Abstract
Gemifloxacin is a recently developed fluoroquinolone with potent activity against Streptococcus pneumoniae. We show that the drug is more active than moxifloxacin, gatifloxacin, levofloxacin, and ciprofloxacin against S. pneumoniae strain 7785 (MICs, 0.03 to 0.06 μg/ml versus 0.25, 0.25, 1, and 1 to 2 μg/ml, respectively) and against isogenic quinolone-resistant gyrA-parC mutants (MICs, 0.5 to 1 μg/ml versus 2 to 4, 2 to 4, 16 to 32, and 64 μg/ml, respectively). Gemifloxacin was also the most potent agent against purified S. pneumoniae DNA gyrase and topoisomerase IV in both catalytic inhibition and DNA cleavage assays. The drug concentrations that inhibited DNA supercoiling or DNA decatenation by 50% (IC50s) were 5 to 10 and 2.5 to 5.0 μM, respectively. Ciprofloxacin and levofloxacin were some four- to eightfold less active against either enzyme; moxifloxacin and gatifloxacin showed intermediate activities. In assays of drug-mediated DNA cleavage by gyrase and topoisomerase IV, the same order of potency was seen: gemifloxacin > moxifloxacin > gatifloxacin > levofloxacin ≈ ciprofloxacin. For gemifloxacin, the drug concentrations that caused 25% linearization of the input DNA by gyrase and topoisomerase IV were 2.5 and 0.1 to 0.3 μM, respectively; these values were 4-fold and 8- to 25-fold lower than those for moxifloxacin, respectively. Each drug induced DNA cleavage by gyrase at the same spectrum of sites but with different patterns of intensity. Finally, for enzymes reconstituted with quinolone-resistant GyrA S81F or ParC S79F subunits, although cleavable-complex formation was reduced by at least 8- to 16-fold for all the quinolones tested, gemifloxacin was the most effective; e.g., it was 4- to 16-fold more active than the other drugs against toposiomerase IV with the ParC S79F mutation. It appears that the greater potency of gemifloxacin against both wild-type and quinolone-resistant S. pneumoniae strains arises from enhanced stabilization of gyrase and topoisomerase IV complexes on DNA.
Gemifloxacin is a novel antibacterial fluoroquinolone with broad-spectrum activity and particular potency against Streptococcus pneumoniae (3, 20), the main cause of community-acquired pneumonia (2). The drug is effective in vitro against penicillin-susceptible and -resistant isolates through its ability to target the essential enzymes DNA gyrase and topoisomerase IV. These ATP-dependent type II topoisomerases, encoded by the gyrA-gyrB and parC-parE genes, respectively, act by a double-stranded DNA break and cooperate to allow chromosome replication and segregation (1, 9, 14, 16, 32, 34). Quinolones are thought to trap a ternary drug-topoisomerase-DNA complex which cellular processes convert into an irreversible lethal lesion, perhaps a double-stranded DNA break (5).
The presence of two topoisomerase targets allows the possibility of quinolone action through one or both enzymes (24). Thus, S. pneumoniae strains need not show cross-resistance to all quinolones (24). Interestingly, gemifloxacin retained activity against certain ciprofloxacin-resistant clinical isolates bearing multiple resistance mutations in parC, parE, and gyrB (12). The drug is also relatively active against S. pneumoniae mutants that express the common combination of quinolone resistance mutations at S81 in GyrA and S79 in ParC (11).
Attempts to understand the greater potencies of quinolones such as gemifloxacin and clinafloxacin are at an early stage (11, 12, 25). Both agents select gyrA mutants in the first step, suggesting that the drugs act preferentially through gyrase in vivo (11, 25). Similar findings have been reported for the 8-methoxyquinolones, gatifloxacin and moxifloxacin (7, 29). However, the difficulty in selecting such mutants suggests that these agents may act substantially through both gyrase and topoisomerase IV, so-called dual targeting (11, 25). There have been relatively few studies of quinolone action on the purified enzymes. Quinolone inhibition of recombinant gyrase and topoisomerase IV enzymes obtained either in His-tagged form (26) or following cleavage of fusion proteins expressed in Escherichia coli (17, 18, 21) has been reported. Unfortunately, the fusion protein-derived enzymes were of low specific activity and the work did not examine drug-induced DNA cleavage arising from stabilization of the cleavable complex, i.e., the relevant cytotoxic lesion. Recently, we showed that gemifloxacin and clinafloxacin cause enhanced stabilization of cleavable complexes by both enzymes (11, 26). However, those studies were limited largely to comparisons with ciprofloxacin, a drug not used for the treatment of pneumococcal infections, and did not examine currently marketed fluoroquinolones. Moreover, the effects of quinolone resistance mutations on cleavable-complex stabilization have yet to be investigated.
In the study described here, we compared the in vivo and in vitro actions of gemifloxacin versus those of moxifloxacin, gatifloxacin, levofloxacin, and ciprofloxacin against wild-type S. pneumoniae gyrase and topoisomerase IV, whose subunit primary sequences have been established. In addition, we examined the actions of the drugs against the same complexes but with a single validated GyrA S81F or ParC S79F resistance mutation.
MATERIALS AND METHODS
Bacterial strains and DNA substrates.
Quinolone-susceptible S. pneumoniae strain 7785 and mutants 1C1, 2C6, 2C7, 1S1, 1S4, 2S1, 2S4, and 3C4 have been described previously (22, 23, 25). E. coli BL21(λDE3)(pLysS) was from our laboratory collection. Conditions for growth of bacterial strains were as described previously (25). Supercoiled pBR322 DNA and relaxed pBR322 DNA were prepared by standard procedures (26). Kinetoplast DNA from Crithidia fasciculata was obtained from Topogen, Inc., Columbus, Ohio.
Drugs and reagents.
Gemifloxacin mesylate was provided by GlaxoSmithKline, Harlow, United Kingdom. Moxifloxacin, gatifloxacin, levofloxacin, and ciprofloxacin hydrochloride were obtained from their respective manufacturers.
Drug susceptibilities.
MICs were determined by twofold agar dilution on brain heart infusion plates containing 10% horse blood (24).
Recombinant gyrase and topoisomerase IV proteins.
S. pneumoniae GyrB, GyrA, ParC, and ParE subunits were expressed as His-tagged proteins in E. coli BL21(λDE3)(pLysS) by isopropyl-β-d-thiogalactopyranoside induction of plasmids pXP9, pXP10, pXP13, and pXP14, which contain the respective gyrB, gyrA, parC, and parE genes cloned from strain 7785 (26). The proteins were purified to >95% homogeneity by nickel chelate column chromatography and exhibited specific activities of >2 × 105 U/mg when they were assayed with an excess of the complementing subunit (26). GyrA S81F and ParC S79F proteins, whose purities and activities were comparable to those of their wild-type counterparts, were obtained similarly (27).
Enzyme assays.
Topoisomerase and DNA cleavage assays were carried out as described previously (26). The DNA products were resolved by electrophoresis in 1% agarose, stained with ethidium bromide, photographed, and quantitated with an Alpha Innotech digital camera and associated software.
RESULTS
Activities of fluoroquinolones against S. pneumoniae 7785 and its isogenic quinolone-resistant mutants.
To allow comparison of in vivo and enzyme inhibition results, we first examined the antibacterial activity of gemifloxacin versus those of moxifloxacin, gatifloxacin, and levofloxacin, three antipneumococcal fluoroquinolones used clinically, and ciprofloxacin, a quinolone used for the treatment of infections caused by gram-negative pathogens. For this purpose, we used quinolone-susceptible S. pneumoniae isolate 7785 and its mutants carrying gyrA, parC, or gyrA and parC resistance mutations (Table 1). The gemifloxacin and ciprofloxacin MICs for parental strain 7785 were 0.03 to 0.06 and 1 to 2 μg/ml, respectively, values that were essentially identical to those reported previously (11). The MICs of moxifloxacin, gatifloxacin, and levofloxacin for strain 7785 were 0.25, 0.25, and 1 μg/ml, respectively. Thus, gemifloxacin was at least 4- to 16-fold more active against 7785 than the other four fluoroquinolones tested.
TABLE 1.
Fluoroquinolone susceptibilities of S. pneumoniae 7785 and its mutants
| Strain | Mutation
|
MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| GyrA | ParC | GEM | MXF | GAT | LVX | CIP | |
| 7785 | 0.03-0.06 | 0.25 | 0.25 | 1 | 1-2 | ||
| 1C1 | 0.06 | 0.12-0.25 | 0.25 | 1 | 2 | ||
| 2C6 | S79Y | 0.12 | 0.5 | 1 | 8 | 8 | |
| 2C7 | S79F | 0.12 | 0.5 | 1 | 4-8 | 8 | |
| 1S1 | S81F | 0.12 | 0.5 | 0.5-1 | 2 | 2 | |
| 1S4 | S81Y | 0.12 | 0.5 | 0.5-1 | 2 | 2 | |
| 2S1 | S81F | S79Y | 1 | 4 | 4 | 16-32 | 64 |
| 2S4 | S81F | D83N | 0.5 | 4 | 2 | 16 | 64 |
| 3C4 | S81Y | S79Y | 1 | 2 | 2 | 16 | 64 |
GEM, gemifloxacin; MXF, moxifloxacin; GAT, gatifloxacin; LVX, levofloxacin; CIP, ciprofloxacin.
Table 1 shows that for each of the mutants, gemifloxacin was the most potent fluoroquinolone and ciprofloxacin was the least potent fluoroquinolone. Thus, the presence of an efflux mutation in strain 1C1 (11) had little effect on susceptibility to gemifloxacin, as did parC changes in strains 2C6 and 2C7 or gyrA alterations in strains 1S1 and 1S4, which in each case increased the gemifloxacin MIC to 0.12 μg/ml. The presence of both gyrA and parC changes (strains 2S1, 2S4, and 3C4) increased the gemifloxacin MICs to 0.5 to 1 μg/ml. For ciprofloxacin and levofloxacin, parC mutations conferred four- to eightfold increases in the MICs, whereas gyrA mutations had little effect. The gyrA-parC mutants were highly resistant to both drugs (MICs, 16 to 64 μg/ml). These data support previous work showing that both ciprofloxacin and levofloxacin act preferentially through topoisomerase IV in S. pneumoniae (7, 10, 13, 19, 22, 28, 31). Moxifloxacin and gatifloxacin showed intermediate activities, being some four- to eightfold more potent than levofloxacin but four- to eightfold less active than gemifloxacin against the various mutants. Similar to their effects on the activity of gemifloxacin, single gyrA or parC mutations elevated the MICs of gatifloxacin and moxifloxacin by only two- to fourfold, an observation suggesting that both gyrase and topoisomerase IV contribute in setting of the in vivo susceptibility to these quinolones.
Gemifloxacin was the most potent catalytic inhibitor of S. pneumoniae gyrase and topoisomerase IV.
To compare the potencies of fluoroquinolones against their enzyme targets in vitro, we used S. pneumoniae GyrB, GyrA, ParC, and ParE subunits overexpressed from strain 7785 genes as His-tagged proteins in E. coli (26). Following one-step nickel chelate column chromatography, each subunit was obtained at high purity (>95% homogeneity) in milligram amounts and was free of contaminating host topoisomerase activities. The proteins were stable (several years at −70°C) and were highly active (specific activities, 2 × 105 U/mg) in topoisomerase assays when they were mixed with an excess of the complementing subunit (26). Fluoroquinolone inhibition of DNA supercoiling by S. pneumoniae DNA gyrase was examined over a range of drug concentrations by using relaxed pBR322 DNA as the substrate. For topoisomerase IV, drug inhibition of enzymatic decatenation of kinetoplast DNA was determined. In each case, drug potencies were expressed as the drug concentration that resulted in 50% inhibition of enzyme activity (IC50s). Each measurement was repeated at least twice, with comparable results. Representative data are shown in Table 2.
TABLE 2.
Inhibitory activities of fluoroquinolones against S. pneumoniae DNA gyrase and topoisomerase IV
| Quinolone | IC50 (μM)
|
|
|---|---|---|
| Gyrasea | Topoisomerase IVb | |
| Gemifloxacin | 5-10 | 2.5-5 |
| Moxifloxacin | 20 | 10 |
| Gatifloxacin | 20-40 | 10-20 |
| Levofloxacin | 80 | 40 |
| Ciprofloxacin | 40 | 20 |
Effects on ATP-dependent supercoiling of relaxed pBR322 DNA.
Measured for decatenation of kinetoplast DNA.
The IC50s for inhibition of gyrase and topoisomerase IV by ciprofloxacin were 40 and 20 μM, respectively, confirming previous work (26). The data presented for the other fluoroquinolones reveal that gemifloxacin was the most effective inhibitor of both gyrase and topoisomerase IV, with IC50s of 5 to 10 and 2.5 to 5 μg/ml, respectively. Moreover, for each enzyme, the order of potency was gemifloxacin > moxifloxacin > gatifloxacin > ciprofloxacin > levofloxacin. Interestingly, as noted previously, topoisomerase IV was more sensitive than gyrase to inhibition by quinolones (17, 18, 26) (Table 2).
Potencies of different quinolones in stabilizing the cleavable complex of gyrase reconstituted with wild-type GyrA or GyrA S81F protein.
More effective stabilization of cleavable complexes with gyrase and topoisomerase IV has been suggested to underlie the greater potency of gemifloxacin against S. pneumoniae (11). To compare the cleavable-complex stabilization achieved with the clinically approved antipneumococcal fluoroquinolones and to examine the effects of GyrA (and ParC) mutations, supercoiled pBR322 DNA was incubated with gyrase (or topoisomerase IV [see below]) in the absence or presence of a wide range of drug concentrations, and DNA breakage was induced by the subsequent addition of sodium dodecyl sulfate (SDS). Following incubation with proteinase K (to remove covalently bound GyrA or ParC proteins), DNA samples were analyzed by agarose gel electrophoresis. The drug concentration that caused 25% linearization of the input DNA (CC25) was determined at least three times in independent experiments (Table 3). Figure 1shows the results of a representative cleavage experiment for S. pneumoniae gyrase, in which drug concentrations were adjusted to yield comparable degrees of DNA cleavage.
TABLE 3.
Potencies of gemifloxacin and comparator quinolones in stabilizing the cleavable complex with gyrase and topoisomerase IV
| Quinolone | CC25 (μM) for DNA breakage
|
||||
|---|---|---|---|---|---|
| Gyrase
|
Topoisomerase IV
|
||||
| Wild type | GyrA Phe-81 mutation | Wild type | ParC Phe-79 mutation | ||
| Gemifloxacin | 2.5 | 160 | 0.1-0.3 | 10-20 | |
| Moxifloxacin | 10 | 160-320 | 2.5-5 | 160 | |
| Gatifloxacin | 10-20 | 160-320 | 5-10 | 80-160 | |
| Levofloxacin | 20-40 | >640 | 5-10 | >320 | |
| Ciprofloxacin | 40 | >640 | 2.5-5 | >160 | |
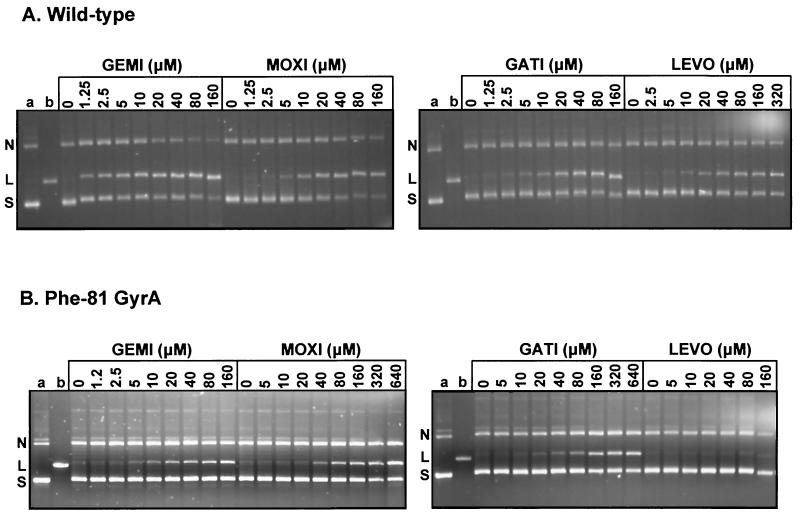
FIG. 1.
Drug-promoted DNA breakage by wild-type S. pneumoniae DNA gyrase (A) and the mutant enzyme reconstituted with GyrA S81F (B). Supercoiled plasmid pBR322 DNA (0.4 μg) was incubated with recombinant S. pneumoniae GyrB (1.7 μg) and either wild-type GyrA (A) or GyrA S81F (B) (in each case, 0.45 μg) in the absence or presence of gemifloxacin (GEMI), moxifloxacin (MOXI), gatifloxacin (GATI), or levofloxacin (LEVO) at the indicated concentrations. Following treatment with SDS and proteinase K, the DNA products were examined in 1% agarose gels. Lanes a and b, supercoiled and linear pBR322, respectively; S, L, and N, supercoiled, linear, and nicked plasmid pBR322 bands, respectively.
For wild-type gyrase, gemifloxacin and ciprofloxacin were the most and least effective in stimulating DNA breakage, with CC25s of 2.5 and 40 μM, respectively (Fig. 1A; Table 3). Moxifloxacin and gatifloxacin were some fourfold and four- to eightfold less effective than gemifloxacin, respectively, with CC25s of 10 and 10 to 20 μM, respectively. Levofloxacin behaved similarly to ciprofloxacin but was marginally more active (Table 3). Thus, the rank order of potency in stabilizing the cleavable complex with gyrase was gemifloxacin > moxifloxacin > gatifloxacin > levofloxacin > ciprofloxacin, an order similar to the respective MICs measured for wild-type strain 7785 (Table 1).
Figure 1B shows the typical results of cleavage experiments performed with gyrase reconstituted with a GyrA protein bearing an S81F alteration, a mutation frequently acquired in quinolone-resistant clinical strains and laboratory isolates (11, 25). All five quinolones mediated dose-dependent DNA cleavage by the mutant enzyme, but drug levels in the range 10 to 160 μM were needed to see an effect. From Fig. 1B, it is clear that gemifloxacin and levofloxacin were the most and the least effective, respectively, in stimulating DNA cleavage by the mutant enzyme. These and other experiments (data not shown for ciprofloxacin) established the CC25s presented in Table 3. Thus, the S81F change in GyrA increased the CC25 at least 16- to 32-fold for all the quinolones tested. Interestingly, strain 1S1, which expressed the same mutant protein, showed only a two- to fourfold increase in resistance (Table 1).
S79F ParC mutation inhibits cleavable-complex formation with topoisomerase IV.
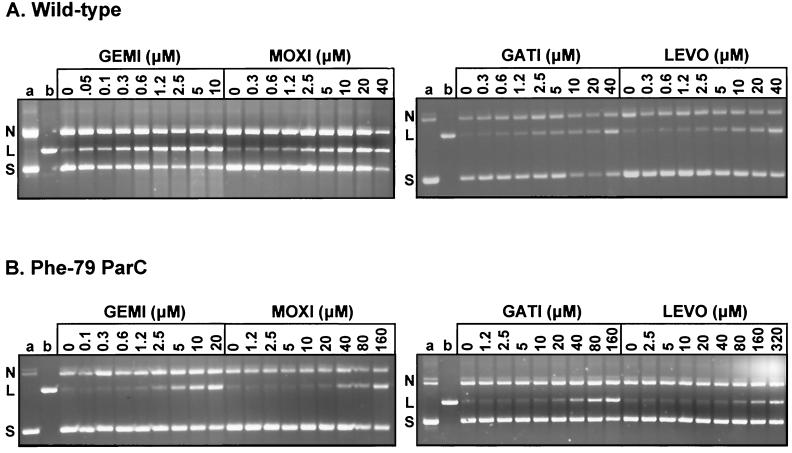
Figure 2presents the results of DNA cleavage experiments for topoisomerase IV reconstituted with wild-type ParC (Fig. 2A) or ParC with the S79F mutation (Fig. 2B). The CC25s determined from these and other experiments (data not shown) are summarized in Table 3. Gemifloxacin was the quinolone most effective at inducing DNA cleavage by the wild-type enzyme, being >10-fold better than the other drugs. However, the CC25s of all the drugs were lower than those measured with DNA gyrase (Table 3). The presence of the S79F mutation elevated the CC25 at least 8- to 16-fold for all the drugs: the order of activity was gemifloxacin ≫ moxifloxacin ≈ gatifloxacin > levofloxacin ≈ ciprofloxacin (Fig. 2B). The observation that high drug levels induce a dose-dependent DNA cleavage with the mutant gyrase and topoisomerase IV enzymes could suggest that the ParC S79F change (and the GyrA S81F change) acts by lowering the level of drug binding to the complex (Fig. 1B and 2B).
FIG. 2.
Quinolone-mediated DNA cleavage by S. pneumoniae topoisomerase IV (A) and by its mutant ParC S79F complex (B). Plasmid pBR322 (0.4 μg) was incubated with ParE (1.7 μg) and either wild-type ParC (A) or ParC S79F (B) (0.45 μg). Samples were processed and analyzed as described in the Fig. 1 legend. The abbreviations and lane contents are as described in the Fig. 1 legend.
Quinolones mediate DNA cleavage by gyrase at the same spectrum of sites.
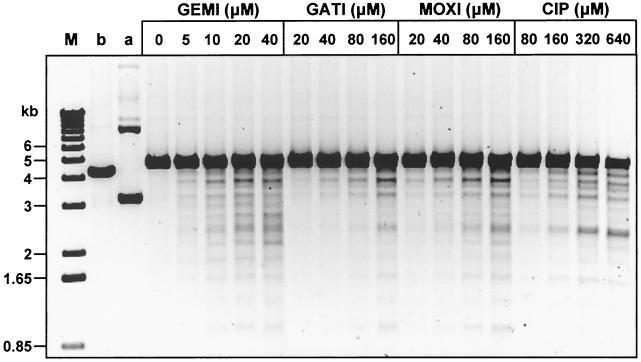
Studies with supercoiled pBR322 as the substrate provide information on the relative efficiency of cleavable-complex formation but do not reveal the sites of enzyme action. An ability of quinolones to direct DNA cleavage at markedly different sites could influence the killing properties of the drugs against S. pneumoniae. To examine this question, pBR322 DNA linearized with EcoRI was used in a DNA cleavage assay with increasing amounts of gemifloxacin, gatifloxacin, and moxifloxacin with a fixed amount of gyrase, the intracellular target of the three drugs (Fig. 3).Ciprofloxacin, which targets topoisomerase IV, was also included for comparison (Fig. 3). Each enzyme cleavage site on linear DNA results in two specific fragments, which allows the spectrum of sites to be compared between quinolones. Each quinolone caused dose-dependent increases in the amount of DNA breakage (Fig. 3). Gemifloxacin at 20 to 40 μM induced substantial DNA breakage. Much higher levels (>160 μM) of the other quinolones were required to generate comparable cleavage levels. The cleavage patterns observed for gemifloxacin, gatifloxacin, and moxifloxacin, which select gyrA mutants in vivo (7, 11, 29), were very similar, although not identical, to each other; and their cleavage patterns differed from the pattern seen for ciprofloxacin. Scrutiny of Fig. 3 reveals that the four quinolones cause subtly different levels of cleavage at particular sites but appear to act at the same overall spectrum of sites.
FIG. 3.
Site specificity of quinolone-promoted DNA cleavage by S. pneumoniae DNA gyrase. EcoRI-cut pBR322 DNA (0.4 μg) was incubated with GyrA (0.45 μg) and GyrB (1.7 μg) in the absence or presence of quinolones at the indicated concentrations (in micromolar). After treatment with SDS and proteinase K, DNA samples were examined by electrophoresis in 1% agarose. Lanes M, b, and a; DNA size markers, linear pBR322, and supercoiled pBR322, respectively. GEMI, gemifloxacin; GATI, gatifloxacin; MOXI, moxifloxacin; CIP, ciprofloxacin.
DISCUSSION
To understand the molecular basis for the greater potency of gemifloxacin vis-à-vis comparator quinolones against S. pneumoniae and its quinolone-resistant strains, we have examined the in vitro activities of the drugs against the purified gyrase and topoisomerase IV targets. Of the five quinolones tested, gemifloxacin was the most active inhibitor of both enzymes. The rank order for catalytic inhibition and cleavable-complex formation with each enzyme was gemifloxacin > moxifloxacin > gatifloxacin > levofloxacin ≈ ciprofloxacin; i.e., similar to that observed for inhibition of growth of wild-type S. pneumoniae strain 7785. Each quinolone induced a similar pattern of gyrase cleavage sites on DNA, although with some differences in cleavage intensities at particular sites. Although GyrA S81F or ParC S79F mutations each reduced the level of cleavable-complex formation by at least 8- to 16-fold for all five quinolones tested, gemifloxacin remained the most effective drug against the mutant complexes. Thus, it was marginally more active against gyrase reconstituted with the GyrA S81F protein. Moreover, compared to the other agents, it retained greater activity against topoisomerase IV containing ParC S79F. Our results on cleavable-complex stabilization have important implications in explaining the action of gemifloxacin against quinolone-resistant strains.
The gyrase and topoisomerase IV proteins used in the present study are identical (except for a His tag) to those produced by strain 7785 and its gyrA and parC mutants. This correspondence allows comparison of drug actions in vitro and in vivo. It is noteworthy that the GyrA S81F or ParC S79F subunits reconstituted enzymes that were much less efficiently trapped by quinolones as a cleavable complex (Table 3). For example, either mutation increased the CC25 of gemifloxacin for gyrase and topoisomerase IV some 60-fold. By contrast, the gemifloxacin MICs for strains 1S1 and 2C7 expressing the GyrA S81F or ParC S79F protein exhibited only two- to fourfold increases (Table 1). These data indicate that the full effect of the resistance mutation is moderated in vivo by drug action on the second wild-type topoisomerase target. Similar behavior was seen for moxifloxacin and gatifloxacin. Consistent with the poison mechanism of quinolone action (15), these data suggest that both gyrase and topoisomerase IV contribute significantly as intracellular targets of gemifloxacin, gatifloxacin, and moxifloxacin in S. pneumoniae.
Our results complement those of another study of recombinant S. pneumoniae gyrase and topoisomerase IV, in which the proteins were expressed by a different method by using topoisomerase genes amplified from quinolone-susceptible and -resistant S. pneumoniae strains (18). Although the exclusive focus of the earlier study was on inhibition of enzyme activity and not on stabilization of the cleavable complex, some comparisons can be made with our data. First, topoisomerase IV was shown to be more sensitive than gyrase to quinolone inhibition, confirming previous observations (17, 21, 26), with gemifloxacin being about fourfold more effective in inhibiting topoisomerase IV than grepafloxacin, moxifloxacin, trovafloxacin, levofloxacin, and ciprofloxacin (i.e., similar to the results presented in Table 1). However, in contrast to our results (Table 1), gyrase was inhibited uniformly by all the quinolones tested. Second, quinolone IC50s for topoisomerase IV and gyrase complexes containing S79F plus A189V mutations in ParC and S81F in GyrA, respectively, were 3- to 7-fold and 1.3- to 2.7-fold higher than those for the wild type (18). These values for the resistant topoisomerase IV and gyrase appear to be rather low: the 6.8- and 2.7-fold increases in the IC50s of ciprofloxacin, respectively, are in contrast to the >16-fold increases in the IC50s of ciprofloxacin and sparfloxacin, which we have measured for complexes containing ParC S79F and GyrA S81F mutations (27). We note that the mutant enzyme complexes studied by Morrissey and George (18) in some instances had multiple mutations in the quinolone resistance-determining region and were derived from clinical isolates that were not isogenic with the wild-type susceptible strain, thereby complicating comparisons between enzyme complexes. Moreover, the recombinant subunits were generated by proteolytic cleavage of fusion proteins, and all had specific activities some 40-fold lower than those of the His-tagged subunits reported here. In any event, given that quinolones act in vivo by stabilizing a cleavable complex (15), it would appear that the CC25 for DNA breakage rather than the IC50 for enzyme inhibition may be a more useful comparative parameter. However, it should be noted that even CC25s are at least 10-fold higher than MICs, suggesting that much lower levels of DNA cleavage in vivo are sufficient to inhibit bacterial growth.
Characterization of the resistant enzyme complexes reported here casts light on the gemifloxacin susceptibilities of quinolone-resistant clinical isolates of S. pneumoniae. Previously, we have reported that two multidrug-resistant isolates with parC S79F, parE D435V, and gyrB E474K alleles and for which the ciprofloxacin MIC was 64 μg/ml were susceptible to gemifloxacin (MICs, 0.12 μg/ml) (12). Although the genetic and enzymatic effects of the ParE D435V and GyrB E474K mutations are poorly characterized, the data presented here indicate that the ParC S79F alteration reduces cleavable-complex formation by topoisomerase IV by 60-fold for both ciprofloxacin and gemifloxacin. Two factors likely account for the 500-fold differences in the ciprofloxacin and gemifloxacin MICs for the parC-parE-gyrB mutants. First, and most important, ciprofloxacin acts through topoisomerase IV in vivo, and therefore, the ParC S79F mutation will exert a resistance phenotype (7, 10, 13, 19, 22, 28, 31). By contrast, gemifloxacin appears to act through both DNA gyrase and topoisomerase IV, perhaps with a slight preference for gyrase, as suggested by stepwise selection experiments (11). Thus, in the presence of wild-type GyrA, the effects of the ParC S79F mutation will be greatly reduced (Table 1). Second, although the ParC D435V and GyrB E474K mutations affect ciprofloxacin susceptibility (12), these alterations appear to have minimal effects on the action of gemifloxacin in vivo. It remains to be seen whether the absence of a gyrA mutation is sufficient to render quinolone-resistant isolates universally susceptible to gemifloxacin.
Finally, we note that gemifloxacin exhibited greater potency than other quinolones tested against S. pneumoniae gyrA-parC mutants expressing the common combination of either S81F or S81Y changes in GyrA and S79F or S79Y changes in ParC (Table 1) (11). Our enzyme data suggest that the enhanced potency of gemifloxacin against these mutant strains likely arises from enhanced stabilization of the mutant gyrase and topoisomerase IV complexes on DNA (Table 3). Interestingly, the gemifloxacin MICs of 0.5 to 1.0 μg/ml for these gyrA-parC strains were lower than the peak concentration of 2.3 μg/ml achieved in the plasma of human subjects following the administration of a 320-mg oral dose (8). In the absence of an approved breakpoint for gemifloxacin (33), further work will be needed to determine if the greater potency of gemifloxacin translates into a therapeutic benefit against the quinolone-resistant mutants now being detected in surveillance studies (4, 6, 30).
Acknowledgments
G.Y. was supported by a visiting fellowship from the Spanish Society for Clinical Microbiology and Infectious Diseases. J.E.M., K.A.G., and part of this study were funded by a project grant from GlaxoSmithKline.
G.Y. and J.E.M. contributed equally to this work.
REFERENCES
- 1.Adams, D. E., E. M. Shekhtman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning DNA replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, J. G., and L. M. Grundy. 1995. Community-acquired pneumonia. N. Engl. J. Med. 333:1618-1624. [DOI] [PubMed] [Google Scholar]
- 3.Cormican, M. G., and R. N. Jones. 1997. Antimicrobial activity and spectrum of LB20304, a novel fluoronaphthyridone. Antimicrob. Agents Chemother. 41:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doern, G. V., M. A. Pfaller, M. E. Erwin, A. B. Brueggemann, and R. N. Jones. 1998. The prevalence of fluoroquinolone resistance among clinically significant respiratory tract isolates of Streptococcus pneumoniae in the United States and Canada--1997 results from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 32:313-316. [DOI] [PubMed] [Google Scholar]
- 5.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felmingham, D., and J. Washington. 1999. Trends in the antimicrobial susceptibility of bacterial respiratory tract pathogens--findings of the Alexander Project 1992-1996. J. Chemother. 11:5-21. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda, H., and K. Hiramatsu. 1999. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:410-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gee, T., J. M. Andrews, J. P. Ashby, G. Marshall, and R. Wise. 2001. Pharmacokinetics and tissue penetration of gemifloxacin following a single oral dose. J. Antimicrob. Chemother. 47:431-434. [DOI] [PubMed] [Google Scholar]
- 9.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gootz, T. D., R. Zaniewski, S. Haskell, B. Schmieder, J. Tankovic, D. Girard, P. Courvalin, and R. J. Polzer. 1996. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob. Agents Chemother. 40:2691-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heaton, V. J., J. E. Ambler, and L. M. Fisher. 2000. Potent antipneumococcal activity of gemifloxacin is associated with dual targeting of gyrase and topoisomerase IV, an in vivo target preference for gyrase, and enhanced stabilization of cleavable complexes in vitro. Antimicrob. Agents Chemother. 44:3112-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heaton, V. J., C. G. Goldsmith, J. E. Ambler, and L. M. Fisher. 1999. Activity of gemifloxacin against penicillin- and ciprofloxacin-resistant Streptococcus pneumoniae displaying topoisomerase- and efflux-mediated resistance mechanisms. Antimicrob. Agents Chemother. 43:2998-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janoir, C., V. Keller, M.-D. Kitzis, N. J. Moreau, and L. Gutmann. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Higara, and H. Suzuki. 1990. New topoisomerase essential for chromosomal segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 15.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuuchi, K., L. M. Fisher, M. H. O'Dea, and M. Gellert. 1980. DNA gyrase action involves the introduction of transient double strand breaks into DNA. Proc. Natl. Acad. Sci. USA 77:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrissey, I., and J. George. 1999. Activities of fluoroquinolones against Streptococcus pneumoniae type II topoisomerases purified as recombinant proteins. Antimicrob. Agents Chemother. 43:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrissey, I., and J. T. George. 2000. Purification of pneumococcal type II topoisomerases and inhibition by gemifloxacin and other quinolones. J. Antimicrob. Chemother. 45(Suppl. S1):101-106. [DOI] [PubMed] [Google Scholar]
- 19.Munoz, R., and A. G. De la Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of quinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh, J.-I, K.-S. Paek, M.-J. Ahn, M.-Y. Kim, C.-Y. Hong, I.-C. Kim, and J.-H. Kwak. 1996. In vitro and in vivo evaluations of LB20304, a new fluoronaphthyridone. Antimicrob. Agents Chemother. 40:1564-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onodera, Y., Y. Uchida, M. Tanaka, and K. Sato. 1999. Dual inhibitory activity of sitafloxacin (DU-6859a) against DNA gyrase and topoisomerase IV of Streptococcus pneumoniae. J. Antimicrob. Chemother. 44:533-536. [DOI] [PubMed] [Google Scholar]
- 22.Pan, X.-S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, X.-S., and L. M. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan, X.-S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan, X.-S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, X.-S., and L. M. Fisher. 1999. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob. Agents Chemother. 43:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan, X.-S., G. Yague, and L. M. Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perichon, B., J. Tankovic, and P. Courvalin. 1997. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1166-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova, E., J. J. Millichap, G. A. Noskin, and L. R. Peterson. 2000. Intracellular targets of moxifloxacin: a comparison with other fluoroquinolones. J. Antimicrob. Chemother. 45:583-590. [DOI] [PubMed] [Google Scholar]
- 30.Piddock, L. J. V. 1994. New quinolones and gram-positive bacteria. Antimicrob. Agents Chemother. 38:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tankovic, J., B. Perichon, J. Duval, and P. Courvalin. 1996. Contribution of mutations in the gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob. Agents Chemother. 40:2502-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 33.Wise, R., and J. M. Andrews. 1999. The in-vitro activity and tentative breakpoint of gemifloxacin, a new fluoroquinolone. J. Antimicrob. Agents 44:679-688. [DOI] [PubMed] [Google Scholar]
- 34.Zechiedrich, E. L., and N. R. Cozzarelli. 1995. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9:2859-2869. [DOI] [PubMed] [Google Scholar]