Abstract
We investigated the phenotypic and genetic properties of metallo-β-lactamase-producing Pseudomonas isolates collected at a tertiary-care hospital in Korea since 1995. The prevalence of imipenem resistance among Pseudomonas aeruginosa isolates reached 16% in 1997, when 9% of the resistant organisms were found to produce VIM-2 β-lactamase, a class B enzyme previously found only in P. aeruginosa isolates from Europe. VIM-2-producing isolates of Pseudomonas putida were also detected. Resistance was transferable from both these species to P. aeruginosa PAO4089Rp by filter mating, although the resistance determinant could not be found on any detectable plasmid. Serotyping showed that many of the VIM-2-producing P. aeruginosa isolates belonged to serotypes O:11 and O:12, and pulsed-field gel electrophoresis of XbaI-digested genomic DNA revealed that many had identical profiles, whereas the P. putida isolates were diverse. Sequencing showed that the blaVIM-2 genes resided as cassettes in class 1 integrons. In contrast to previous VIM-encoding integrons, the integron sequenced from a P. aeruginosa isolate had blaVIM located downstream of a variant of aacA4. blaVIM also lay in a class 1 integron in a representative P. putida strain, but the organization of this integron was different from that sequenced from the P. aeruginosa strain. In conclusion, the metallo-β-lactamase produced by these imipenem-resistant Pseudomonas isolates was VIM-2, and the accumulation of producers reflected clonal dissemination as well as horizontal spread. Strict measures are required in order to control a further spread of resistance.
Carbapenems such as imipenem are stable to most β-lactamases. Nevertheless, the first isolate of Pseudomonas aeruginosa with transferable imipenem resistance due to a metallo-β-lactamase production was reported in Japan in 1991 (26). The enzyme produced by this isolate almost certainly was IMP-1, which subsequently has been repeatedly found in Japan, not only in P. aeruginosa and Acinetobacter baumannii (23), but also in Serratia marcescens (14) and other members of Enterobacteriaceae (5). Rasmussen and Bush (18) predicted that the increasing use of carbapenems would select for more carbapenem-hydrolyzing organism on a wider scale, and this prediction is now proving correct. Several close relatives of IMP-1 have been reported during the past 3 years: IMP-2 from an isolate of A. baumannii in Italy (19), IMP-3 (MET-1) from Shigella flexneri in Japan (6), and IMP-4 from Acinetobacter and Citrobacter spp. from Hong Kong, China (3).
Descriptions of IMP-5 to IMP-8 enzymes are understood to be in press. IMP-1 to IMP-4 have at least 80% homology to each other, but in 1999, a new type of acquired metallo-β-lactamase, VIM-1, was reported to have been found in an isolate of P. aeruginosa collected in Italy (8). Soon afterwards, isolates of P. aeruginosa with a related enzyme, VIM-2, were reported in France (16, 17). Since then, outbreaks of VIM β-lactamase-producing P. aeruginosa have been reported in Greece (25) and Italy (4).
Although the VIM and IMP metallo-β-lactamase have only 30 to 40% amino acid homology, both are often encoded by mobile gene cassettes inserted into integrons (1, 8, 17), which are sometimes located on plasmids. Most of these integrons belong to class 1, but their structures vary among isolates.
In the present study, we determined the phenotypes and genetic properties of metallo-β-lactamase-producing Pseudomonas strains collected at one hospital in Korea since 1995.
(This study was presented, in part, at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy [K. Lee, Y. Chong, H. B. Shin, and D. Yong, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-85, 1998] and at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy [K. Lee, J. B. Lim, J. H. Yum, D. Yong, J. R. Choi, Y. Chong, and J. M. Kim, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2003, 2000].)
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
Nonduplicate isolates of Pseudomonas spp. were from clinical specimens from patients in a tertiary-care hospital in Seoul, Korea. The species were identified by conventional methods (7) and with the ID 32 GN system (bioMerieux, Marcy-l'Etoile, France). Imipenem resistance was initially detected by a disk diffusion method (13), and the resistant isolates were kept in skim milk at −76°C until used for further testing. The MICs of antimicrobial agents were determined by an agar dilution method (12). The agents and β-lactamase inhibitors used were ampicillin and cephalothin (Sigma, St. Louis, Mo.), cefoxitin and imipenem (Merck/Sharp & Dohme, Rahway, N.J.), meropenem (Sumitomo, Tokyo, Japan), cefotaxime (Aventis, Frankfurt, Germany), ceftazidime and clavulanic acid (GlaxoSmithKline, Greenford, United Kingdom), piperacillin and tazobactam (Wyeth, Pearl River, N.Y.), aztreonam (Bristol-Myers Squibb, Princeton, N.J.), amikacin and kanamycin (Dong-A Pharmaceutical, Seoul, Korea), gentamicin (Chong Kun Dang Pharmaceutical, Seoul, Korea), streptomycin (Meiji Seika, Tokyo, Japan), tobramycin (Daewoong-Lilly, Seoul, Korea), and ciprofloxacin (Bayer Korea, Seoul, Korea). Metallo-β-lactamase production was screened with modified Hodge and EDTA disk synergy tests (9).
Conjugation.
The filter mating method (20) was used to determine the transferability of the carbapenem resistance determinant using the rifampin-resistant mutant P. aeruginosa, PAO4089Rp (26) as a recipient. Mueller-Hinton agar containing 100 μg of rifampin per ml and 2 μg of imipenem per ml was used to select transconjugants.
Serotyping, plasmid preparation, PFGE of genomic DNA, and hybridization.
O serotypes of the P. aeruginosa isolates were determined by a slide-agglutination method of the International Antigenic Typing Scheme. Plasmids were isolated by alkaline lysis (21). For pulsed-field gel electrophoresis (PFGE), XbaI-digested genomic DNA was prepared according to the instruction of Bio-Rad (Hercules, Calif.) and fragments were separated for 20 h at 6 V/cm at 11°C using a CHEF-DR II System (Bio-Rad), with initial and final pulse times of 0.5 and 30 s, respectively. DNA fingerprints were compared visually and were interpreted as recommended by Tenover et al. (24).
For Southern hybridization, plasmid or genomic DNA was transferred from electrophoresis gels to nylon membranes using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). A DIG DNA Labeling and Detection Kit (Roche Diagnostics, Mannheim, Germany) was used to label the blaVIM-2 probe and for hybridization.
β-Lactamase assays and isoelectric focusing.
The β-lactamase activities of cell sonicates from overnight Luria-Bertani broth cultures were determined using a spectrophotometer (UV-1601PC; Shimadzu Corp., Tokyo, Japan) with a 100 μM solution of β-lactams in 50 mM phosphate buffer (pH 7.0) at 30°C. The wavelengths used were 235 nm for benzylpenicillin (Sigma) and 297 nm for both imipenem and meropenem (11). The protein content of the sonicates was determined with a commercial assay reagent (Bio-Rad).
The isoelectric points of β-lactamases were determined by loading cell sonicates to precast pH 3 to 10 gels (Novel Experimental Technology, San Diego, Calif.). These were electrophoresed on a ThermoFlow Electrophoresis Temperature Control System (Novel Experimental Technology). After electrophoresis, the gels were overlaid with filter papers soaked in 0.7 mg of nitrocefin per ml in a 30 mM N-(2-acetamido)-2-aminoethanesulfonic acid-NaOH buffer (pH 7.0) (8). Sonicates of Escherichia coli and P. aeruginosa VR-143/97 (8) producing TEM-1 (pI 5.4) and VIM-1 (pI 5.3) β-lactamases, respectively, were used as controls. Imipenem-hydrolyzing bands were confirmed by observation for growth when the gel was overlaid with Mueller-Hinton agar containing E. coli ATCC 25922 and 2 μg of imipenem per ml and then incubated overnight at 37°C. The gels were overlaid with a filter paper soaked in 20 mM EDTA for 5 min before the imipenem-containing agar was added so that inhibition of imipenem-hydrolyzing activities could be observed.
Detection of metallo-β-lactamase genes by PCR.
Previously reported PCR primers were used to determine the presence of blaIMP (2) and blaVIM-2 (17). PCR was carried out with 1 μl of heat-extracted template DNA, 20 pmol of each primer, and PreMix (Bioneer, Cheongwon, Korea) containing 1 U of Taq DNA polymerase in a total volume of 20 μl. A thermal cycler (Eppendorf-Netheler-Hinz, Hamburg, Germany) was used under the following conditions: 94°C for 5 min, 25 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s, followed by 72°C for 7 min. The bands were confirmed by hybridization with a blaVIM-2 probe.
Sequencing the blaVIM-containing integron.
PCR products were obtained by the hot-start method in a total volume of 100 μl, with 5 μl of heat-extracted template DNA, 20 pmol of primers INT/5′CS and INT/3′CS (19), and 3 U of LA Taq DNA polymerase (Takara, Shiga, Japan). The reaction conditions consisted of a predenaturation at 94°C for 12 min and then 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 5 min. The extension time was increased by 5 s at each cycle, as described by Levesque and Roy (10).
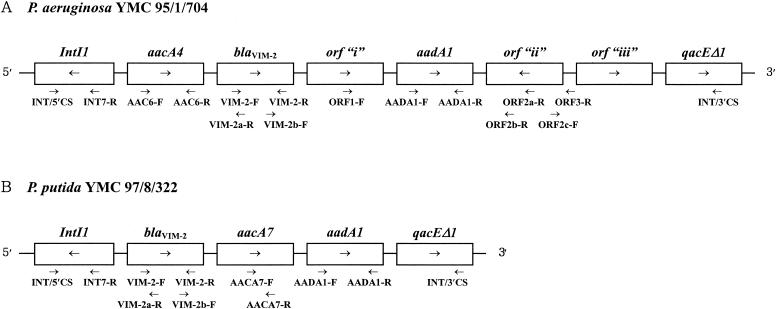
The PCR products were ligated in the pGEM-T Easy Vector (Promega, Madison, Wis.) and transformed into E. coli DH5α by electroporation using an E. coli Pulser (Bio-Rad) (22). Successful clones were selected by the observation of decreased imipenem disk zones. Plasmids from cloned strains were used to sequence blaVIM and the other cassettes in the integrons, using the dideoxy-chain termination method with an ABI 3700 Autosequencer and the reagents supplied by its manufacturer (Perkin-Elmer, Foster City, Calif.). Primers INT/5′CS and INT/3′CS were those reported previously (19). The nucleotides of custom-designed primers were as follows: INT7-R, 5′-GTT CTT CTA CGG CAA GGT GC-3′ (bp 261 to 280); VIM-2-F, 5′-ATT GGT CTA TTT GAC CGC GTC-3′ (bp 55 to 75); VIM-2-R, 5′-TGC TAC TCA ACG ACT GAG CG-3′, (bp 784 to 803); VIM-2b-F, 5′-CAA CAA CAC TAC CCG GAA GC-3′, (bp 679 to 698), VIM-2a-R, 5′-ATC TGG TAA AGC CGG ACC TC-3′ (bp 127 to146); AAC6-F, 5′-ACT GAG CAT GAC CTT GCA AT-3′ (bp 4 to 23), AAC6-R, 5′-CTG GCG TGT TTG AAC CAT GT-3′ (bp 470 to 489); AACA7-F, 5′-AGC ATT GGG CTC GTG GTC G-3′ (bp 323 to 341); AACA7-R, 5′-GAC ACC TCC GTG AAT CCA G-3′ (bp 410 to 428); AADA1-F, 5′-TGA TTT GCT GGT TAC GGT GAC-3′ (bp 144 to 164); AADA1-R, 5′-CAC TAC ATT TCG CTC ATC G-3′ (bp 543 to 561); ORF1-F, 5′-GAC TTC TGG GCG TTC ATT GG-3′ (bp 257 to 276); ORF2a-R (5′-TGG CGA CAA GGT GAA AGA C-3′ (bp 486 to 504); ORF2b-R, 5′-TGG CTA TCA GCA CTG CGT TGA G-3′ (bp 1002 to 1023); ORF2c-F, 5′-ATC CAG CTC AAC GCA GTG CTG AT-3′ (bp 1007 to 1029); ORF3-R, 5′-AGA AGC CAA ACG CCC ATC GTA-3′ (bp 1 to 21 upstream from orf “iii”) (Fig. 2). The determination of the sequences was repeated with more than two clones from independent amplicons. Also, the sequences on both strands were determined.
FIG. 2.
Schematic structure (not to scale) of integrons in isolates of P. aeruginosa YMC 95/1/704 and P. putida YMC 97/8/322, which had approximate sizes of 5.6 and 3 kb, respectively. The ORFs are boxed. The names of unknown orfs “i,” “ii,” and “iii” are arbitrary. Arrows indicate their transcriptional orientation. orf “ii” had a reversed orientation. Primers used for sequencing are shown under the integron structures (see text for primer sequences).
Nucleotide sequence accession numbers.
The nucleotide sequences reported have been assigned the GenBank accession nos. as follows: for the integron from P. aeruginosa YMC 95/1/704, AY029772; for P. putida YMC 97/8/322, AF327064.
RESULTS
Prevalence of imipenem resistance and metallo-β-lactamase producer.
In 1995, 3% of P. aeruginosa isolates at the hospital were imipenem resistant, and among the resistant isolates examined, five had metallo-β-lactamases. Of eight imipenem-resistant P. aeruginosa collected in 1996, none had metallo-β-lactamases. In 1997, however, the prevalence of imipenem resistance rose to 16% and among the total of 130 imipenem-resistant isolates 11.5% had metallo-β-lactamases (Table 1). The prevalence rate of imipenem resistance in P. aeruginosa then stabilized, as did the proportion with metallo-β-lactamases. Metallo-β-lactamase-producing P. putida isolates were first detected in 1997. PCR showed that all of the metallo-β-lactamase producers had blaVIM-2 but none had blaIMP-1.
TABLE 1.
Trend of isolation of imipenem-resistant Pseudomonas spp. and detection of blaVIM-2-positive isolates
| Yr |
P. aeruginosa
|
P. putida
|
||||
|---|---|---|---|---|---|---|
| No. of isolates (% imipenem resistance) | No. of blaVIM-2 isolates
|
No. of isolates (% imipenem resistance) | No. of blaVIM-2 isolates
|
|||
| Tested | Positive (%) | Tested | Positive (%) | |||
| 1994 | 1,939 (4) | 1 | 0 (0) | 38 (8) | 0 | NAa |
| 1995 | 966 (3) | 20 | 5 (25.0) | 6 (0) | 0 | NA |
| 1996 | 2,189 (6) | 8 | 0 (0) | 37 (16) | 0 | NA |
| 1997 | 2,078 (16) | 130 | 15 (11.5) | 17 (6) | 1 | 1 (100) |
| 1998 | 2,934 (15) | 147 | 12 (8.2) | 35 (29) | 5 | 5 (100) |
| 1999 | 3,129 (15) | 187 | 11 (5.9) | 35 (17) | 3 | 3 (100) |
| Total | 13,235 (11) | 493 | 43 (8.7) | 168 (15) | 9 | 9 (100) |
NA, not applicable.
Isoelectric focusing of extracts of the blaVIM-2-positive isolates detected a β-lactamase with a pI of ca. 5.3, which was no longer apparent if the gels were overlaid with EDTA. A biological assay showed that this pI 5.3 band inactivated imipenem, allowing the growth of the E. coli indicator strain in an agar overlay. Almost all of the blaVIM-2-positive isolates also had other β-lactamase bands with pIs of ca. 7 to >8.
Among the total of 52 blaVIM-2-positive P. aeruginosa and P. putida isolates collected, 35 (67%) were from urines, 10 (19%) were from sputa, and 2 (4%) were from blood samples; among the 9 P. putida isolates all except one were from urine. Most of the source patients had underlying malignancy, recent brain surgery, indwelling urinary catheters, prior therapy with various antimicrobial agents, and/or admission to intensive care units (data not shown).
Conjugative transfer of resistance.
Carbapenem resistance was transferred from 6 of the 43 P. aeruginosa isolates and 2 of the 9 P. putida isolates with blaVIM-2 to a P. aeruginosa recipient. However, repeated attempts failed to detect the presence of plasmids capable of hybridizing with the blaVIM-2 probe, while the fragments of the XbaI-digested genomic DNA of clinical isolates showed hybridization. Resistances to ciprofloxacin and amikacin were not transferred to the transconjugants.
Susceptibilities of isolates and transconjugants.
MIC ranges of imipenem for blaVIM-2-positive clinical isolates and transconjugants of P. aeruginosa were 8 to >128 and 16 to 32 μg/ml, respectively; those for P. putida isolates and transconjugants were 16 to 128 and 16 to 32 μg/ml, respectively (Table 2). MICs of imipenem and meropenem for the P. aeruginosa recipient were 0.25 and 0.5 μg/ml, respectively. MICs of all of the other β-lactams for clinical isolates were ≥32 μg/ml, except for aztreonam, whose MICs were as low as 4 μg/ml for some isolates.
TABLE 2.
Antimicrobial susceptibilities of blaVIM-2-positive clinical isolates and their transconjugants
| Antimicrobial agenta |
P. aeruginosa
|
P. putida
|
||||
|---|---|---|---|---|---|---|
| CIb (n = 43)
|
TCb (n = 6)
|
CI (n = 9)
|
TC (n = 2)
|
|||
| MIC rangec | MIC50c | MIC90c | MIC range | MIC range | MIC range | |
| Imipenem | 8->128 | 16 | 128 | 16-32 | 16-128 | 16-32 |
| Meropenem | 8->128 | 16 | 128 | 8-32 | 32-128 | 8-16 |
| Piperacillin | 64->256 | 256 | >256 | 8-128 | 64->256 | 256 |
| Piperacillin-tazobactamd | 32->128 | 128 | >128 | 8-128 | 32->256 | 128 |
| Cefotaxime | >128 | >128 | >128 | 64->128 | >128 | >128 |
| Cefotaxime-clavulanic acidd | >128 | >128 | >128 | 32->128 | >128 | >128 |
| Ceftazidime | 32->128 | 128 | >128 | 8-64 | 32->128 | 128 |
| Aztreonam | 4-64 | 16 | 32 | 2-4 | 16-32 | 4 |
| Amikacin | 2->128 | >128 | 16 | 4 | 16->128 | 4 |
| Gentamicin | 8->128 | >128 | 128 | 64 | 32->128 | 64 |
| Kanamycin | 64->128 | >128 | >128 | >128 | 16->128 | >128 |
| Streptomycin | 128->128 | >128 | >128 | >128 | >128 | 128 |
| Tobramycin | 2->128 | >128 | 32 | 16-32 | 16->128 | 32 |
| Ciprofloxacin | 0.12->128 | 32 | 16 | 0.12-0.25 | 16 | 0.12 |
MICs of ampicillin, cephalothin, and cefoxitin for all of the strains were >128 μg/ml.
Abbreviations: CI, clinical isolates; TC, transconjugants.
Values are in micrograms per milliliter; MIC50 and MIC90, MICs for 50 and 90% of strains, respectively.
Piperacillin-tazobactam: tazobactam at a fixed concentration of 4 μg/ml; cefotaxime- clavulanic acid: a 2:1 combination.
A slight reduction in the MICs of piperacillin was achieved in the presence of tazobactam (4 μg/ml), but the MICs of cefotaxime were not reduced by clavulanic acid. Most of the VIM-2-producing P. aeruginosa isolates were also resistant to gentamicin, tobramycin, and ciprofloxacin.
Serotype and PFGE profile.
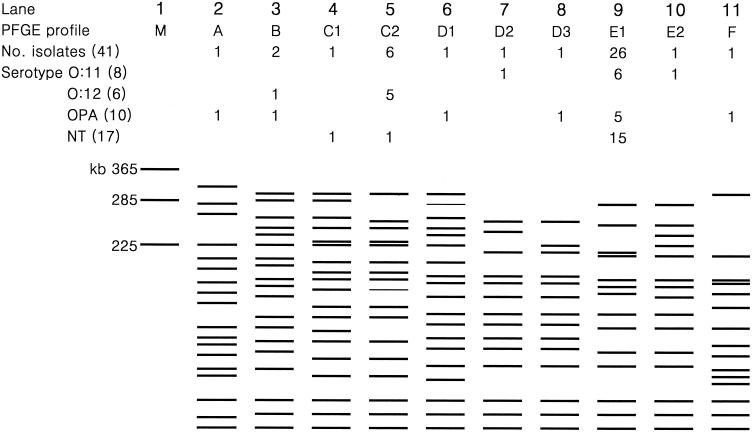
PFGE of XbaI-digested genomic DNA from blaVIM-2-positive P. aeruginosa showed that 26 isolates gave an identical pattern, E1 (Fig. 1). Among these, 11 strains were isolated from 1997 to 1999: 6 serotype O:11 isolates were from two each of pediatric and neurosurgery patients and from one each of gynecology and rehabilitation patients; 5 O polyagglutinating (OPA) isolates were from two internal medicine patients and from one each of pediatric, rehabilitation, and neurosurgery patients. Among the six isolates with PFGE pattern C2, five were serotype O:12 and were isolated from four ICU patients in 1997 and one pediatric patient in 1995. Among the six isolates of P. aeruginosa that were gene transfer positive, four had PFGE pattern C2 with serotype O:12. The remaining two serotype OPA isolates had PFGE patterns A and D1. Eight different PFGE profiles were seen among the nine blaVIM-2-positive P. putida isolates.
FIG. 1.
Schematic presentation of PFGE patterns of XbaI-digested genomic DNA of blaVIM-2-positive isolates of P. aeruginosa. M in lane 1 is a size marker (a chromosome of Saccharomyces cerevisiae). The numbers of isolates with serotypes O:11, O:12, and OPA are shown under each PFGE profile, and total numbers are in parentheses. OPA, O polyagglutination; NT, serotype not tested.
Biochemical detection of VIM β-lactamases.
Cell sonicates of all blaVIM-2-positive isolates hydrolyzed imipenem in spectrophotometric assays; this activity was inhibited by EDTA, but not by clavulanic acid. Using extracts of a representative P. aeruginosa strain, relative hydrolysis rates of 100 μM benzylpenicillin, imipenem, and meropenem were in a 100:21:7 ratio. With extracts of a P. putida isolate this ratio was 100:2:<1, reflecting a relatively higher background penicillinase activity. Imipenem-hydrolyzing activities of the P. aeruginosa and P. putida extracts were 96 and 61 U per mg of protein, respectively (data not shown).
Sequences of blaVIM-2 and flanking genes.
PCR products of the integron carrying blaVIM-2 were detected from 41 of the 43 P. aeruginosa and all of the 9 P. putida isolates. The sizes of the integrons carrying the blaVIM-2 varied among the isolates, from ca. 3 to 6 kb, but in 23 of 50 isolates they were ca. 5 kb. The sizes in the 25 P. aeruginosa strains with PFGE pattern E1 were ca. 4 kb in 6 isolates, 5 kb in 15 isolates, and 6 kb in 4 isolates. The first isolates of P. aeruginosa and P. putida were chosen to analyze the nucleotide sequence of the blaVIM allele and its flanking genes. Analysis revealed sequences of blaVIM-2 in both cases. The structure of the integron from the P. aeruginosa isolate was aacA4 [Asp171→Val variant of aac(6′)-II], followed immediately downstream by blaVIM-2; in addition aadA1 was present. The integron from the P. putida isolate differed in that the first cassette was blaVIM-2 followed by aacA7 and aadA1 (Fig. 2).
DISCUSSION
The frequency of imipenem resistance among P. aeruginosa isolates at the hospital rose to 16% in 1997, thereafter stabilizing. Approximately 9% of the resistance isolates were metallo-β-lactamase-producers, and all these had blaVIM-2 (Table 1). In another study in 1999 (K. Lee, H. S. Lee, S. H. Kang, M. H. Lee, W. K. Song, S. J. Kim, and Y. Chong, Abstr. 7th Western Pacific Cong. Chemother. Infect. Dis., abstr. FP2.8, 2000), it was found that blaVIM-2-positive P. aeruginosa isolates were widespread in other Korean hospitals too, being detected in 9 of 29 hospitals surveyed, with an overall prevalence rate of 11.9% among imipenem-resistant P. aeruginosa isolates. As reported here, blaVIM-2 has also spread to P. putida in a Korean hospital.
It is striking that the metallo-β-lactamase found in Korea is not the IMP-1 type which is scattered in Japan, but the VIM type, which was previously reported only from Europe (8, 17). Moreover, although VIM-1 and VIM-2 β-lactamase-producing P. aeruginosa isolates were collected in 1996 and 1997 in Europe, it is now clear that VIM-2 producers have existed in Korea since 1995 and have become much more widespread than has been observed elsewhere. Most of the present blaVIM-2-positive isolates were P. aeruginosa, but nine were P. putida. The P. aeruginosa isolates were mostly from urines and sputa, indicating that these were important sources of the spread. PFGE of XbaI-digested genomic DNA showed that many of these organisms clustered to patterns C2 and E1. Serotyping showed that all members of the former group tested were serotype O:12, whereas those of the latter were either serotype O:11 or serotype OPA. These data imply a major role for clonal spread. It is interesting that VIM-producing P. aeruginosa isolates reported in Greece (25) were also serotype O:12, which is otherwise a rare type. The occurrence of blaVIM-2 in different PFGE and serotypes, as well as in two different Pseudomonas species in the present study, indicated horizontal spread, too. Only two of nine P. putida isolates shared an identical PFGE profile, suggesting a much greater role for horizontal spread in the latter species (data not shown). Moreover, an ongoing study has revealed the presence of blaVIM-2 in Acinetobacter and Serratia spp. from Korea.
In earlier studies, blaIMP has been found to be located on conjugative plasmids (26), but blaVIM-2 has not previously been found on self-transferable elements (17), while the blaVIM-1 gene was located on the chromosome in the sole producer studied (8). Initial attempts, using an E. coli recipient, to transfer blaVIM-2 from the present isolates were unsuccessful. Nevertheless, we were able to transfer the resistance from six P. aeruginosa and two P. putida isolates by using P. aeruginosa PAO4089Rp as a recipient. Repeated attempts to detect blaVIM on plasmids were unsuccessful with both the isolates and the transconjugants. Rather, fragments of genomic DNA separated by PFGE hybridized with a gene probe for blaVIM-2, suggesting a chromosomal location. We cannot, however, exclude possible carriage by a large plasmid that associated strongly with the chromosome. MICs of imipenem for the present isolates of P. aeruginosa ranged from 8 to >128 μg/ml, with an MIC50 of 16 μg/ml. Variations in the level of resistance may depend on the amount of VIM enzyme or on the permeability of the isolates. Not all VIM-2-producing isolates therefore would be categorized as imipenem resistant when using the NCCLS resistance breakpoint of ≥16 μg/ml. This situation recapitulates that seen with some extended-spectrum β-lactamase-producing isolates of E. coli and Klebsiella pneumoniae, which appear susceptible to extended-spectrum cephalosporins at NCCLS breakpoints (12).
Most of our VIM-2-producing P. aeruginosa isolates were also resistant to most aminoglycosides and ciprofloxacin, creating a serious problem for the choice of therapy. However, carbapenemase-producing P. aeruginosa isolates are susceptible to aztreonam because there is poor hydrolysis (18).
It was reported elsewhere that the pI of VIM-2 β-lactamase was 5.6 rather than 5.3 (17); in our experience, however, it was difficult to obtain sharp metallo-β-lactamase bands from cell sonicates as reported also for Stenotrophomonas maltophilia (15) and repeated analysis found that the pI of VIM-2 was indistinguishable from that of the VIM-1 control, which was run in parallel.
Our initial attempts directly to sequence the PCR products of integrons were unsuccessful due to the presence of another integron. Therefore, we used cloned strains. The nucleotide sequences of the blaVIM-2 alleles from our P. aeruginosa and P. putida strains were identical to each other and to that of the original VIM-2 producer, P. aeruginosa COL-1 (17). The structure of the integron in our P. aeruginosa strain was different from that in strain COL-1, which contained only the VIM-2 cassette (17), and was more similar to those in strains RON-1 and RON-2 (16), which also contained other cassettes besides blaVIM-2. However, the first cassette at the 5′ end of the integron from our P. aeruginosa strain was different from previously described aacA7 (strain RON-1) or aacA29a (RON-2) (16) in that it was a variant of aacA4, conferring resistance to gentamicin, tobramycin, and kanamycin, but not to amikacin. Also, downstream of the blaVIM-2, aadA1 was present rather than aacC1 and aacA4 (strain RON-1) or aacA29b (RON-2). Furthermore, unknown orf “i,” orf “ii,” and orf “iii” occupied a large space between blaVIM-2 and qacEΔ1. Strangely, repeated sequencing showed that the unknown orf “ii” lay in a reversed orientation, which would preclude its expression under the single promoter of the integron structure. The integron from the P. putida strain was different in that the first cassette was blaVIM-2 and that aacA7 was present along with aadA1 and qacEΔ1.
In conclusion, the metallo-β-lactamase produced by these P. aeruginosa isolates, collected in Korea since 1995, was VIM-2 and this enzyme and its contingent resistance have also reached P. putida. This is the first report of VIM-2 β-lactamase outside Europe and the first report for P. putida worldwide. The blaVIM-2 gene resided on two novel class 1 integrons. The persistence and spread of VIM-2-mediated resistance reflected clonal as well as horizontal spread. Stricter measures are required to control its further spread in Korea and elsewhere.
Acknowledgments
We are grateful to the Laboratory of Hospital Infection, Central Public Health Laboratory, for serotyping P. aeruginosa isolates, and to G. M. Rossolini, Universita di Siena, Siena, Italy, for a VIM-1-producing P. aeruginosa strain. We thank Young Hee Suh for technical assistance.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chong, Y., K. Lee, R. Okamoto, and M. Inoue. 1997. Characteristics of extended-spectrum β-lactam hydrolyzing activity of Klebsiella pneumoniae and Escherichia coli strains isolated from clinical specimens. Korean J. Infect. Dis. 29:477-485. [Google Scholar]
- 3. Chu, Y.-W., M. Afzal-Shah, E. T. S. Houang, M.-F. I. Palepou, D. J. Lyon, N. Woodford, and D. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornaglia, G., A. Mazzariol, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 5.Hirakata, Y., K. Izumikawa, T. Yamaguchi, H. Takemura, H. Tanaka, R. Yoshida, J. Matsuda, M. Nakano, K. Tomono, S. Maesaki, M. Kaku, Y. Yamada, S. Kamihara, and S. Kohno. 1998. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 42:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyobe, S., H. Kusadokoro, J. Ozaki, N. Matsumura, S. Minami, S. Haruta, T. Sawai, and K. O'Hara. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:2023-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiska, D. L., and P. H. Gilligan. 1999. Pseudomonas, p. 517-525. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 8.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Yong, and J. H. Yum. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88-91. [DOI] [PubMed] [Google Scholar]
- 10.Levesque, C., and P. H. Roy. 1993. PCR analysis of integrons, p. 590-594. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 11.Livermore, D. M., and J. D. Williams. 1996. β-Lactams: mode of action and mechanisms of bacterial resistance, p. 502-578. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 12.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed., M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed., M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne, D. J., R. Cramp, J. H. Bateson, J. Neale, and D. Knowles. 1994. Rapid identification of metallo- and serine β-lactamases. Antimicrob. Agents Chemother. 38:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., T. Lambert, S. Tuerkoglue, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen, B., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice, L. B., and R. A. Bonomo. 1996. Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents. p. 453-501. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 21.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning, a laboratory manual, 3rd ed., p. 1.32. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, a laboratory manual, 3rd ed., p. 15.14. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Takahashi, A., S. Yomoda, I. Kobayashi, T. Okubo, M. Tsunoda, and S. Iyobe. 2000. Detection of carbapenemase-producing Acinetobacter baumannii in a hospital. J. Clin. Microbiol. 38:526-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsakris, A., S. Pournaras, N. Woodford, M.-F. I. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]