Abstract
A genomic library from a strain of Salmonella enterica serovar Paratyphi B that exhibits multiple drug resistance (MDR) was constructed in Escherichia coli. Two of the recombinant plasmids, pNOR5 and pNOR5, conferred resistance only to fluoroquinolones in E. coli, whereas the third, pNCTR4, conferred the MDR phenotype. Sequence and subcloning analysis showed that it is the presence of RecA on the first two plasmids which confers resistance to fluoroquinolones in E. coli. A similar analysis established that the MDR phenotype conferred by pNCTR4 is due to a gene, rma (resistance to multiple antibiotics), which encodes a 13.5-kDa polypeptide. The derived sequence for Rma exhibits a high degree of similarity to those of a group of MarA-like activators that confer MDR in E. coli. A MalE-Rma fusion protein was purified to near homogeneity and was shown to interact with a DNA fragment carrying a MarA operator sequence. Furthermore, overexpression of rma in E. coli caused changes in the outer membrane protein profile that were similar to those reported for MarA. These results suggest that Rma might act as a transcriptional activator of the marA regulon.
The emergence of clinical isolates of salmonellae that exhibit multiple drug resistance (MDR) represents an increasing threat to human health. While the use of antibiotics in livestock feed is considered a primary cause of this phenomenon, the extensive use of fluoroquinolones in the treatment of typhoid also led to the frequent isolation of typhoid strains that exhibit MDR in numerous outbreaks in developing countries in Africa and south Asia (26, 32). Recently, the first recognized outbreak of fluoroquinolone-resistant Salmonella enterica serovar Paratyphi infection in the United States was reported in two nursing homes and one hospital in Oregon (23).
The molecular mechanisms of MDR have been studied extensively in Escherichia coli. These include the induction of efflux pumps to promote extrusion (the acrRAB and emrRAB loci) as well as the down regulation of outer membrane porins (e.g., OmpF) to reduce the inflow of a variety of structurally unrelated antibiotics (16, 18, 22). The marRAB (5) and soxRS (21, 35) systems exert overlapping effects on the regulation of efflux pumps and porin synthesis in E. coli via a sophisticated control circuit (18). While the natural stimulants for the mar and sox systems overlap and some of their derivatives are structurally related, they contribute independently to the control network of MDR through direct interactions with the affected operons. A recent paper by Barbosa and Levy (2) reported on the differential expressions of over 60 chromosomal genes in the MarA regulon of E. coli. RamA of Klebsiella pneumoniae and Enterobacter cloacae and PqrA of Proteus vulgaris, which have moderate sequence similarities to the E. coli MarA protein, have also been reported to confer MDR when they are overexpressed in E. coli (11, 13, 15).
Understanding of the molecular basis of MDR is important for the design and use of antimicrobial agents. One of the laboratories collaborating on the present study has previously reported on the isolation of a derivative of S. enterica serovar Paratyphi B that exhibits the MDR phenotype (8). We initiated the present study in order to identify the genetic determinants of this resistance. The study described in the present paper indicates that the rma (resistance to multiple antibiotics) gene of serovar Paratyphi B confers the MDR phenotype in E. coli. We report on an analysis of the outer membrane proteins in a recombinant strain of E. coli which overexpresses a MalE-Rma fusion protein as well as the results of gel retardation assays with the purified fusion protein. The results indicate that the Rma protein is a new member of the MarA-SoxS family of transcriptional regulators.
MATERIALS AND METHODS
Strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. Unless otherwise indicated, cultures were grown in Luria-Bertani (LB) enriched medium under aerobic condition at 37°C with the following supplements, as required: ampicillin at 50 μg/ml and norfloxacin at 2 μg/ml. Cell growth was monitored optically at 600 nm.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E coli DH5α | supE44 lacU169 (80lacM15) hsdR1 recA1 endA1 gyrA96 thi-1 relA1 | New England BioLabs |
| S. enterica serovar Paratyphi B | Wild-type strain; a clinical isolate | 8 |
| S. enterica serovar Paratyphi B M95 | Mutant strain with MDR | 8 |
| Plasmids | ||
| pUC19 | Multicopy vector, AMPr | New England BioLabs |
| pNCTR4 | NORr CHLr TETr GENs | This study |
| pGEM-T | PCR cloning vector | Promega |
| pNCTR44 | NORr CHLr TETr GENs | This study |
| pNCTR42 | NORr CHLr TETr GENs | This study |
| pNOR5 | NORr | This study |
| pNOR6 | NORr | This study |
| pNOR7 | recA subclone, NORr | This study |
| pMal-c2X | Purification and expression vector | New England BioLabs |
| pMal-Rma | MalE-Rma protein fusion | This study |
NOR, norfloxacin; CHL, chloramphenicol; TET, tetracycline, GEN, gentamicin; AMP, ampicillin.
Construction of a shotgun genomic library.
Chromosomal DNA was prepared from a derivative of S. enterica serovar Paratyphi B that exhibits MDR with an AquaPure genomic DNA isolation kit (Bio-Rad Laboratories). The DNA was partially digested with Sau3AI, and fragments of 1 to 10 kb were separated by electrophoresis on 0.5% agarose and purified with a QIAEX II gel extraction kit (Qiagen). The vector plasmid, pUC19, was treated with restriction endonuclease BamHI and was subsequently dephosphorylated with bacterial alkaline phosphatase. The ligation mixture was incubated with T4 DNA ligase (Boehringer Mannheim GmbH, Mannheim, Germany) overnight at 14°C. For transformation, competent E. coli DH5α cells were prepared by the method of Inoue et al. (12).
Drug susceptibility testing.
The MICs of the antimicrobial agents were determined by the broth microdilution technique (19). Twofold serial dilutions of antimicrobial agents in 100 μl of antibiotic medium 3 with an inoculum of 103 to 104 CFU of logarithmically grown cells were prepared in 96-well microtiter plates. The range of antibiotic concentrations tested was 0 to 128 μg/ml. The MIC was defined as the lowest concentration of the antimicrobial agent that inhibits visible growth after 18 to 24 h of incubation at 37°C. The MICs reported here represent the means for quadruplicate experiments.
Preparation of outer membrane proteins and their analysis by polyacrylamide gel electrophoresis (PAGE).
Crude outer membrane proteins were prepared by the procedure described by Spratt (29). Cells were grown in 1 liter of LB broth at 37°C with vigorous aeration and were harvested in the mid-log phase to the early stationary growth phase. After the cells were cooled on ice, they were centrifuged at 6,000 × g for 6 min in a GSA rotor and were resuspended in 25 ml of ice-cold 10 mM sodium phosphate (pH 7.0). The cells were disrupted by passage twice through a French pressure cell (Sim-Aminco; Spectronic Unicam, Rochester, N.Y.) at 15,000 lb/in2. Large cell debris and unbroken cells were removed by centrifugation at 8,000 × g for 20 min at 4°C in the SS-34 rotor of a Sorvall RC2B centrifuge. The cell membranes were pelleted out of the supernatant by centrifugation at 100,000 × g for 40 min at 4°C in the Ti 70 rotor of an XL-90 Beckman ultracentrifuge. The membranes were suspended in 6 ml of ice-cold phosphate buffer, washed twice by centrifugation at 100,000 × g, and finally stored in the same buffer at a concentration of 40 mg/ml at −80°C.
The crude outer membrane proteins were subjected to further purification by the procedure described by Sawai et al. (28). The crude outer membrane fraction obtained from 1 liter of bacterial culture was suspended in 3 ml of 10 mM phosphate (pH 7.0) containing 2.0% sodium lauroyl sarcosinate (Sigma), and the mixture was incubated at 20°C for 10 min. The suspension was then centrifuged at 100,000 × g for 40 min at 10°C in a 50 Ti rotor (Beckman-Coulter). The pellet was resuspended, washed twice with 1% lauroyl sarcosine in phosphate buffer, and then used as the purified outer membrane preparation. The concentration of the protein was measured by a protein assay (Bio-Rad Laboratories, Hercules, Calif.).
Sodium dodecyl sulfate (SDS)-PAGE of the outer membranes was performed in a 10% acrylamide gel by the method of Sambrook et al. (27). The purified outer membrane protein, solubilized in the same volume of 2× SDS gel-loading buffer (1× buffer is composed of 50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol), was heated in boiling water for 2 min before application. The gel was stained with Coomassie brilliant blue G (Sigma).
Western blot analysis of TolC.
The level of TolC expression was determined with antiserum specific for the TolC protein and purified outer membranes. Equal amounts (30 μg each) of outer membrane proteins were subjected to separation by SDS-PAGE on a 10% gel. The gel was then equilibrated in transfer buffer containing 10 mM Tris base, 200 mM glycine, and 10% methanol for 5 min before it was electroblotted onto a polyvinylidene difluoride membrane (Applied Biosystems). The standard protocol for Western blotting was followed (31), and the immunocomplex was detected by the presence of alkaline phosphatase-conjugated secondary antibody and the chromogenic substates nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate.
Expression and purification of MalE-Rma fusion protein.
The pMAL protein fusion system (New England BioLabs) was used for expression of the MalE-Rma fusion protein. The structural gene of rma was amplified by PCR with Pfu polymerase (Promega) and two oligonucleotide primers whose sequences were specific for regions flanking rma. The purified PCR product was ligated to XmnI-digested vector pMAL-c2X so that the first methionine codon was fused in frame to the C terminus of a modified MalE without the signal peptide. After introduction of the plasmid into competent E. coli DH5α cells, successful transformants were selected by the protocol suggested by the manufacturer. The resulting recombinant plasmid, pMAL-Rma, was confirmed by restriction analysis and DNA sequencing. Synthesis of the MalE-Rma protein in the recombinant E. coli strain harboring pMAL-Rma was induced by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Following growth and induction, the cells were harvested by centrifugation at 5,000 × g at 4°C. The cells were resuspended in 50 mM Tris-HCl (pH 8.0) containing 1 mM phenylmethylsulfonyl fluoride and were disrupted as described above. Ruptured cells were centrifuged at 12,000 × g for 10 min at 4°C to eliminate cell debris and unbroken cells. The supernatant was loaded at a flow rate of 1.0 ml/min onto a 10-ml heparin column (Pharmacia), which was preequilibrated with 50 mM Tris-HCl (pH 8.0) at 4°C. Proteins were eluted with a linear gradient of KCl in the same buffer. The 55-kDa MalE-Rma fusion protein, monitored by SDS-PAGE and mobility shift assays, was eluted at 0.2 M KCl. Fractions containing the MalE-Rma fusion protein were combined and passed onto a 20-ml amylose affinity column (New England BioLabs). The MalE-Rma fusion protein was released from the column by elution with 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-10 mM maltose. The protein concentration was determined by the method of Bradford (3).
Mobility shift assays.
A 26-bp fragment of double-stranded DNA containing the MarA operator sequence, 5′-GGGATTTAGCAAACGTGGCATCGG-3′, was generated from the annealing of two oligonucleotide primers with complementary sequences. For use as a probe in mobility shift assays, the 26-bp double-stranded DNA fragment was labeled with [γ-32P]ATP by use of the DNA polymerase Klenow fragment, and the excess labeled nucleotide was eliminated with Sephadex G-25.
The binding reaction mixture (20 μl) contained various concentrations of the purified MalE-Rma fusion protein in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA in the presence of 0.2 μg of nonspecific sheared chromosomal DNA of Bacillus subtilis. The reaction mixtures were incubated at room temperature for 15 min before they were loaded onto a 10% native polyacrylamide gel equilibrated with running buffer containing 40 mM Tris-acetate and 1 mM EDTA. The gels were prerun for 30 min before they were loaded. After the gels were loaded, they were run at 24 mA until the bromophenol blue dye front had migrated at least 50% of the gel length. The gels were transferred to a piece of filter paper, dried, and exposed to a phosphorimager plate (Fuji) for image analysis.
Nucleotide sequence accession number.
The nucleotide sequence was determined with an ABI Prism 377 DNA sequencer. The sequence of rma and its flanking regions has been deposited in the GenBank database under accession number AF411103.
RESULTS
Cloning of S. enterica serovar Paratyphi B MDR genetic determinants.
A previous report described the isolation and characterization of a norfloxacin-resistant derivative of S. enterica serovar Paratyphi B that exhibited an MDR phenotype (8). The MICs of a number of antibiotics were determined for mutant strain M95 (Table 2). The results show that the MICs of norfloxacin, tetracycline, and chloramphenicol for M95 are significantly higher than those for the wild-type strain, while the MIC of gentamicin was unchanged.
TABLE 2.
Effect of Rma expression on antibiotic susceptibility
| Strain | MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| Nor-floxacin | Chloram-phenicol | Tetra-cycline | Genta-micin | |
| E. coli | 0.05 | 2.0 | 0.5 | 1.0 |
| S. enterica serovar Paratyphi | 0.1 | 2.0 | 0.5 | 1.0 |
| S. enterica serovar Paratyphi M95 | 8.0 | 16.0 | 4.0 | 1.0 |
| E. coli/pNOR5 | 16.0 | 2.0 | 0.5 | 1.0 |
| E. coli/pNCTR4 | 8.0 | 16.0 | 4.0 | 1.0 |
| E. coli/pMal-c2X | 0.05 | 2.0 | 0.5 | 1.0 |
| E. coli/pMal-Rma | 8.0 | 16.0 | 4.0 | 1.0 |
Determined by the broth microdilution technique (19). MICs represent the means for quadruplicate experiments.
To identify the genetic determinants for the MDR phenotype of S. enterica serovar Paratyphi B M95, a shotgun genomic library of this mutant strain was constructed by use of plasmid pUC19 as the cloning vector. The resulting shotgun library cocktail was introduced into E. coli DH5α by transformation, and transformants were selected on LB agar plates containing 100 μg of ampicillin per ml and 2 μg of norfloxacin per ml. Three clones were obtained from this experiment, and the presence of recombinant plasmids in these clones was confirmed by restriction enzyme digestion. The MICs of norfloxacin, tetracycline, chloramphenicol, and gentamicin for the three E. coli recombinants and parent strain E. coli DH5α were determined. Two E. coli recombinants, carrying pNOR5 and pNOR6, respectively, exhibited a phenotype of resistance to norfloxacin only. Only one E. coli recombinant, harboring plasmid pNCTR4, displayed the MDR phenotype, with the MICs for the transformant being similar to those for serovar Paratyphi B M95 (Table 2).
Overexpression of RecA causes norfloxacin-specific resistance in E. coli DH5α.
As described above, three recombinant strains of E. coli DH5α showing specific resistance to norfloxacin were identified from shotgun cloning experiments. Sequence analysis indicated that two of the three plasmids, pNOR5 and pNOR6, contained the recA gene. Earlier reports have indicated that the RecA protein plays a role in the repair of DNA damage following exposure to quinolones (14, 24, 33). Accordingly, the recA gene of S. enterica serovar Paratyphi B along with its putative ribosomal binding site was amplified by PCR and cloned into pUC19. When the resulting plasmid, pNOR7, was introduced into E. coli DH5α, which possesses the recA1 mutation, the resulting recombinant strain retained resistance to norfloxacin. These results show that the recA gene alone confers resistance to norfloxacin in E. coli.
Identification of the rma gene conferring MDR in E. coli.
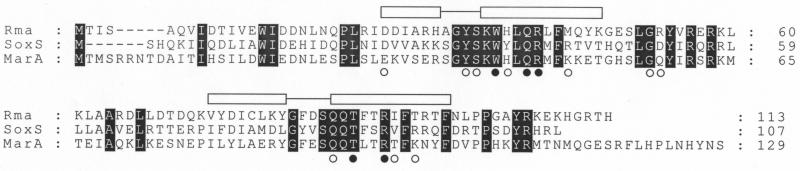
The 14-kb chromosomal insert of pNCTR4 was digested with restriction endonucleases EcoRI and HindIII, and the resulting three restriction fragments were cloned separately into pUC19 for subcloning analysis. The MICs for E. coli recombinants harboring the resultant plasmids (pNCTR41, pNCTR42, and pNCTR43) were determined. The results (Table 2) revealed that plasmid pNCTR42 harbors the gene that confers MDR in E. coli. The nucleotide sequence of the 5.6-kb EcoRI insert of pNCTR42 was determined and was subsequently analyzed by a search for homology with the sequences in the nonredundant database as well as the unfinished genomic sequences of S. enterica serovar Paratyphi A and S. enterica serovar Typhimurium at the National Center for Biotechnology Information (NCBI) with the BLAST program. The results of the sequence comparison indicate that the 5.6-kb insert of pNCTR42 is most likely the ligation product of two noncontiguous chromosomal fragments. Importantly, the chromosomal insert in pNCTR42 contains an intact open reading frame (ORF) that appeared to be a particularly promising candidate as a genetic determinant of MDR on the basis of a comparison of its sequence with the sequences in the protein database at NCBI. This ORF encodes a putative polypeptide of 113 amino acids (13.5 kDa) which exhibits 75 to 88% sequence identity to a group of transcriptional activators that have been reported to confer MDR (11, 13, 15): RamA of K. pneumoniae (GenBank accession number Q48413), Enterobacter aerogenes (GenBank accession number CAB95659), and E. cloacae (GenBank accession number P55922). The amino acid sequence of this ORF possesses a moderate 38% sequence identity to those of MarA (GenBank accession number L06966) and SoxS (GenBank accession number M60111) of E. coli (Fig. 1).
FIG. 1.
Multiple-sequence alignment of Rma, SoxS, and MarA. Sequences were aligned by use of the Clustal W program (30). Residues that are conserved among these proteins are shaded. The locations of two helix-turn-helix motifs of MarA are marked with open boxes (helices) connected by lines (turns). Black circles, MarA residues that are involved in determination of sequence specificity; open circles, residues in contact with the phosphate backbone of the operator DNA (25).
The sequence containing this ORF along with the preceding putative ribosomal binding site was amplified from pNCTR42 by PCR with Taq DNA polymerase and two flanking oligonucleotide primers. The PCR product was purified and cloned into vector plasmid pGEM-T. When it was introduced into E. coli DH5α, the recombinant strain harboring the resulting plasmid, pNCTR44, displayed the MDR phenotype (Table 2), as was the case with pNCTR42. The corresponding gene was therefore designated rma (resistance to multiple antibiotics).
The rma gene and its upstream regulatory region were also amplified by PCR from the genomic DNA of wild-type S. enterica serovar Paratyphi B. After this region was cloned into pGEM-T, the nucleotide sequence of this region was determined and compared with that of pNCTR42, which originated from the mutant strain of S. enterica serovar Paratyphi B with MDR. The two nucleotide sequences were completely identical.
In vitro binding of the MalE-Rma fusion protein to a MarA operator of E. coli.
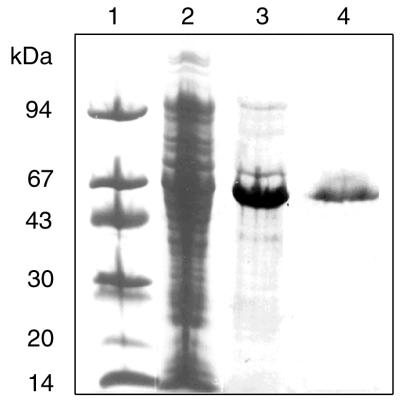
As indicated above, the Rma protein of S. enterica serovar Paratyphi B exhibited a very high level of sequence identity to the RamA protein of K. pneumoniae and a moderate degree of identity to the MarA and SoxS proteins of E. coli. Furthermore, most of the residues in two helix-turn-helix structures that were shown to be important for sequence recognition and base contacts in MarA (25) are conserved in Rma of S. enterica serovar Paratyphi B (Fig. 1). This conservation suggested the possibility of cross interactions between these regulatory proteins and their cognate operators. To investigate this possibility, a MalE-Rma fusion protein was constructed to determine if Rma binds to MarA operator sequences. The structural gene of rma was amplified by PCR and fused in frame to the C terminus of the maltose binding protein (MalE) in expression vector pMAL-c2X. After induction with IPTG, the MalE-Rma fusion protein was purified from E. coli cells harboring the recombinant plasmid. SDS-PAGE analysis indicated that the 55-kDa MalE-Rma fusion protein (42 kDa of MalE plus 13 kDa of Rma) was purified to near homogeneity (Fig. 2).
FIG. 2.
Purification of the MalE-Rma fusion protein. A cell extract was prepared from a recombinant strain of E. coli harboring pMal-Rma and was subjected to fractionation as described in Materials and Methods. Protein samples were separated by SDS-PAGE and stained with Coomassie brilliant blue. Lane 1, polypeptide molecular size standards; lane 2, crude extract after induction; lane 3, amylose affinity column fraction; lane 4, HiTrap heparin affinity column fraction.
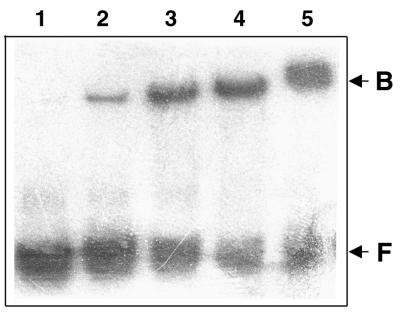
Binding of the MalE-Rma protein to a MarA operator DNA sequence was analyzed by mobility shift assays with a labeled synthetic 26-bp double-stranded DNA fragment representing the MarA binding sequence, 5′-GGGGATTTAGCAAAACGTGGCATCGG-3′ (25). The results (Fig. 3) showed specific retardation of the DNA probe by the MalE-Rma fusion protein, which indicated that the MarA operator of E. coli is recognized by the Rma protein of S. enterica serovar Paratyphi B.
FIG. 3.
Gel retardation experiments. The radioactive 32P-labeled MarA operator DNA was incubated with increasing concentrations of the purified MalE-Rma fusion protein (lanes 5 to 2). Lane 1, probe control without protein. F, free DNA; B, bound DNA.
The presence of rma in E. coli causes induction of TolC synthesis and reduction of OmpF synthesis.
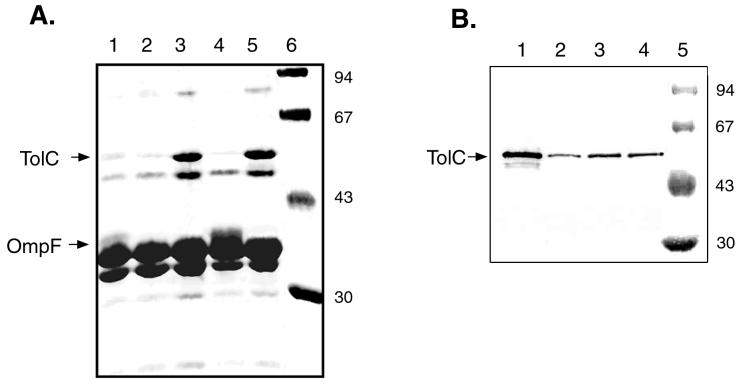
In a previous report (8) on the characterization of the mutant strain of S. enterica serovar Paratyphi B with MDR, the levels of expression of several outer membrane proteins were found to be different from those of the proteins of the wild-type strain. To further investigate the effect of rma in E. coli, the outer membrane protein profiles of E. coli strains with and without the MDR phenotype were analyzed by SDS-PAGE. As shown in Fig. 4A, the presence of rma-containing plasmids (pNCTR4 and pNCTR42) in E. coli DH5α affects the levels of several protein bands, particularly two polypeptides with estimated molecular masses of 50 and 40 kDa. The presence of rma increases the level of a 50-kDa polypeptide and represses that of a 40-kDa polypeptide. The estimated molecular masses of these two polypeptides correspond quite well to those of TolC and OmpF of E. coli, respectively, (1). Western blot analysis with anti-TolC antibody confirmed that the 50-kDa polypeptide is TolC (Fig. 4B). Similarly, experiments with outer membranes prepared from cells grown in the presence of sodium salicylate supported the conclusion that the 40-kDa polypeptide is indeed OmpF. Exogenous salicylate has been shown specifically to suppress the synthesis of outer membrane porin OmpF (7). Such salicylate repression is evident with the 40-kDa band (Fig. 4A, lanes 2 and 5). We therefore conclude that the presence of the S. enterica serovar Paratyphi B rma gene in E. coli induces and represses the synthesis of TolC and OmpF, respectively.
FIG. 4.
Analysis of outer membrane proteins. (A) Equal amounts of outer membrane proteins were subjected to SDS-PAGE and stained with Coomassie brilliant blue. Lane 1, E. coli host cells grown in LB broth; lane 2, host cells grown in LB broth in the presence of 10 mM sodium salicylate; lanes 3 to 5, cells harboring pNCTR4, pUC19, and pNCTR42, respectively; lane 6, polypeptide molecular size standards. The locations of polypeptide bands for TolC and OmpF are marked. (B) Western blot of TolC. Lane 1, cells harboring pNCTR42; lane 2, E. coli host cells grown in the presence of 10 mM sodium salicylate; lane 3, E. coli host cells; lane 4, cells harboring pUC19; lane 5, polypeptide molecular size standards. The numbers to the right of both gels are in kilodaltons.
It has been reported that salicylate also induces the mar operon (7). Since the tolC gene is activated by the mar system (2), one might expect TolC induction by exogenous salicylate through activation of the mar system. However, such induction was not observed in the present study, suggesting that induction of the mar system by salicylate is not at a level high enough to activate the tolC gene.
DISCUSSION
The results reported here indicate that overexpression of RecA confers resistance to norfloxacin but not MDR in E. coli. The involvement of RecA in norfloxacin resistance has been implied by the increased sensitivities of recA mutants to this compound (24, 33). This report shows that overexpression of RecA increases the level of resistance of E. coli to norfloxacin. This effect is likely mediated through the function of RecA in the SOS response. It has been suggested that norfloxacin induces a certain degree of DNA damage or interference with DNA replication (14). This, in turn, could serve as the signal to activate the coprotease activity of RecA to trigger the cascade of the SOS regulatory system (34), resulting in a higher level of resistance to norfloxacin.
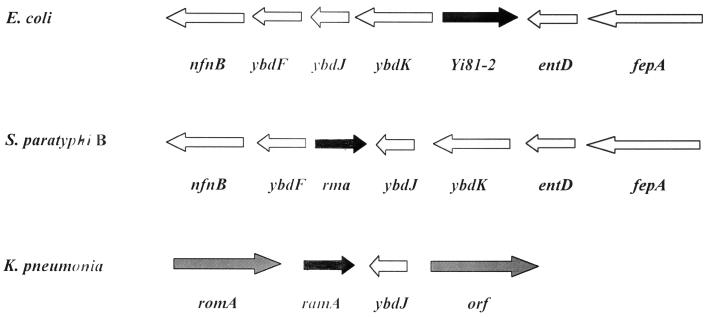
In contrast, the rma gene of S. enterica serovar Paratyphi B, identified in the present study, was shown to cause an MDR phenotype in E. coli. The Rma protein of serovar Paratyphi B shares significant sequence identity (75 to 88%) with the RamA proteins of several other gram-negative bacteria, which have previously been reported to cause the MDR phenotype in E. coli (11). The unpublished sequence for S. enterica serovar Typhimurium shows a gene organization identical to that reported here for S. enterica serovar Paratyphi B (Fig. 5) with roxA in place of rma. The genes rma and roxA should be considered identical since their derived amino acid sequences are identical. By extending the comparison to other gram-negative bacteria, the gene organization in the ramA region of K. pneumoniae differs considerably from that of rma in S. enterica serovar Paratyphi B, but several factors suggest that ramA and rma are orthologues. These factors are the 88% identity of the derived polypeptide sequences, the similarities of the MDR phenotype when Rma or RamA is overexpressed in E. coli, and the presence of the ybdJ locus downstream of either gene (Fig. 5). Interestingly, no Rma counterpart can be found in E. coli by sequence comparison. The results of genomic comparison (Fig. 5) indicate that the gene organization surrounding rma of S. enterica serovar Paratyphi B and the corresponding region in E. coli are similar, with two exceptions: the absence of rma and the presence of Yi81-2 of unknown function in E. coli.
FIG. 5.
Genomic comparison of the flanking regions surrounding rma loci. The nucleotide sequences for E. coli, S. enterica serovar Paratyphi B (S. paratyphi B), and K. pneumoniae were taken from the database at NCBI. Arrows indicate the relative sizes, locations, and transcriptional orientations of genes in the regions; and the designation for each gene is marked under the arrows.
The results of sequence comparison indicate that the Rma protein is a new member of the MarA-SoxS family of transcriptional regulators. Furthermore, the results of the mobility shift experiments reported here provide strong evidence that the Rma protein of S. enterica serovar Paratyphi B interacts with the MarA operator sequence. Although Rma of serovar Paratyphi B and MarA of E. coli share only 38% identity in their amino acid sequences, the residues of MarA that have been shown to be important for DNA interactions (25) are mostly conserved in Rma (Fig. 1). These results support the hypothesis that the Rma protein of S. enterica serovar Paratyphi B can substitute for MarA in affecting the expression of large number of genes in the MarA regulon and that this effect is the likely basis of its ability to cause a similar MDR phenotype in E. coli.
The overexpression of TolC synthesis and the reduction in OmpF synthesis, which are associated with the rma-dependent MDR phenotype in E. coli, as reported here (Fig. 4), are similar to the changes in the outer membrane protein profile that were previously reported for the MDR phenotype conferred by the MarA proteins of other gram-negative bacteria (6, 9, 18). A recent report of a study that used microarray analysis (2) described similar changes as a result of the constitutive expression of MarA in E. coli. Repression of ompF by MarA is thought to occur indirectly through its activation of micF, whose RNA product interacts with ompF mRNA to prevent translation (4, 18). The OmpF protein is one of the major porin proteins in gram-negative bacteria (20), and TolC is required for the function of the AcrAB efflux pump, which is an essential player in the development of the MDR phenotype by MarA (9, 17, 18, 22). The similarities of the effects of rma of S. enterica serovar Paratyphi B and the marA genes of other gram-negative bacteria on the outer membrane protein profiles provide further support for the hypothesis that these two genes might exert their effects via similar mechanisms.
Miller and Sulavik (18) have proposed that the development of the MDR phenotype by different MarA-like regulatory proteins reflects an overlap of adaptive responses to different environmental stimuli. In general, these MarA-like proteins are small, and they are categorized as a subfamily of the AraC-XylS family (10). On the basis of the three-dimensional structure of MarA (10, 25), it is reasonable to assume that these MarA-like regulatory proteins are also monomeric in nature and have two helix-turn-helix domains for DNA binding. Most likely, they do not possess domains for oligomerization and ligand binding like other larger AraC-like regulatory proteins do. Therefore, expression of these MarA-like molecules is likely to be subject to control by other regulatory proteins in response to their cognate environmental stimulants. The MarA-MarR and SoxS-SoxR pairs represent two such examples, with marR and soxR genes located immediately upstream of the corresponding marA and soxS genes. Comparison of the genomes with the finished and unfinished sequences from NCBI showed that the marA-marR and the soxS-soxR configurations are conserved in the genome of S. enterica serovar Paratyphi B. However, analysis of the gene organization in the regions flanking the rma locus of S. enterica serovar Paratyphi B revealed no candidate for a cognate regulator of Rma (Fig. 5).
Although overexpression of Rma causes MDR in E. coli, the nucleotide sequences of the rma structural gene and its putative regulatory region are identical between S. enterica serovar Paratyphi B mutant M95 with MDR and its parent strain. These results suggest that it is overexpression of the rma gene that results in the MDR phenotype and raise the possibility that the phenotype of M95 is the result of a mutation that causes the induction of rma on the chromosome. Such a mutation could be in a cognate regulatory protein that is yet to be identified.
Acknowledgments
M.A.Y. and H.E.E. contributed equally to this work.
We thank Phang C. Tai for the gift of TolC antiserum and valuable suggestions.
This work was supported in part by National Science Foundation grant MCB-9985660.
REFERENCES
- 1.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa, T., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 4.Chou, J. H., J. T. Greenberg, and B. Demple. 1993. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J. Bacteriol. 175:1026–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antiobiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, S. P., L. M. McMurry, and S. B. Levy. 1988. marA locus causes decreased expression of OmpF protein in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856–7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faesal, W., N. A. Hassouna, and M. Yassien. 2000. Effect of nicardipine, verapamil, and reserpine on the sensitivity of efflux-mediated multidrug resistant Salmonella paratyphi B mutant selected by ciprofloxacin. Egypt. J. Biotechnol. 8:208–221. [Google Scholar]
- 9.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George, A. M., R. M. Hall, and H. W. Stokes. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141(Pt. 8):1909–1920. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28. [DOI] [PubMed] [Google Scholar]
- 13.Ishida, H., H. Fuziwara, Y. Kaibori, T. Horiuchi, K. Sato, and Y. Osada. 1995. Cloning of the multidrug resistance gene pqrA from Proteus vulgaris. Antimicrob. Agents Chemother. 39:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668–27677. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu, T., M. Ohta, N. Kido, Y. Arakawa, H. Ito, T. Mizuno, and N. Kato. 1990. Molecular characterization of an Enterobacter cloacae gene (romA) which pleiotropically inhibits the expression of Escherichia coli outer membrane proteins. J. Bacteriol. 172:4082–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomovskaya, O., and K. Lewis. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, D., D. N. Cook, M. Albert, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 26:45–55. [DOI] [PubMed] [Google Scholar]
- 18.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441–448. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Nikaido, H. 1996. Outer membrane, p. 29–47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, and B. Magasanik (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 21.Nunoshiba, T., E. Hidalgo, C. F. Amabile-Cuevas, and B. Demple. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS gene. J. Bacteriol. 174:6054–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic resistance (Mar) mutants. J. Bacteriol. 178:306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen, S. J., E. E. DeBess, T. E. McGivern, N. Marano, T. Eby, S. Mauvais, V. K. Balan, G. Zirnstein, P. R. Cieslak, and F. J. Angulo. 2001. A nosocomial outbreak of fluoroquinolone-resistant Salmonella infection. N. Engl. J. Med. 344:1572–1579. [DOI] [PubMed] [Google Scholar]
- 24.Piddock, L. J., and R. N. Walters. 1992. Bactericidal activities of five quinolones for Escherichia coli strains with mutations in genes encoding the SOS response or cell division. Antimicrob. Agents Chemother. 36:819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413–10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe, B., L. R. Ward, and E. J. Threlfall. 1997. Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin. Infect. Dis. 24:106–109. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sawai, T., R. Hiruma, N. Kawana, M. Kaneko, F. Taniyasu, and A. Inami. 1982. Outer membrane permeation of β-lactam antibiotics in Escherichia coli, Proteus mirabilis, and Enterobacter cloaacae. Antimicrob. Agents Chemother. 22:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of E. coli K12. Eur. J. Biochem. 72:341–352. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gilson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4674–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umasankar, S., R. A. Wall, and J. Berger. 1992. A case of ciprofloxacin-resistant typhoid fever. Commun. Dis. Rep. Rev. 2:132–140. [PubMed] [Google Scholar]
- 33.Uriose, A., G. Herrera, V. Aleixandre, and M. Blanco. 1991. Influence of recA mutations on gyrA dependent quinolone resistance. Biochimie 73:519–521. [DOI] [PubMed] [Google Scholar]
- 34.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400–1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, and B. Magasanik (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 35.Wu, J., and B. Weiss. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]