glycerate | 3′ | 3.55, 3.61 (1H, dd) | 63.87 |
| 2′ | 3.76 (1H, m) | 72.66 | |
| 1′ | 3.89 (4H, m) | 67.58 | |
| Glycerol | 3 | 3.92 (1H, m) | 67.61 |
| 2 | 3.96 (1H, m) | 69.93 | |
| 1 | 4.10, 4.17 (1H, dd) | 66.27 |
Antimicrobial activity of bacilysocin.
Although bacilysocin was isolated by using S. aureus 209P as the test organism, we examined the activity of this antibiotic against various bacteria (35 strains) and fungi (22 strains) as test organisms. The results obtained with only a limited number of organisms are shown in Table 2. Bacilysocin displayed apparent activity against a couple of strains of S. aureus, but showed no activity (MIC, >100 μg/ml) against the other gram-positive bacteria examined. Instead, its activity was more pronounced against the eucaryotic organism Saccharomyces cerevisiae, in addition to the fungi Candida pseudotropicalis and Cryptococcus neoformans, which are characterized by nonfilamentous growth (Table 2).
TABLE 2.
Antibiological spectrum of bacilysocin
| Test organism | MICa (μg/ml) |
|---|---|
| Candida albicans 3147 IMC F-4 | >100 |
| Candida pseudotropicalis IMC F-2 | 25 |
| Cryptococcus neoformans var. neoformans sero AIMC F-185 | 25 |
| Cryptococcus neoformans IMC F-10 | 25 |
| Saccharomyces cerevisiae IMC F-7 | 5 |
| Trichophyton rubrum IMC F-184 | >100 |
| Aspergillus niger IMC F-16 | >100 |
| Staphylococcus aureus FDA209P | 50 |
| Staphylococcus aureus Smith | 25 |
| Staphylococcus aureus MS9610 | >100 |
| Micrococcus luteus FDA16 | 100 |
| Bacillus subtilis NRRL B-558 | >100 |
| Escherichia coli K-12 | >100 |
| Pseudomonas aeruginosa A3 | >100 |
| Mycobacterium smegmatis ATCC 607 | >100 |
The MICs for the bacteria were determined by the agar dilution method with Mueller-Hinton agar (Difco). The MICs for the yeast and fungi were determined by the same method with synthetic amino acid medium for fungi agar.
Effect of disruption of ytpA gene.
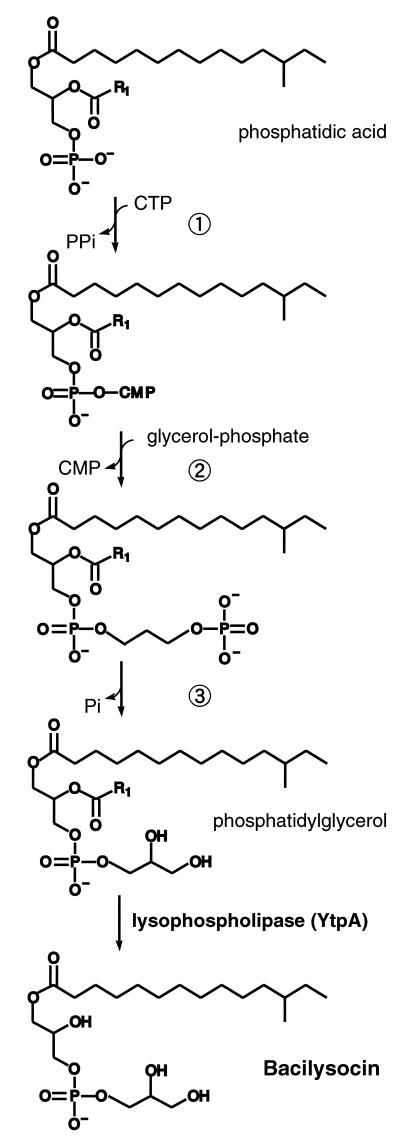
According to the structure assigned as described above, it was possible to assume that certain kinds of lipolytic enzymes may play a role in the synthesis of bacilysocin. We postulated that the final step in the biosynthesis of bacilysocin involves the direct cleavage of an acyl ester bond at the sn-2 position for phosphoglycerides (see Fig. 7). If this is the case, a phospholipase A2-like enzyme (specific cleavage at position sn-2) or a phospholipase B-like enzyme (nonspecific cleavage at position sn-1 or sn-2) may be a key enzyme for bacilysocin synthesis. After searching the genome project database, we found a phospholipase-like gene, ytpA, which encodes a protein homologous to lysophospholipase (LPL) (Fig. 5A). LPL is known to catalyze the hydrolysis of monoacyllysophospholipids. Two LPLs, L1 and L2, are reported to exist in Escherichia coli (4). However, the discrimination between LPL and phospholipase B is unclear, since a number of enzymes termed LPLs can hydrolyze diacylphospholipids and therefore fall into the category for phospholipase B. A search for an enzyme with homology to ytpA revealed the E. coli phospholipase B, and the YtpA protein was found to have a motif conserved in the lipase (Fig. 5B). Thus, it is possible that the ytpA gene is involved in the hydrolysis of the 2-sn-acyl of phosphatidylglycerol in B. subtilis, leading to the synthesis of bacilysocin. To assess this possibility we constructed a ytpA disruptant. Construction of a strain harboring a disrupted allele in ytpA was carried out as described in Materials and Methods (see also Fig. 5A). Strikingly, a ytpA disruptant strain thus generated, strain JJ25, was found to entirely lose the ability to produce bacilysocin, as demonstrated by TLC-bioautography (Fig. 1A, lane b). The growth was affected only slightly. These results indicate that ytpA is responsible for bacilysocin synthesis in B. subtilis. In addition to antibiotic production, the mutant displayed a 10-fold reduction in the ability to form spores (5 × 107 spores/ml) compared to the ability of the parent strain (6 × 108 spores/ml). As expected, the disruptant strain, JJ25, actually lacked phospholipase activity, as determined by using phosphatidylcholine as a substrate (Fig. 6). It is notable that maximum phospholipase activity in the parent strain (detected at 24 to 28 h; Fig. 6) coincides in time with active bacilysocin synthesis (Fig. 2).
FIG. 7.
Scheme for the bacilysocin biosynthetic pathway. The biosynthetic steps represented by the circled numbers 1, 2, and 3 have been well elucidated in E. coli and B. subtilis (for a review, see reference 5) and are catalyzed by the enzymes CDP diglyceride synthase, phosphatidylglycerol-phosphate synthase, and phosphatidylglycerol-phosphate phosphatase, respectively.
FIG. 6.
Changes in phospholipase activity during growth of B. subtilis 168 (parent strain) and the ytpA disruptant, strain JJ25. PC, phosphatidylcholine (substrate); lyso-PC, lysophosphatidylcholine (product). The numbers beneath each lane are times (in hours).
Effect of relA mutation on bacilysocin production.
The production of a certain antibiotic (not yet identified) by strain 168 is known to be blocked by introduction of the relA mutation (8), which is a point mutation that alters Gly240 to Glu in the RelA protein (T. Inaoka and K. Ochi, unpublished data), resulting in a severe reduction in the level of ppGpp accumulation (7). In contrast, bacilysocin production was not affected when the relA mutation was introduced into strain 168, indicating that the biosynthesis of bacilysocin is not under the control of ppGpp (data not shown).
DISCUSSION
B. subtilis strains have been known to produce lipopeptide-type antibiotics, the conformation of which is either a cyclopeptide (i.e., iturinics) or a macrolactone (i.e., surfactins, fengycins, and plipastatins), which contains a long hydrophobic tail (for a review, see reference 16). Current genome projects provide us with a lot of information. On the basis of total genome sequences, even the structures of metabolites such as polyketides can be predicted. To our knowledge, bacilysocin is the first antibiotic with a phospholipid structure to be found to be produced by B. subtilis. Bacilysocin (lysophosphatidylglycerol) may be derived from phosphatidylglycerol through acyl ester hydrolysis. Phosphatidylglycerol is the major component of phospholipids in B. subtilis, which constitutes about 75% of the total phospholipids (5). One lipolytic enzyme may be sufficient for the conversion of phosphatidylglycerol to bacilysocin. As mentioned above, LPL often shows phospholipase B-like activity, e.g., hydrolysis of 1- or 2-acyl bonds. Gene disruption experiments show that YtpA actually has the ability to catalyze the hydrolysis reaction of one acyl bond in phosphatidylcholine, resulting in lysophosphatidylcholine (Fig. 6), although the actual cleavage point is unknown. Therefore, it is conceivable that the YtpA protein is a key enzyme for the synthesis of bacilysocin by the hydrolysis of phosphatidylglycerol (Fig. 7). It is an intriguing notion that bacilysocin somehow plays a role in the sporulation of B. subtilis, since bacilysocin nonproducers (due to ytpA disruption) displayed an impaired ability to form spores.
Acknowledgments
This work was supported by a grant from the Organized Research Combination Systems of the Science and Technology Agency of Japan.
We are grateful to Yoshiaki Ohashi and Koji Kasai for advice concerning the gene manipulation experiments.
REFERENCES
- 1.Babasaki, K., T. Takao, Y. Shimonishi, and K. Kurahashi. 1985. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. (Tokyo) 98:585–603. [DOI] [PubMed] [Google Scholar]
- 2.Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi. 1998. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42:2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hrafnsdottir, S., and A. K. Menon. 2000. Reconstitution and partial characterization of phospholipid flippase activity from detergent extracts of the Bacillus subtilis cell membrane. J. Bacteriol. 182:4198–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karasawa, K., and S. Nojima. 1991. Lysophospholipases from Escherichia coli. Methods Enzymol. 197:437–445. [DOI] [PubMed] [Google Scholar]
- 5.Mendoza, D., R. Grau, and J. E. Cronan Jr. 1993. Biosynthesis and function of membrane lipids, p. 411–421. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 6.Nishijima, M., Y. Akamatsu, and S. Nojima. 1974.Purification and properties of a membrane-bound phospholipase A1 from Mycobacterium phlei. J. Biol. Chem. 249:5658–5667. [PubMed] [Google Scholar]
- 7.Ochi, K., J. C. Kandala, and E. Freese. 1981. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J. Biol. Chem. 256:6866–6875. [PubMed] [Google Scholar]
- 8.Ochi, K., and S. Ohsawa. 1984. Initiation of antibiotic production by the stringent response of Bacillus subtilis Marburg. J. Gen. Microbiol. 130:2473–2482. [DOI] [PubMed] [Google Scholar]
- 9.Ozcengiz, G., N. G. Alaeddinoglu, and A. L. Demain. 1990. Regulation of biosynthesis of bacilysin by Bacillus subtilis. J. Ind. Microbiol. 6:91–100. [DOI] [PubMed] [Google Scholar]
- 10.Paik, S. H., A. Chakicherla, and J. N. Hansen. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273:23134–23142. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Stover, A. G., and A. Driks. 1999. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J. Bacteriol. 181:1664–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuge, K., T. Ano, M. Hirai, Y. Nakamura, and M. Shoda. 1999. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother. 43:2183–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vollenbroich, D., G. Pauli, M. Ozel, and J. Vater. 1997. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis. Appl. Environ. Microbiol. 63:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada, K. 1989. Ph.D. thesis. Hiroshima University, Hiroshima, Japan.
- 16.Zuber, P., M. M. Nakano, and M. A. Marahiel. 1993. Peptide antibiotics, p. 897–916. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.