Abstract
The oxazolidinones are a novel class of antimicrobial agents that target protein synthesis in a wide spectrum of gram-positive and anaerobic bacteria. The oxazolidinone PNU-100766 (linezolid) inhibits the binding of fMet-tRNA to 70S ribosomes. Mutations to oxazolidinone resistance in Halobacterium halobium, Staphylococcus aureus, and Escherichia coli map at or near domain V of the 23S rRNA, suggesting that the oxazolidinones may target the peptidyl transferase region responsible for binding fMet-tRNA. This study demonstrates that the potency of oxazolidinones corresponds to increased inhibition of fMet-tRNA binding. The inhibition of fMet-tRNA binding is competitive with respect to the fMet-tRNA concentration, suggesting that the P site is affected. The fMet-tRNA reacts with puromycin to form peptide bonds in the presence of elongation factor P (EF-P), which is needed for optimum specificity and efficiency of peptide bond synthesis. Oxazolidinone inhibition of the P site was evaluated by first binding fMet-tRNA to the A site, followed by translocation to the P site with EF-G. All three of the oxazolidinones used in this study inhibited translocation of fMet-tRNA. We propose that the oxazolidinones target the ribosomal P site and pleiotropically affect fMet-tRNA binding, EF-P stimulated synthesis of peptide bonds, and, most markedly, EF-G-mediated translocation of fMet-tRNA into the P site.
A novel class of antimicrobial agents, the oxazolidinones, target a wide spectrum of gram-positive and anaerobic bacteria (4, 6, 9, 28). These compounds act by inhibiting protein synthesis and have no effect on replication or transcription (8). Cell extracts exposed to oxazolidinones are impaired in protein synthesis when programmed by native mRNAs but do not appear to be inhibited when programmed by synthetic mRNAs that lack the signals required for initiation and termination of translation (7, 8, 26). This suggested that these compounds may target the initiation reaction. The oxazolidinone PNU-100766 (linezolid; Fig. 1) inhibits binding of the initiator fMet-tRNA to the 70S ribosomal particle programmed with a synthetic mRNA that harbors a Shine-Dalgarno sequence and a properly spaced initiation codon (29).
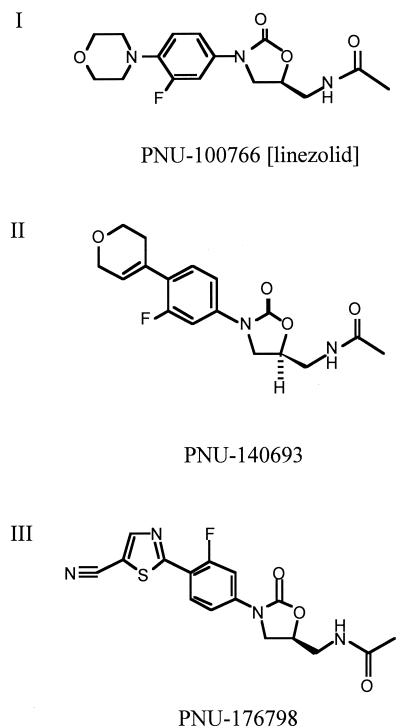
FIG. 1.
Structures of oxazolidinones PNU-100766 (I), PNU-140693 (II), and PNU-176798 (III).
Mutations to oxazolidinone resistance map to domain V of the 23S rRNA in Halobacterium halobium (18), Staphylococcus aureus (27), and the enterococci while mapping to domains IV and V in Escherichia coli (33). The position of these PNU-100766 resistance mutations suggested to us that the oxazolidinones may target peptidyl transferase indirectly by affecting the binding of the initiator tRNA. Since the P site accommodates the initiator tRNA and the nascent protein, these drugs could also affect the affinity of the peptidyl-tRNA for the ribosome.
Recent studies have indicated that the oxazolidinones bind to 70S ribosomes, as well as to 50S subunits (19), but not to 30S subunits. In contrast, a report by Matassova et al. (20) demonstrated that oxazolidinone footprints map to the central domain of the 16S rRNA whereas the 23S rRNA footprints map to domain V (20). Domain V is known to be involved in the peptidyl transferase reaction and in the binding of the 3′ terminus of the tRNA substrates (23). However, efforts to demonstrate an effect of the oxazolidinones on the in vitro activity of the peptidyl transferase of ribosomes derived from E. coli (this work) or from H. halobium (18) were unsuccessful.
Here, we studied the effect of oxazolidinones on peptide bond formation in the presence of a protein, elongation factor P (EF-P), that is essential for the optimum efficiency and specificity of peptide bond synthesis. We found that the oxazolidinones inhibit EF-P-dependent peptide bond synthesis. To examine this further, the fMet-tRNA was bound to the A site and subsequently translocated to the P site with EF-G. The oxazolidinones were found to markedly inhibit the EF-G-mediated translocation of the fMet-tRNA from the ribosomal A site to the P site. We propose that these antibiotics target the ribosomal P site and inhibit initiation of protein synthesis by preventing the proper binding of fMet-tRNA.
MATERIALS AND METHODS
Bacteria and reagent preparation.
[35S]methionine (1,175 μCi/μmol) was purchased from ICN. Mid-log-phase strain MRE600 cells were purchased from the University of Alabama Fermentation Facilities Center, Birmingham. The AUG triplet was synthesized by using polynucleotide phosphorylase as previously described (30). PNU-100766, PNU-140693, and PNU-176798 were obtained from Pharmacia Corp. Puromycin was labeled with 3H2O by incubation at 75°C for 12 h and then extracted into ethyl acetate.
Assay of cell-free translation.
Inhibition of cell-free translation was demonstrated by using the E. coli S30 Extract System for Circular DNA (Promega Corp., Madison, Wis.). Each reaction mixture contained 2.5 μl of S30 premix, 7.5 μl of S30 extract, and 2.5 μl of 10% drug-dimethyl sulfoxide (DMSO). Drugs were dissolved in 30% DMSO and used in a final DMSO concentration of 5% during the assay. Control reaction mixtures received 5% DMSO and no drug. Reactions were initiated by the addition of 1 μg (2.5 μl) of plasmid pBestLuc and incubated at 37°C for 30 min. Placement on ice for at least 5 min stopped the reactions. A 1:8 dilution of the reaction mixture in dilution buffer was prepared, and 10 μl was added to 50 μl of luciferase reagent for reading of luminescence at 542 nm on a SpectraMAX Gemini (Molecular Devices). All reaction mixtures were prepared in triplicate. An E. coli tolC::Tn 10 HN814 mutant from H. Nikaido, University of California, Berkeley, was used to determine the effect of the oxazolidinones in vivo. The tolC mutant is defective in the efflux pump that prevents access of the drugs to the cell (27).
Preparation of ribosomes and ribosomal subunits.
Ribosomes (70S) were isolated from mid-log-phase E. coli cells. The procedures were performed at 4°C. The cells (50-g lots) were broken by grinding with 100 g of alumina (Alcoa) and suspended in 100 ml of buffer A (0.01 M Tris [pH 7.4], 0.001 M dithiothreitol, 0.03 M NH4Cl, 0.01 M MgCl2). DNase (1.0 μg/ml) was added, and the mixture was incubated for 5 min. The suspension was centrifuged for 20 min at 13,000 rpm (GSA rotor; Sorvall) to remove unbroken cells, debris, and alumina. The centrifugation was repeated at 15,000 rpm for 30 min, and the supernatant was centrifuged at 20,000 rpm for 18 h in a Ti 70 rotor (Beckman). The resulting ribosome pellet was suspended by gentle stirring for 2 h in 2 to 3 ml of buffer A containing 6 mM MgCl2. The suspension was then layered onto a 0 to 40% sucrose gradient that was centrifuged for 18 h at 21,000 rpm in a swinging-bucket rotor (SW 40 Ti). One-milliliter fractions were collected from the top of the tube, and the A260 was monitored. The fractions containing the 70S ribosomes were combined and centrifuged at 24,000 rpm for 24 h in the Ti 70 rotor. The pellet was suspended in buffer A containing 6 mM MgCl2 and centrifuged again on a second 0 to 40% sucrose gradient for 18 h at 18,000 rpm in a swinging-bucket rotor (SW 40 Ti). The fractions were identified by A260 and collected as described above, and the 70S peak was isolated by ultracentrifugation for 24 h at 18,000 rpm in the Ti 70 rotor. The 70S ribosomes were suspended in buffer A containing 6 mM MgCl2 and stored in small aliquots at −80°C.
Ribosomal subunits from the first centrifugation were isolated after dialysis of the 70S ribosomes in buffer A containing 1 mM MgCl2. The subunits were isolated on sucrose gradients as described above and then concentrated by ultracentrifugation. The subunits were also stored in buffer A with 6 mM MgCl2 at −80°C.
Initiation complex formation.
Binding of the f[35S]Met-tRNA to E. coli 70S ribosomes was conducted as previously described (8), except that initiation reaction mixtures were prepared without initiation factors and contained 6 mM magnesium acetate [Mg(Ac)2], 0.08 μM AUG, 30 mM NH4Cl, 10 mM Tris (pH 7.4), and 20 pmol of 70S ribosomes in a final volume of 60 μl. The reaction mixtures were incubated for 15 min at 35°C, and the reactions were terminated by addition of cold buffer A, washed with buffer A through Millipore filters, and counted by liquid scintillation.
Assay of peptidyl transferase and purification of EF-P.
EF-P was purified as previously described (1). EF-P-dependent peptidyl transferase activity was assayed as described by Chung et al. (5) with the following modifications. During the first step, the initiation complex was prepared with f[35S]Met-tRNA, AUG, and 70S ribosomes as described above for 20 min at 30οC and then the samples were cooled to 0οC. Unless otherwise specified, 1 μM puromycin and 15% methanol were added in the second step and peptide bond formation was allowed to proceed for 5 min at 30οC in the presence or absence of antibiotics and/or EF-P. PNU-176798 was first dissolved in 30% DMSO before addition to the assay mixture. Control reaction mixtures contained DMSO instead of a drug.
Peptidyl transferase fragment reaction.
Peptidyl transferase was assayed in the presence of ethanol and 60 mM MgC12 as described by Monro (21). Protein concentration was measured by the method of Bradford (3). The fMet-tRNA was prepared as previously described (11).
EF-G-dependent translocation.
Initiation complexes were formed at 4°C for 20 min as described above, by using 2.5 mM MgCl2, 20 mM Tris (pH 7.5), 50 mM NH4Cl, 0.10 mM dithiothreitol, and 0.1 mM GTP. Reaction mixtures were subsequently incubated for 20 min at 4°C in the absence or presence of 1.5 μg of EF-G. The EF-G recombinant protein was purified as previously described for the RbbA protein (16). The reactions were stopped by chilling on ice prior to the addition of 0.4 μM [3H]puromycin. Synthesis of f[35S]Met-[3H]puromycin was measured by extraction of each reaction mixture with ethyl acetate, and the radioactivity of the doubly labeled product was measured after addition of scintillation fluid.
RESULTS
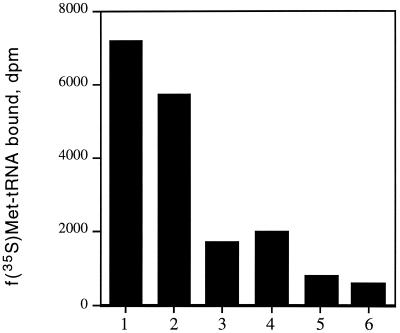
In order to further examine the ability of oxazolidinones to inhibit the binding of fMet-tRNA to the ribosome, we used three different compounds varying in potency against bacteria and cell-free translation (Fig. 1). Table 1 demonstrates that the potency of these oxazolidinones against bacteria correlated well with their ability to inhibit cell-free translation, with PNU-176798 proving to be approximately ninefold more potent than PNU-100766. Both compounds exhibited dose-dependent inhibition of fMet-tRNA binding to 70S ribosomes; the 50% inhibitory concentrations (IC50s) of PNU-176798 and PNU-100766 for inhibition of 70S initiation complex formation were 32 and 152 μM, respectively (Table 1). Incubation of preformed initiation complexes with 80 μM PNU-176798 did not result in destabilization (Fig. 2). Binding of fMet-tRNA was inhibited by kanamycin, a well-established inhibitor of the ribosomal P site (29). Conversely, fMet-tRNA binding was insensitive to the action of tetracycline, which impairs the ribosomal A site (data not shown).
TABLE 1.
Antibacterial activity and translation inhibition by oxazolidinones
| Oxazolidinone | MIC for E. coli (μM)a | IC50 (μM) for:
|
||
|---|---|---|---|---|
| Translation | 70S initiation | Translocation | ||
| PNU-100766 | 24 | 4.7 | 152 | 110 |
| PNU-140693 | 6 | 1.74 | NAb | 41 |
| PNU-176798 | 1.4 | 0.53 | 32 | 8 |
E. coli tolC knockout.
NA, not applicable.
FIG. 2.
Effect of oxazolidinone PNU-176798 on the dissociation of initiation complexes. The initiation complexes were formed as described in Materials and Methods and were undiluted (lanes 1 and 2) or diluted 10-fold (lanes 3 and 4) or 20-fold (lanes 5 and 6) in buffer A containing 6 mM Mg(Ac)2 in the presence of 80 μM PNU-176798 (lanes 2, 4, and 6) or in its absence (lanes 1, 3, and 5). Reactions were continued for 5 min at 35°C. The f[35S]Met-tRNA that remained bound to the ribosomes was then measured by filtration on nitrocellulose filters. Determinations were performed in duplicate.
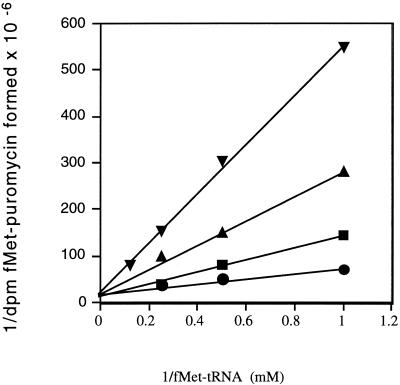
The kinetics of PNU-176798 inhibition were further examined as a function of the fMet-tRNA concentration added to the initiation complex assay mixture. Figure 3 shows that the Km for fMet-tRNA increased with the antibiotic concentration, while the Vmax values remained relatively constant. For example, in the presence of 66 μM PNU-176798, the Km for fMet-tRNA increased from about 0.1 μM (no drug) to 0.8 μM, indicating that PNU-176798 inhibition was competitive.
FIG. 3.
Lineweaver-Burke plots of the inhibition by oxazolidinone PNU-176798 of the fMet-puromycin formation. Formation of fMet-puromycin was measured as described in Materials and Methods, by using different concentrations of f[35S]Met-tRNA. Each reaction mixture contained the following concentrations of the antibiotic PNU-176798: •, no antibiotic; ▪, 16.6 μM; ▴, 50.0 μM; ▾, 66 μM.
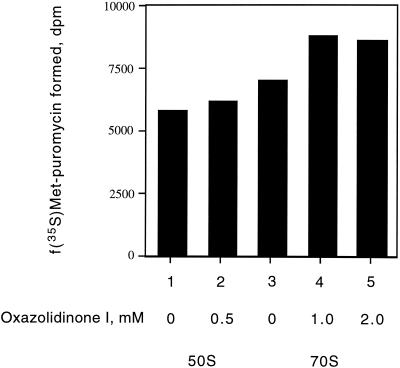
Inhibition of fMet-tRNA binding results in inhibition of the peptidyl transferase reaction if the substrate is, indeed, bound to the ribosomal P site and is in proper juxtaposition with the peptidyl transferase center of the 50S subunit. This reaction is generally measured in the presence of organic solvents that presumably help to bind the tRNA substrates to the ribosome, as well as to activate the dormant peptidyl transferase. Under such conditions, it was demonstrated that the peptidyl transferase of both the 50S subunit and 70S ribosomes is efficiently inhibited by lincomycin and chloramphenicol, two well-known inhibitors of peptidyl transferase (data not shown). However, Fig. 4 shows that the reaction was impervious to the addition of up to 2 mM PNU-100766 or the more potent compound PNU-176798 (data not shown).
FIG. 4.
Effect of oxazolidinone PNU-100766 on peptide bond formation using the fragment reaction. The reactions were conducted at 4°C as described by Monroe (21), and the reaction mixtures contained 60 mM MgCl2, 1 mM puromycin, 33% ethanol, and 50 pmol 70S or 50S subunits. Determinations were performed in duplicate.
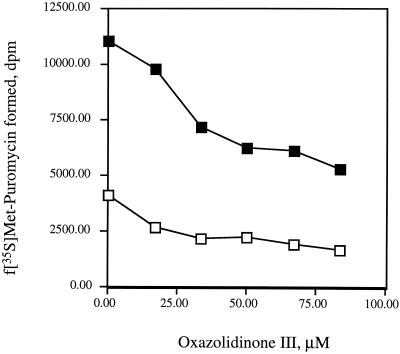
Several of the proteins known to be required for reconstitution of synthesis (10, 12) were examined in attempts to increase the efficiency of the peptidyl transferase reaction. As shown in Fig. 5, addition of antibiotic to the reaction mixture inhibits the activity of peptidyl transferase and results in discernible inhibition of peptidyl transferase by the oxazolidinone PNU-176798, resulting in IC50s on the order of 40 μM.
FIG. 5.
Effect of oxazolidinone PNU-176798 on EF-P-stimulated synthesis of peptide bonds. Reactions were conducted with 15% methanol, 1 μM puromycin, and EF-P as described in Materials and Methods. The antibiotic was added in the first incubation during formation of the initiation complex. □, reaction mixtures without added EF-P. Where indicated, 0.4 μg of EF-P was added to each reaction mixture (▪).
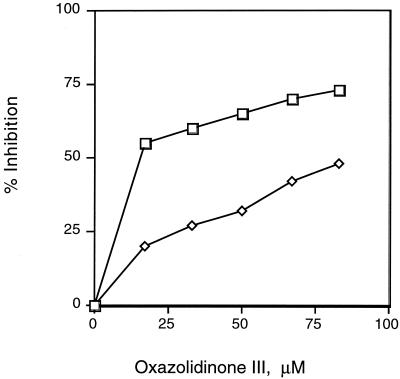
Figure 6 shows that in the presence of EF-P, more-pronounced peptidyl transferase inhibition occurs when PNU-176798 is added after formation of the initiation complex. Less inhibition is observed when the antibiotic is added during binding of the substrate to the ribosome in the presence or absence of EF-P.
FIG. 6.
Preferential effect of oxazolidinone PNU-176798 on the initiation step preceding peptide bond synthesis. Initiation complex formation was allowed to proceed for 20 min at 30°C with the indicated concentrations of the antibiotic, and then 1 μM puromycin and 0.4 μg of EF-P were added (⋄). The antibiotic was added after formation of the initiation complex prior to the addition of puromycin and EF-P (□).
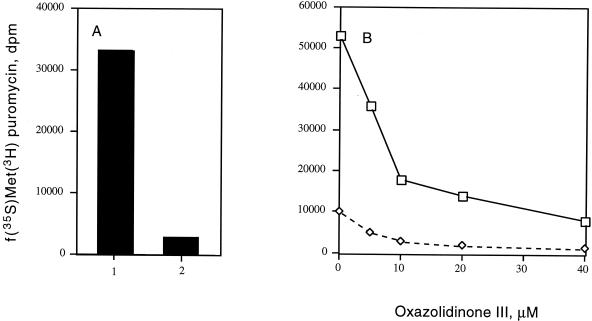
To further examine whether the oxazolidinones preferentially affect the ribosomal P site, fMet-tRNA was bound to the A site of the ribosome at 4οC. Addition of EF-G and GTP stimulated translocation of the initiator tRNA from the ribosomal A site to the P site, allowing peptide bond synthesis to occur in the presence of puromycin. The reaction was demonstrated to be dependent upon EF-G (Fig. 7A) and GTP (data not shown), resulting in a 40-fold increase in fMet-puromycin synthesis under the conditions described. Addition of PNU-176798 markedly inhibited the EF-G-mediated translocation of fMet-tRNA (Fig. 7B). Interestingly, examination of the kinetics of oxazolidinone inhibition of the translocation reaction revealed that this translocation reaction is particularly sensitive to inhibition, resulting in PNU-100766, PNU-140693, and PNU-176798 IC50s of 110, 41, and 8 μM, respectively (Table 1).
FIG. 7.
Effect of EF-G on the translocation of fMet-tRNA from the ribosomal A site to the P site. Initiation complexes were formed as described in Materials and Methods. The reactions were performed at 0°C and, where indicated, contained 1.5 pmol of EF-G and/or the designated concentrations of the oxazolidinone PNU-176798. (A) Reactions in the presence (column 1) or absence (column 2) of EF-G. (B) Oxazolidinone concentration required to inhibit EF-G-dependent translocation. The f[35S]Met-puromycin product was measured by ethyl acetate extraction as described in Materials and Methods. Symbols: □, reaction mixtures containing EF-G; ⋄, reaction mixtures without EF-G.
DISCUSSION
The oxazolidinones selectively interfere with the protein synthetic process (7, 8, 26), and PNU-100766 has been reported to inhibit the formation of 70S initiation complexes in vitro (29). The inhibition of initiation is consistent with a number of biochemical experiments indicating that PNU-100766 does not interfere with protein chain elongation directed by synthetic random polymers or with the codon-dependent termination reaction (19, 26). Here, we report that the initiation reaction is also impaired by PNU-176798, a compound that is a potent inhibitor of translation in vivo. In contrast to its inhibitory effect on the forward initiation reaction, this oxazolidinone exhibits no discernible effect on the stability of the initiation reaction. The oxazolidinones acted as competitive inhibitors of fMet-tRNA binding to the ribosome.
Oxazolidinones have been reported to bind much more strongly to 50S and 70S particles than to 30S subunits of ribosomes (19). Footprinting studies with a photoreactive oxazolidinone demonstrated cross-linking to both the 16S and 23S rRNAs, binding to base A864 of the central domain of the 16S rRNA in a region that is not highly conserved (20). The footprints found on the 23S rRNA mapped to U2113, A2114, U2118, A2119, and C2153 (20), encompassing part of domain V and extending into the region that binds ribosomal protein L1 and comprise the tRNA exit (E) site (20). The oxazolidinone footprints that neighbor the E site may not define the precise binding site, as cross-linking agents do not necessarily target the exact site of ligand interaction. Alternatively, if the antibiotics, indeed, bind to the E site, this binding could affect the A site by a negative allosteric mechanism, as proposed for protein chain elongation (24). The E site has also been clearly demonstrated to influence the translocation reaction by affecting the position of the 3′ terminus of the P site-bound substrate (32).
By using a mutagenized plasmid bearing the rrn operon and either an acrAB or a tolC mutant, Xiong et al. mapped oxazolidinone resistance in E. coli to base G2032 in a part of domain IV of the 23S rRNA that interacts with domain V (33). All of the other reported mutants map to several bases of domain V of the 23S rRNAs of H. halobium, E. faecalis, and S. aureus (18, 27, 33). One simple interpretation of this finding is that the oxazolidinones target the ribosomal P site in a region that has several points of contact spanning both the 30S and 50S subunits. For the 50S subunit, the P site maps to G2252, A2451, U2506, and U2585 (2), most of which are adjacent to the site of oxazolidinone resistance mutations (18, 27, 33). One of these bases, A2451, has also been proposed to be the catalytic residue of the peptidyl transferase (22).
The oxazolidinones cross-link to the 50S E site of domain V, and mutations to resistance involve several bases within the ring structure of domain V that are part of the ribosomal P site (18, 33). Here we demonstrate that binding of fMet-tRNA to ribosomes, EF-P-stimulated synthesis of peptide bonds, and the translocation reaction are impaired by the oxazolidinones. It is possible that these drugs bind with various affinities to each ribosomal site or to a hybrid site of these particles. However, the simplest interpretation is that the oxazolidinones principally target the P site of the ribosomes and subsequently influence other ribosomal sites. The P site harbors the nascent chain that bears many amino acids that must be positioned within the tunnel that spans both subunits. Thus, it is entirely possible that the oxazolidinones also alter the P site in such a fashion as to prevent entry of the peptide chain into the exit tunnel. It has, in fact, been reported that oxazolidinones decrease the chain length of the nascent peptide (13), in keeping with the idea that they interfere with their effect on the ribosomal P site.
If the P site of the ribosome is, indeed, affected by the oxazolidinones, these drugs would be expected to inhibit the peptidyl transferase when the tRNA substrate is properly positioned within the peptidyl transferase center. However, efforts to detect peptidyl transferase inhibition by the oxazolidinones have been unsuccessful in studies utilizing either E. coli or H. halobium ribosomes (18, 27, 33). It is clear from both previous reports and this work that the oxazolidinone IC50 for the initiation reaction is significantly higher than that measured for inhibition of cell-free translation (29). Therefore, in this study, we sought to enhance the sensitivity of the peptidyl transferase assay by using the potent oxazolidinone PNU-176798 and EF-P. EF-P strongly stimulates peptide bond synthesis (13, 14), and this synthesis was significantly inhibited in this study by PNU-176798. EF-P has no direct effect on the binding of fMet-tRNA to the ribosome as measured by filtration assays (10). However, this protein could potentially help to position fMet-tRNA in proper proximity to the peptidyl transferase center.
EF-P accelerates peptide bond synthesis from aminoacyl-tRNAs or from CCA amino acids, which are poor acceptors of ribosomal peptidyl transferase (10, 14). EF-P binds to the 30S subunit and to 70S ribosomes and also interacts with the 50S particle. Discernible footprints can be detected in domain V of the 50S subunit as a result of its interaction with EF-P (unpublished data). Interestingly, EF-P protects U2555, A2564, and C2576, which are near the site of eperezolid resistance in S. aureus (27).
The crystal structure of EF-P from homologous protein eIF-5A of Methanococcus jannaschii indicates that the protein has an elongated shape and that the N- and C-terminal ends of the molecule are charge polarized (17). EF-P-stimulated synthesis of peptide bonds is inhibited by streptomycin, which acts on the 30S subunit, as well as lincomycin and chloramphenicol, which impair the peptidyl transferase activity of the 50S subunit (1). These properties imply that EF-P could bind to both subunits as it activates peptide bond synthesis. This action of the protein might be brought about by its interactions with the ribosome, which may then properly position the 3′ terminus of the fMet-tRNA on the peptidyl transferase cavity prior to peptide bond synthesis. Alternatively, EF-P could enhance the affinity of the amino-acyl-tRNA (or the puromycin analogue) by binding to the ribosome.
Study of the effect of the antibiotic during the initiation reaction or after its completion, followed by addition of EF-P, suggests that this oxazolidinone has an effect apart from inhibition of initiation. Experiments reported here, in which the fMet-tRNA was bound to the A site and then translocated to the P site by the action of EF-G (15), indicate that the antibiotic may, indeed, be capable of inhibiting translocation into the ribosomal P site. Indeed, the IC50s of the oxazolidinone PNU-176798 are about 10-fold lower for translocation than for initiation (Table 1).
If the oxazolidinones target the ribosomal P site, one would expect the synthesis of all polymers to be affected. However, it has been reported that synthesis directed by poly(U) (26) and other synthetic polymers (7, 8) is impervious to the action of these drugs. Thus, the oxazolidinones clearly inhibit translation programmed by native templates, which harbor initiation signals, but do not inhibit synthesis directed by random mRNAs lacking these signals.
The translocation step from fMet-tRNA is preceded by a number of reactions that differ during synthesis of the first peptide bond (25, 31). The initiator tRNA structural properties must be recognized in order to permit the joining of the subunits, as well as the approximation of the 3′ end of fMet-tRNA to the 3′ terminus of the incoming aminoacyl-tRNA, that must occur prior to peptide bond synthesis. Unlike the elongation step that follows, the entrance of the second aminoacyl-tRNA into the ribosomes is not necessarily influenced by the filling of the E site with a tRNA that must exit the ribosome after peptide bond formation. Also, the first translocation may involve dislodging of the mRNA-ribosome-fMet-tRNA complex from the interaction of the 3′ terminus of the 16S rRNA with the mRNA. The strong inhibition by the oxazolidinones of the translocation step could target one or more of these intermediate steps that underlie the special nature of the synthesis of the first peptide bond. However, the simplest explanation for the mode of inhibition of the oxazolidinones is that they target the ribosomal P site, thus exhibiting pleiotropic effects on several intermediate steps of translation.
Acknowledgments
We are grateful to K. Nierhaus for a generous gift of the overexpressing plasmid harboring the fusA gene encoding EF-G.
This work was supported by a grant from the Pharmacia Corp.
REFERENCES
- 1.Aoki, H., S.-L. Adams, M. A. Turner, and M. C. Ganoza. 1997. Molecular characterization of the efp gene product involved in a peptidyl transferase reaction. Biochimie 79:7-11. [DOI] [PubMed] [Google Scholar]
- 2.Bocchetta, M., L. Xiong, and A. S. Mankin. 1998. 23S rRNA positions essential for tRNA binding in ribosomal functional sites. Proc. Natl. Acad. Sci. USA 95:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. Rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brickner, S. J., M. R. Hutchinson, M. R. Barbachyn, P. R. Manninen, D. A. Ulaniwicz, S. A. Garmon, K. C. Grega, S. K. Hendges, D. S. Toops, C. W. Ford, and G. E. Zurenko. 1996. Synthesis and antimicrobial activity of U-100592 and U-100766, two oxazolidinone antibacterial agents for the potential treatment of multidrug-resistant gram-positive bacterial infections. J. Med. Chem. 39:673-679. [DOI] [PubMed] [Google Scholar]
- 5.Chung D.-G., N. D. Zahib, R. M. Baxter, and M. C. Ganoza. 1990. Peptidyl transferase: the soluble protein EFP restores the efficiency of 70S ribosome catalyzed peptide bond synthesis, p. 69-80. In G. Spedding (ed.), Ribosomes and protein synthesis: a practical approach. IRL Press, Oxford Publishing Co., Oxford, United Kingdom.
- 6.Daly, J. S., G. M. Eliopoulos, S. Willey, and R. C. Moellering. 1988. Mechanism of action and in vitro and in vivo activities of S-6123, a new oxazolidinone compound. Antimicrob. Agents Chemother. 32:1341-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eustice, D. C., P. A. Feldman, and A. M. Slee. 1988. Mechanism of action of DuP 721, a new antibacterial agent: effect on macromolecular synthesis. Biochem. Biophys. Res. Commun. 150:965-971. [DOI] [PubMed] [Google Scholar]
- 8.Eustice, D. C., P. A. Feldman, I. Zajac, and A. M. Slee. 1988. Mechanism of action of DuP 721: inhibition of an early event during initiation of protein synthesis. Antimicrob. Agents Chemother. 32:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford, C. W., J. C. Hamel, D. M. Wilson, J. K. Moerman, D. Stapert, R. J. Yancey, D. K. Hutchinson, M. R. Barbachyn, and S. J. Brickner. 1996. In vivo activities of U-100592 and U-100766, novel oxazolidinone antimicrobial agents, against experimental bacterial infections. Antimicrob. Agents Chemother. 40:1508-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganoza, M. C., and H. Aoki. 2000. Peptide bond synthesis: function of the efp gene product. Biol. Chem. 381:553-559. [DOI] [PubMed] [Google Scholar]
- 11.Ganoza, M. C., N. Barraclough, and J.-T. Wong. 1976. Purification and properties of an N-formylmethionyl-tRNA hydrolase. Eur. J. Biochem. 65:613-617. [DOI] [PubMed] [Google Scholar]
- 12.Ganoza, M. C., C. Cunningham, and R. M. Green. 1985. Isolation and point of action of a factor from E. coli required to reconstruct translation. Proc. Natl. Acad. Sci. USA 82:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glick, B. R., and M. C. Ganoza. 1975. Identification of a soluble protein that stimulates peptide bond synthesis. Proc. Natl. Acad. Sci. USA 72:4257-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glick, R. B., S. Chladek, and M. C. Ganoza. 1979. Peptide bond formation stimulated by protein synthesis factor EF-P is dependent on the aminoacyl moiety of the acceptor. Eur J. Biochem. 97:23-28. [DOI] [PubMed] [Google Scholar]
- 15.Kevin, S., K. S. Wilson, and H. F. Noller. 1998. Mapping the position of elongation factor EF-G in the ribosome by hydroxyl radical probing. Cell 92:131-139. [DOI] [PubMed] [Google Scholar]
- 16.Kiel, M. C., and M. C. Ganoza. 2000. Functional interactions of an Escherichia coli ribosomal ATPase. Eur. J. Biochem. 268:1-10. [DOI] [PubMed] [Google Scholar]
- 17.Kim, K. K., H. Yokota, R. Kim, and S.-H. Kim. 1997. Cloning, expression and crystallization of a hyperthermophilic protein that is homologous to the eukaryotic initiation factor, eIF-5A. Protein Sci. 6:2268-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloss, P., L. Xiong, D. L. Shinabarger, A. S. Mankin. 1999. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 19.Lin, A. H., R. W. Murray, T. J. Vidmar, and K. R. Marotti. 1997. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 41:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matassova, N. B., M. V. Rodnina, R. Endermann, H.-P. Kroll, V. Pleiss, H. Wild, and W. Wintermeyer. 1999. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA 5:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monroe, R. E. 1967. Catalysis of peptide bond formation by 50S ribosomal subunits from Escherichia coli. J. Mol. Biol. 26:147-151. [DOI] [PubMed] [Google Scholar]
- 22.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 23.Noller, H. F. 1993. Peptidyl transferase: protein, ribonucleoprotein, or RNA? J. Bacteriol. 175:5297-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rheinberger, H. J., and K. H. Nierhaus. 1980. Simultaneous binding of three tRNA molecules by the ribosome of Escherichia coli. Biochem. Int. 1:297-303. [Google Scholar]
- 25.Rodnina, M. V. 1999. Dynamics of translation on the ribosome: molecular mechanisms of translocation. FEMS Microbiol. Rev. 23:317-333. [DOI] [PubMed] [Google Scholar]
- 26.Shinabarger, D. L., K. R. Marotti, R. W. Murray, A. H. Lin, R. P. Melchior, S. M. Swaney, D. S. Dunyak, W. F. Demyan, and J. M. Buysse. 1997. Mechanism of action of oxazolidinones. Effect of linezolid and eperezolid on translation reactions. Antimicrob. Agents Chemother. 41:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinabarger, D. L. 1999. Mechanism of action of the oxazolidinone antibacterial agents. Exp. Opin. Investig. Drugs 8:1195-1202. [DOI] [PubMed] [Google Scholar]
- 28.Slee, A. M., M. A. Wuanola, R. J. McRipley, I. Zajac, P. T. Bartolomew, W. A. Gregory, and M. Forbes. 1987. Oxazolidinones, a new class of synthetic antibacterial agents: in vitro and in vivo activities of DuP105 and Dup721. Antimicrob. Agents Chemother. 31:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaney, S. M., H. Aoki, M. C. Ganoza, and D. L. Shinabarger. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents and Chemother. 42:3251-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thach, R. E. 1966. Enzymatic synthesis of oligonucleotide of defined sequence, p. 520-534. In G. L. Cantoni and D. R. Davies (ed.), Procedures in nucleic acid research. Harper & Row, New York, N.Y.
- 31.Wilson, S. W., and H. F. Noller. 1998. Molecular movement inside the translational engine. Cell 92:337-349. [DOI] [PubMed] [Google Scholar]
- 32.Wintermeyer, W., R. Lill, and J. M. Robertson. 1990. Role of the tRNA exit site in ribosomal translocation, p. 348-357. In W. E. Hill, A. Dahlberg, Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome. American Society for Microbiology, Washington, D.C.
- 33.Xiong, L., P. Kloss, S. Douthwaite, N. M. Anderson, S. Swaney, D. C. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]