Clarithromycin resistance, the major cause of Helicobacter pylori treatment failure (2, 7), is attributed to point mutations within the peptidyltransferase-encoding region in domain V of the 23S rRNA gene (10, 15). Three distinct point mutations have been found to be associated with macrolide resistance in H. pylori strains (14, 15). Mutations A2142G and A2143G are the most frequently reported, whereas mutation A2142C is less common, e.g., it was found in 5 of 129 resistant strains (11). Two additional mutations, A2115G and G2141A, have been described, occurring in the same strain (4), but neither has been reported subsequently.
The three most common mutations in the 23S rRNA gene can be detected in a two-step process using different molecular methods, PCR-DNA enzyme immunoassay (5, 9), and PCR-line probe assay (12). Recently, real-time PCR-based methods have been developed to detect these mutations (1, 3, 6), but none of them can distinguish among the three common mutations. PCR-restriction fragment length polymorphism (PCR-RFLP), another two-step process, allows the identification of mutations A2142G and A2143G using the BbsI (8) and BsaI (14, 15) restriction enzymes, respectively.
In this study, detection of mutation A2142C using the enzyme BceAI (BcefI), a type IIS restriction endonuclease which recognizes the two nonpalindromic sequences 5′-ACGGC-3′ and 5′-GCCGT-3′(13) is reported for the first time. Novel PCR and RFLP protocols developed to detect the three most frequent H. pylori 23S rRNA gene mutations are also reported.
H. pylori genomic DNA was isolated using the QIAamp DNA mini kit (Qiagen SA), and a 267-bp fragment was amplified by PCR using primers 5′-AGGTTAAGAGGATGCGTCAGTC-3′ (HPY-S) and 5′-CGCATGATATTCCCATTAGCAGT-3′ (HPY-A), corresponding to nucleotides 1931 to 1952 and 2197 to 2175, respectively, of the 23S rRNA gene of H. pylori (GenBank accession number U27270). The amplification reaction was carried out in a final volume of 50 μl containing 1× buffer, 1.5 mM MgCl2, 200 μM each of the four deoxynucleoside triphosphates, 0.2 μM each of the primers, 1 U of Taq polymerase (Eurobio), and 3 μl of extracted DNA. Amplification was performed in a Perkin Elmer GeneAmp 9700 thermocycler (Norwalk, Conn.) with the following conditions: 1 cycle at 94°C for 5 min, 40 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and 1 cycle at 72°C for 7 min.
The 267-bp PCR products were precipitated and suspended in 15 μl of H2O, and 5 μl was digested overnight in a final volume of 15 μl with the restriction enzymes BbsI (5 U), BsaI (5 U) (8), and BceAI (0.5 U) (New England Biolabs).
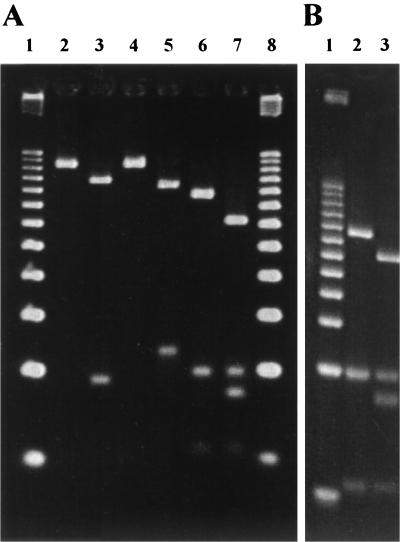
PCR-RFLP allowed the identification of mutations A2142G and A2143G using the BbsI and BsaI restriction enzymes, respectively (Fig. 1A, lanes 3 and 5), as previously described (8, 14, 15). The enzyme BceAI recognized the two restriction sites 5′-ACGGC(N)12↓N-3′ and 5′-N↓(N)12GCCGT-3′ on the 267-bp products obtained from the wild type and the A2142G and A2143G mutants, yielding fragments of 48, 24, and 195 bp (Fig. 1A, lane 6; Fig. 1B, lane 2). On the other hand, mutation A2142C created an additional BceAI recognition site, 5′-ACGGC(N)12↓N-3′, yielding four fragments, with the 195-bp fragment cleaved into 153- and 42-bp products (Fig. 1A, lane 7, and Fig. 1B, lane 3). For rapid detection, either the 195-bp or the 153-bp fragment could be detected on a 2% agarose gel (data not shown). These BceAI cleavages occurred at positions 1979, 2002 and 2155 of the 23S rRNA gene.
FIG. 1.
Detection of mutation A2142C by BceAI-mediated restriction digestion. The restriction fragments of the 267-bp PCR products were analyzed by electrophoresis on a 5% agarose Resophor gel (A) or on a 12% polyacrylamide gel (B) stained with ethidium bromide. (A) PCR-RFLP analysis of mutations A2142G, A2143G, and A2142C occurring in domain V of the 23S rRNA gene of H. pylori. Lanes 1 and 8, 25-bp DNA Step Ladder molecular size markers (Promega). Lanes 2 and 3, PCR products of the wild-type and A2142G H. pylori strains digested with BbsI, respectively. Lanes 4 and 5, PCR products of the wild-type and A2143G H. pylori strains digested with BsaI, respectively. Lanes 6 and 7, PCR products of the wild-type and A2142C H. pylori strains digested with BceAI, respectively. (B) PCR product of the H. pylori strain with mutation A2142C digested with BceAI. Lanes 2 and 3, amplified wild-type PCR product and amplified PCR product presenting the A2142C mutation, respectively. Lane 1, 25-bp DNA Step Ladder (Promega). The wild-type H. pylori reference strain CIP 101260 and strains 683, 677, and 825, with mutations A2142G, A2142C, and A2143C, respectively, in the 23S rRNA gene were used as controls in this study (8).
The PCR protocol developed here could also be used to amplify sequences containing the two rare mutations A2115G and G2141A, described by Hultén et al. (4). RFLP with BbsI, BsaI, and BceAI would lead to the same restriction patterns as obtained with the amplified wild-type sequence, but RFLP with EcoRI and BplI would allow the identification of the A2115G and A2141T mutations, respectively. Lack of access to strains containing these rare mutations precluded obtaining their restriction profiles.
In conclusion, it is possible to detect by PCR-RFLP the three most frequent point mutations of the 23S rRNA gene conferring clarithromycin resistance on H. pylori. This method can be applied to H. pylori strains and also to gastric biopsy samples, allowing the detection of the three genotypes with one PCR sample: 7 of 200 H. pylori-positive biopsies with the A2142C mutation were detected (data not shown).
Real-time PCR-based methods have enormous potential, but limited availability of LightCycler instruments restricts their implementation in many clinical laboratories. For this reason, although more time-consuming, a simple method with the same potential for detection is highly desirable. By extending the highly specific PCR-RFLP technique to the detection of the A2142C mutation, the protocol proposed here achieves this goal.
REFERENCES
- 1.Chisholm, S. A., R. J. Owen, E. L. Teare, and S. Saverymuttu. 2001. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J. Clin. Microbiol. 39:1217-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Helicobacter pylori Study Group. 1997. Current European concepts in the management of Helicobacter pylori infection: the Maastricht consensus report. Gut 41:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson, J. R., N. A. Saunders, B. Burke, and R. J. Owen. 1999. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J. Clin. Microbiol. 37:3746-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hultén, K., A. Gibreel, O. Skold, and L. Engstrand. 1997. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob. Agents Chemother. 41:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marais, A., L. Monteiro, A. Occhialini, M. Pina, H. Lamouliatte, and F. Mégraud. 1999. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction DNA enzyme immunoassay in gastric biopsy specimens. Gut 44:463-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumura, M., Y. Hikiba, K. Ogura, G. Togo, I. Tsukuda, K. Ushikawa, Y. Shiratori, and M. Omata. 2001. Rapid detection of mutations in the 23S rRNA gene of Helicobacter pylori that confers resistance to clarithromycin treatment to the bacterium. J. Clin. Microbiol. 39:691-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mégraud, F. 1997. Resistance of Helicobacter pylori to antibiotics. Aliment. Pharmacol. Ther. 11:43-53. [DOI] [PubMed] [Google Scholar]
- 8.Occhialini, A., M. Urdaci, F. Doucet-Populaire, C. M. Bebear, H. Lamouliatte, and F. Mégraud. 1997. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob. Agents Chemother. 41:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pina, M., A. Occhialini, L. Monteiro, H. P. Doermann, and F. Mégraud. 1998. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J. Clin. Microbiol. 36:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor, D. E., Z. Ge, D. Purych, T. Lo, and K. Hiratsuka. 1997. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Doorn, L. J., Y. Glupczynski, J. G. Kusters, F. Megraud, P. Midolo, N. Maggi Solca, D. M. M. Queiroz, N. Nouhan, E. Stet, and W. G. V. Quint. 2001. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob. Agents Chemother. 45:1500-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Doorn, L.-J., Y. J. Debets-Ossenkopp, A. Marais, R. Sanna, F. Mégraud, J. G. Kusters, and W. G. V. Quint. 1999. Rapid detection, by PCR and reverse hybridization, of mutations in the Helicobacter pylori 23S rRNA gene, associated with macrolide resistance. Antimicrob. Agents Chemother. 43:1779-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venetianer, P. O. A. 1988. BcefI, a new type IIS restriction endonuclease. Nucleic Acids Res. 16:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Versalovic, J., M. S. Osato, K. Spakovsky, M. P. Dore, and R. Reddy. 1997. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J. Antimicrob. Chemother. 40:283-286. [DOI] [PubMed] [Google Scholar]
- 15.Versalovic, J., D. Shortridge, K. Kibler, M. V. Griffy, J. Beyer, R. K. Flamm, S. K. Tanaka, D. Y. Graham, and M. F. Go. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477-480. [DOI] [PMC free article] [PubMed] [Google Scholar]