Abstract
Isolates of Vibrio harveyi, a prawn pathogen, have demonstrated multiple antibiotic resistance to commonly used antimicrobial agents, such as oxytetracycline. In this paper, we describe the cloning and characterization of two tetracycline resistance determinants from V. harveyi strain M3.4L. The first resistance determinant, cloned as a 4,590-bp fragment, was identical to tetA and flanking sequences encoded on transposon Tn10 from Shigella flexneri. The second determinant, cloned as a 3,358-bp fragment in pATJ1, contains two open reading frames, designated tet35 and txr. tet35 encodes a 369-amino-acid protein that was predicted to have nine transmembrane regions. It is a novel protein which has no homology to any other drug resistance protein but has low levels of homology (28%) to Na+/H+ antiporters. Transposon mutagenesis showed that tet35 and txr were required for tetracycline resistance in a heterologous Escherichia coli host. Tetracycline accumulation studies indicate that E. coli carrying tet35 and txr can function as an energy-dependent tetracycline efflux pump but is less efficient than TetA.
Tetracyclines are broad-spectrum antibiotics which are effective not only against gram-positive and gram-negative bacteria but also against mycoplasmas, mycobacteria, and protozoan parasites (27). In bacteria, there are three known mechanisms of tetracycline resistance. The first is mediated by ribosomal protection proteins (TetM-TetS) (4). These are large (72.5-kDa) cytoplasmic proteins that have N-terminal homology to the elongation factors Tu and G (33). TetM and TetO catalyze the release of tetracycline from the ribosome in a GTP-dependent manner (7).
The tetX gene product from Bacteroides is the only example of enzymatic inactivation of tetracycline that has been described so far. The tetX gene has been found in two closely related Bacteroides transposons, and its enzyme, TetX, requires the presence of oxygen and NADPH for activity. Ironically, TetX is functional not in Bacteroides but in aerobically grown Escherichia coli cells (30, 31).
Tetracycline efflux genes, designated tetA-E, tetG, and tetH in gram-negative bacteria and tetK, tetL, tetP, and otrB in gram-positive bacteria, encode membrane-associated proteins which transport tetracycline out of cells (27). The efflux of tetracycline is an energy-dependent process that involves the exchange of a proton for a tetracycline-cation complex in an antiport fashion (37). Therefore, compounds that block the electrochemical gradient also inhibit the transport process (11). These Tet proteins are approximately 46 kDa and have either 12 (gram-negative) or 14 (gram-positive) transmembrane-spanning segments (TMS) (27). Tet efflux proteins belong to the major facilitator superfamily (MFS) of transport proteins.
Membrane topologies of tetracycline efflux proteins have common structural motifs with other drug exporters in the MFS, such as CmlA from Streptomyces lividans, which mediates chloramphenicol resistance (3), and MdfA, a multidrug transporter from E. coli (8). Other than the MF family, there are two families of proton-motive-force (PMF)-dependent drug transporters (25, 26). The small multidrug resistance (SMR) family is composed of proteins that are about 110 amino acids in length and have four TMS, such as the staphylococcal multidrug efflux protein Smr (9, 25). The third family of PMF-dependent drug efflux proteins is known as the resistance/nodulation/cell division (RND) family. These proteins are normally around 1,000 amino acid residues in size and have 12 TMS. One such member is the multidrug exporter ArcB from E. coli (17).
Luminous vibrios like Vibrio harveyi are the main causative agent of luminous vibriosis in farm-reared penaeid shrimp (28). The bacterial infection often results in mass mortality of the affected shrimp and leads to extensive commercial losses (13). Consequently, antibiotics like streptomycin, erythromycin, and chloramphenicol are widely used to treat the infections, whereas oxytetracycline is commonly used as a prophylactic agent. Vibrios isolated from shrimp hatcheries in Java island (Indonesia) have demonstrated multiple antibiotic resistance to antimicrobials like ampicillin, tetracycline, amoxicillin, and streptomycin (35). A recent paper described two novel β-lactamases isolated from V. harveyi that originated from waters around Java (34).
The genetic basis of tetracycline resistance in V. harveyi has not been studied. In this paper, we describe the cloning and characterization of two tetracycline determinants from V. harveyi M3.4L. One of these determinants was found to be identical to tetA, which is present on transposon Tn10 in Shigella flexneri. TetA mediates active efflux of tetracycline from host cells in an antiport fashion (19). The other determinant encodes a putative novel membrane protein that is postulated to mediate tetracycline resistance by also functioning as an efflux pump.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are detailed in Table 1. The V. harveyi strain used in this study was isolated from an Indonesian prawn farm (32, 34). Recombinant plasmids were constructed with the cloning vector pUC18 (39). E. coli TOP10 (Invitrogen Corp., Carlsbad, Calif.) was used as the host strain for the recombinant plasmids. Strains were grown in Luria-Bertani (LB) medium or on LB agar supplemented with 10 μg of tetracycline per ml where appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli TOP 10 | F−mcrA(mrr-hsdRMS-mcrBC)φ80 lac2ΔM15Δlac74 recA1 deoR araD139Δ(ara-leu)7697 galU galK rpsL endA1 nupG u) Smr | Invitrogen |
| V. harveyi | ||
| M3.4L | Tcr Ampr | 32 |
| W3B | Tcs Ampr Kanr | |
| Plasmids | ||
| pUC18 | AmprlacZα | 39 |
| pATJ1 | pUC18 with 3,358-base HindIII Tcr genomic fragment of M3.4L | This study |
| pCT71 | pUC18 with 4,890-base HindIII Tcr genomic fragment of M3.4L | This study |
| pJKM10 | pATJ1 carrying a transposon disruption in tet35 | This study |
| pJKM115 | pATJ1 carrying a transposon disruption in txr | This study |
| pCKM12 | pCT71 carrying a transposon disruption in tetA | This study |
Smr, streptomycin resistance; Tcr, tetracycline resistance; Tcs sensitivity; Ampr, ampicillin resistance; Kmr, kanamycin resistance.
Antimicrobial agents and susceptibility testing.
MICs were determined by an agar dilution technique on Mueller-Hinton agar plates (Oxoid Ltd., Basingstoke, England) with an inoculum of 104 CFU/spot. All the plates were read after an 18-h incubation at 37°C for E. coli and at 30°C for V. harveyi. The antibiotics used were tetracycline, oxytetracycline, minocycline, chloramphenicol, nalidixic acid, rifampin, trimethoprim, spectinomycin, kanamycin, and gentamicin (Sigma Chemical Co., St. Louis, Mo.). Testing of susceptibility to ciprofloxacin was performed by the disk diffusion method with 1-μg ciprofloxacin disks (Oxoid Ltd). All antimicrobial susceptibility tests were set up and the results were interpreted according to the National Committee for Clinical Laboratory Standards (20). Antimicrobial cationic dyes used were ethidium bromide and crystal violet (Sigma).
Enzymes and chemicals.
Chemicals used were of the highest grade commercially available. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs, Inc. (Beverly, Mass.) and used according to the manufacturer's recommendations.
DNA cloning and sequencing of recombinant plasmids.
Genomic DNA from V. harveyi M3.4L was extracted with phenol-chloroform (29). Genomic DNA was digested with HindIII, and the resulting fragments were ligated into the HindIII site of the pUC18 vector. The ligation mixture was transformed into E. coli TOP10 cells, and transformants were selected for tetracycline resistance. Plasmids were prepared with a Wizard Plus Miniprep DNA purification system (Promega, Madison, Wis.). Sequencing of the DNA insert was performed with the ABI Prism Big Dye terminator cycle sequencing ready reaction kit and ABI cycle sequencer A377 (Applied Biosystems/Perkin-Elmer, Foster City, Calif.).
DNA and protein sequence analysis.
DNA sequence analysis was performed with DNASIS (Hitachi Software Engineering Co. Ltd., San Bruno, Calif.). Database similarity searches for both the nucleotide and deduced protein sequences were carried out with BLAST at the National Center of Biotechnology Information website (http://www.ncbi.nlm.nih.gov./BLAST). Multiple sequence alignments of protein sequences were made with CLUSTALW (http://www.ebi.ac.uk/clustalw). The predictions of transmembrane regions were carried out with PRED-TMR2 (24) at http://www.o2.biol.uoa.gr/PRED-TMR2.
Detection of Tn10 by PCR analysis.
In order to determine if V. harveyi M3.4L harbors a complete Tn10, PCR was carried out with primers that were designed based on the complete nucleotide sequence of Tn10 (GenBank accession number AF162223). PCR primers T1 (5′-CTGATGAATCCCCTAATGAT-3′) and T2 (5′-AACACTTGGATTAGTGTTGG-3′) were designed based on the IS10 left element and the jemA gene, whereas T3 (5′-AGCCCGCGGTAAATAGCAAT-3′) and T4 (5′-GATGAATCCCCTAATGATTT-3′) were based on the ORF-L gene and the IS10 right sequence (Fig. 1). These primers were designed to amplify regions that were not cloned in pCT71. Each PCR mixture consisted of 2 μg of V. harveyi M3.4L genomic DNA per ml in 1× PCR buffer, 0.2 mM deoxynucleoside triphosphates, 50 pmol of each primer, and 1.5 U of DyNazyme EXT DNA polymerase (Finnzymes OY, Espoo, Finland) in a 100-μl volume. PCR amplification consisted of an initial incubation of 2 min at 95°C, followed by 25 cycles of 1 min of denaturation at 94°C, 1 min of primer annealing at 50°C, and 4 min of extension at 72°C. The cycles were terminated with a final 10-min extension at 72°C. Reaction products were analyzed on a 1% agarose gel in Tris-borate-EDTA buffer.
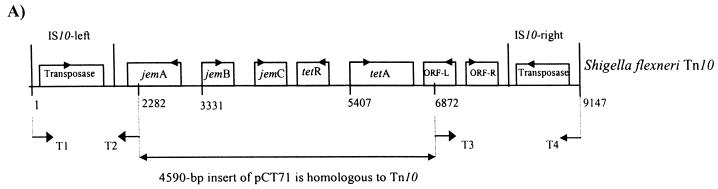
FIG. 1.
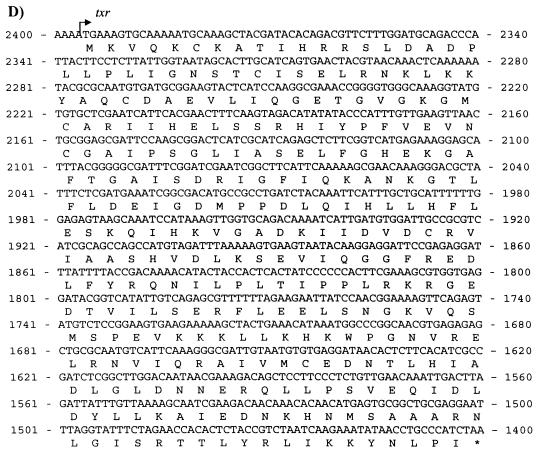
(A) Genetic map of Tn10. The pCT71 insert has complete homology to regions extending from jemA to ORF-L of Tn10. Primers T1-T2 and T3-T4 were used for PCR amplification. The maps are not drawn to scale. (B) Gene organization of pATJ1. The nucleic acid sequence of the insert was used to construct the ORF map. Transcriptional direction is shown by small arrows on the ORF box. The vertical arrows indicate the sites on tet35 and txr, respectively, of EZ::TN transposon transposition. (C) The putative amino acid sequence of Tet35 is given below its nucleic acid sequence. The protein is predicted to have nine TMS regions (bold). (D) Putative amino acid sequence of Txr and its nucleic acid sequence.
Bacterial conjugations.
Mating experiments were carried out to investigate the potential transfer of tetracycline resistance from V. harveyi M3.4L to various tetracycline-sensitive recipients. Plate matings were carried out overnight at 32°C with a donor-to-recipient ratio of 1:1. Cells were scraped off the plate the following day and resuspended in 1 ml of saline. Transconjugants were selected on media supplemented with the appropriate antibiotics. When V. harveyi W3B (Tcs Kmr) was used as the recipient, transconjugants were selected on LB agar plates with 10 μg of tetracycline and 25 μg of kanamycin per ml. In mating experiments with E. coli TOP10 cells (Smr), transconjugants were selected on LB agar plates supplemented with 10 μg of tetracycline and 100 μg of streptomycin per ml, and when Pseudomonas alcaligenes NCIB 9867 (strain P25X) was used as the recipient, transconjugants were selected on minimal medium plates supplemented with 10 μg of tetracycline per ml. Transfer frequency was expressed as the number of transconjugants per recipient cell obtained after plating on selective medium.
Southern blotting and hybridization for PFGE.
DNA fragments of pulsed-field gel electrophoresis (PFGE) gels were run as previously described (34) and transferred onto nylon membranes (Hybond-N+; Amersham, Little Chalfont, Buckinghamshire, England) by capillary transfer and fixed by baking. The hybridization probes used were the 4.6-kb HindIII-HindIII fragment from pCT71 and the tet35 portion of pATJ1 amplified by PCR. Probe labeling was carried out with the ECL nonradioactive detection kit (Amersham Life Science). Hybridization was performed overnight under high-stringency conditions as described by the manufacturer.
Tetracycline uptake and efflux studies in intact cells.
Strains harboring the cloned tetracycline resistance determinants (recombinant plasmids pATJ1 and pCT71) were grown in LB broth supplemented with 10 μg of tetracycline per ml until an A600 of 0.7 was reached. The cells were harvested by centrifugation and washed once in 50 mM potassium phosphate buffer (pH 7.0), 1 mM MgSO4, and 0.25% glucose and resuspended in the same buffer to an optical density at 600 nm of 2 (approximately 2 mg of protein per ml). The cell suspension was preincubated at 37°C in a shaking water bath, and the assay was started with the addition of [3H]tetracycline (1.2 Ci/mmol; New England Nuclear, Boston, Mass.) to a final concentration of 5 μM. At 5-min intervals, 50 μl of the cell sample was removed and placed on top of a mixture of separating oils (melting point bath oil and mineral oil overlaid on 37% saturated sodium chloride solution) in an Eppendorf tube and centrifuged at room temperature for 10 min. The resulting cell pellet was solubilized in 3 ml of Ready Safe liquid scintillation cocktail (Beckman, Fullerton, Calif.) and the radioactivity of [3H]tetracycline accumulated in cells was determined by liquid counting. An energy inhibitor, 0.2 mM carbonyl cyanide m-chlorophenylhydrazone (CCCP), was added after 15 min of incubation with [3H]tetracycline to analyze the energy dependence of the accumulation process.
Transposon mutagenesis.
In vitro transposon mutagenesis was carried out with the EZ::TN <KAN-2> insertion system (Epicentre Technologies, Madison, Wis.), which uses the EZ::TN <KAN-2> transposon carrying a kanamycin resistance marker. The kit was used according to the manufacturer's instructions. Plasmids pATJ1 and pCT71 were used as the target DNA for mutagenesis, and insertion clones were selected on kanamycin plates. The clones were screened by restriction endonuclease digestions with insertion sites accurately mapped by sequencing outwards from both ends of the transposon.
RT-PCR analysis.
Total RNA was prepared from E. coli TOP 10 strains carrying pATJ1 and pJKM115 as previously described (36). The isolated RNA was treated with RNase-free DNase I (Roche, Mannheim, Germany) to remove contaminating DNA. The following primer pairs were used: for amplification of the tet35 gene, tet35F (5′-AGCTAACTACGCGTTCTGGC-3′) and tet35R (5′-GCTGCACCAAT GTGTGTGATC-3′), and for amplification of the txr gene, txrF and txrR (5′-GTAGGCTTGTTAGATGGGCA-3′ and 5′-AATGAAAGTGCAAAAATGCAAAGC-3′). First-strand cDNA was reverse transcribed from 1 μg of total RNA with Expand reverse transcriptase (RT) (Roche) and 20 pmol of forward primers. The enzyme was used according to the manufacturer's instructions. The cDNA was then used for RT-PCR with the addition of 20 pmol each of the appropriate forward and reverse primers. The PCRs were run for 35 cycles of 1 min at 95°C for denaturing, 1 min at 50°C for annealing, and 1 min at 72°C for extension. Samples without RT were also run to ensure that there was no contaminating DNA, and these were used as negative controls. The amplification of 16S rRNA with 16S rRNA gene-specific primers served as a positive control.
Nucleotide sequence accession number.
The nucleotide sequences of tet35 and txr have been deposited in GenBank under accession number AF353562.
RESULTS
Cloning and sequence analysis of the DNA insert in pCT71.
The first tetracycline resistance determinant was cloned in the recombinant plasmid pCT71. Sequencing through the 4,591-bp HindIII fragment of pCT7l revealed that it was identical to nucleotides 2282 to 6872 of Tn10 of Shigella flexneri (GenBank accession number AF162223) (Fig. 1). This region encompasses parts of jemA and jemC and the entire sequences of jemB, tetR, and tetA (5). JemA is predicted to be a sodium-dependent glutamate permease, JemB is a protein of unknown function, and JemC showed sequence homology to bacterial transcriptional regulators that repress arsenic and mercury operons (5). tetA encodes a protein (TetA) known to be responsible for the active efflux of tetracycline from host cells (10, 12), and tetR functions as the repressor of tetA expression (11).
Cloning and sequence analysis of the pATJ1 insert.
The DNA insert present in plasmid pATJ1 carrying the second tetracycline resistance determinant was sequenced. Analysis of the 3,358-bp HindIII genomic fragment of pATJ1 revealed the presence of two open reading frames (ORFs) (Fig. 1) that are convergently transcribed from opposite directions and designated tet35 and txr. tet35 encodes a protein of 369 amino acid residues, designated Tet35, according to the nomenclature for new tetracycline resistant determinants (16). Tet35 has an estimated molecular mass of 39,121 Da. This protein was predicted to have nine α-helical transmembrane-spanning regions (Fig. 1). The protein was found to have significant homology to a number of hypothetical integral membrane proteins, such as HI 1586 of Haemophilus influenzae (49% identity) (GenBank accession number U32832), and, to a lesser extent, to Na+/H+ antiporters of Bacillus firmus (28% identity) (GenBank accession number U61539) and Neisseria meningitidis (25%) (GenBank accession number AE0024090). Tet35 did not share sequence similarities with any other drug transporters in the database or with members of the MF, RND, or SMR family (1, 25). Highly conserved regions or motifs present in these drug efflux families could not be identified in Tet35.
The 957-bp txr gene encoded a putative protein of 318 amino acids, designated Txr, which shared sequence similarities with various transcriptional regulatory proteins. The highest degree of homology (45%) was with the Pseudomonas aeruginosa PA1945 transcriptional regulator (GenBank accession number AE004621). Thirty-five percent homology was found with σ54-dependent transcription factor VC2137 of Vibrio cholerae (GenBank accession number AE004286). These similarity searches suggest that Txr may function as a putative transcription regulator.
Antibiotic susceptibility.
E. coli TOP10 cells harboring recombinant plasmids pATJ1 and pCT71 had increased levels of resistance to tetracycline, oxytetracycline, and the tetracycline derivative minocycline (Table 2), indicating that the insert fragments present on pATJ1 and pCT71 were responsible for tetracycline resistance. It was observed that pATJ1 could not confer resistance to other classes of antibiotics (data not shown). Antimicrobials such as kanamycin, spectinomycin, gentamicin (aminoglycoside), nalidixic acid (quinolone), trimethoprim (diaminopyrimidine), rifampin (rifamycin), ciprofloxacin (fluoroquinolone), chloramphenicol, and ethidium bromide and crystal violet (cationic dyes) were tested. However, the level of resistance of cells containing pATJ1 to all antibiotics tested remained the same as that of susceptible cells. Therefore, pATJ1 has only a narrow drug specificity to tetracyclines and does not produce a multidrug resistance phenotype.
TABLE 2.
MICs for V. harveyi M3.4L and E. coli TOP10 cells carrying pATJ1, pCT71, and transposon insertion plasmids
| Antibiotic | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|
| V. harveyi M3.4L |
E. coli TOP10 carrying:
|
||||||
| No plasmid | pATJ1 | pCT71 | pJKM10 | pJKM115 | pCKM12 | ||
| Tetracycline | 32 | 4 | 256 | 512 | 4 | 4 | 4 |
| Oxytetracycline | 128 | 4 | 512 | 256 | 4 | 4 | 4 |
| Minocycline | 16 | 0.5 | 32 | 32 | 4 | 4 | 4 |
Detection of Tn10 by PCR analysis.
PCR primers based on the IS10 left and right elements of Tn10 as well as the internal jemA and ORF-L sequences were used for amplification, to determine if strain M3.4L harbors the complete Tn10 in its genome. Primer pairs T1-T2 and T3-T4 were designed so that they would amplify away from each end of the pCT71 insert to the ends of the insertion element. If Tn10 is present in the genome, then T1-T2 and T3-T4 would be able to amplify a band of approximately 2.2 kb. However, no band was detected when the reaction mixture from the PCR was analyzed, indicating that strain M3.4L either possessed an incomplete copy of Tn10, i.e., without the IS10 ends, or had flanking ends that were dissimilar to the IS10.
Bacterial conjugation and analysis of transconjugants.
In mating studies using E. coli TOP10 cells or P. alcaligenes NCIB 9867 (strain P25X) as recipients, no transconjugants were obtained. When V. harveyi W3B as used as the recipient, tetracycline-resistant transconjugants were obtained at a frequency of <10−7. Five transconjugants were picked at random, and PFGE of genomic DNA confirmed that the transconjugants were indeed V. harveyi W3B (data not shown). Analysis of these transconjugants also showed that tetA had transferred into the transconjugants, because a hybridizing signal was obtained when the transconjugants were probed with tetA (data not shown).
Distribution of tetracycline resistance determinants in different isolates of V. harveyi.
Six other environmental isolates of V. harveyi were probed with either tetA from clone pCT71 or tet35. Only a large (500-kb) band in strain M3.4L hybridized to the tetA probe. However, with tet35, a hybridizing band of approximately 180 kb was observed not only in strain M3.4L but in the other strains as well (data not shown).
Transposon mutagenesis of the ORFs in pATJ1.
In vitro transposon mutagenesis was used to determine which of the two ORFs, tet35 and txr, was needed for tetracycline resistance. Clone pJKM10 had a single transposon inserted in tet35 (nucleotide position 829), whereas pJKM115 was disrupted by a transposon at nucleotide position 1488 within txr (Fig. 1). As a result of the insertions in either of the ORFs, both clones were rendered susceptible to tetracycline (Table 2), indicating that the two ORFs were both likely to be important in mediating tetracycline resistance. When each of the ORFs was cloned individually by PCR amplification and after ligation to pGEM-T Easy vector (Promega), neither of the recombinant plasmids could confer tetracycline resistance, suggesting that both ORFs may be required for resistance.
Transposon mutagenesis of pCT71.
In vitro transposon mutagenesis using the EZ::TN <KAN-2> transposon was also used to generate a mutant plasmid designated pCKM12. This clone had an insertion at nucleotide position 3287 of pCT71, resulting in the disruption of the tetA ORF. The MIC of tetracycline for the clone dropped to 4 μg/ml (Table 2), indicating that TetA was indeed responsible for tetracycline resistance in plasmid pCT71.
Transcriptional analysis of tet35.
Homology searches suggest that the second ORF, txr, may be a putative transcriptional regulator; hence, RT-PCR was used to determine whether txr could regulate the transcription of tet35. RT-PCR of total RNA isolated from TOP10 cells carrying pATJ1 shows that the tet35F/R and txrF/R primer pairs correctly amplify tet35 and txr, producing bands of the predicted size (Fig. 2). RT-PCR analysis of total RNA from TOP10 cells carrying pJKM115 (transposon disruption in the txr ORF) shows that txrF/R did not amplify a detectable band corresponding to the disrupted gene (Fig. 2). However, tet35F/R was still able to amplify a tet35 band, indicating that the tet35 mRNA was still generated. These results show that txr probably does not act as a transcriptional activator of tet35, as its transcripts could still be detected even after disruption of txr.
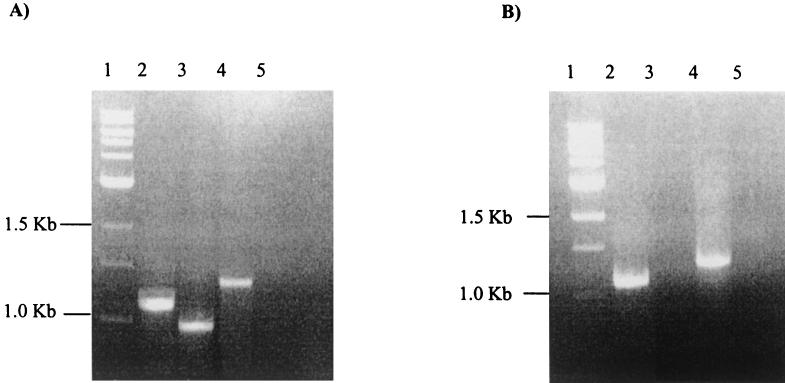
FIG. 2.
RT-PCR analysis of tet35 expression. (A) RT-PCR of total RNA from E. coli TOP10 cells carrying pATJ1. Lanes: 1, 1-kb ladder; 2, 1.1-kb amplicon obtained with tet35F/R primers; 3, 900-base amplicon obtained with txrF/R primers; 4, positive control using 16S rRNA-specific primers, amplifying a 1.3-kb amplicon; 5, negative control (no RT). (B) RT-PCR of total DNA from E. coli TOP10 cells carrying pJKM115. Lanes 1, 1-kb ladder; 2, 1.1-kb PCR amplicon obtained with tet35F/R primers; 3, no detectable amplicon obtained with txrF/R primers; 4; positive control using 16S rRNA specific primers, amplifying a 1.3-kb amplicon; 5, negative control (no RT).
Accumulation of [3H]tetracycline by cells with and without pATJ1.
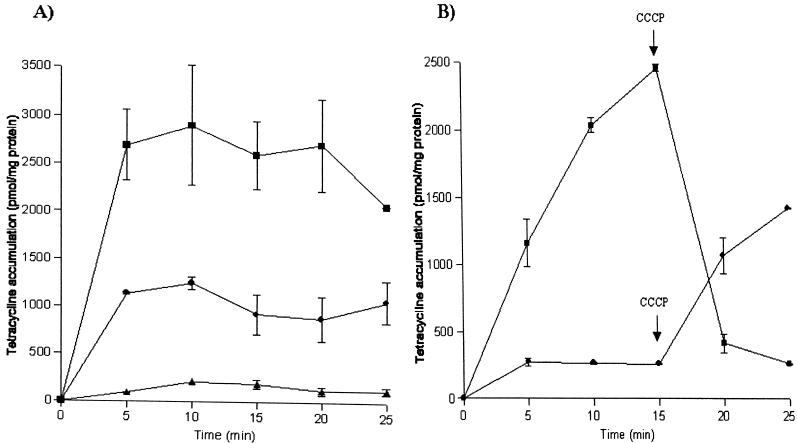
Susceptible E. coli TOP10 cells accumulated more tetracycline than cells carrying pATJ1 or pCT71 (Fig. 3). E. coli cells with pCT71 carrying tetA from Tn10 should express the tetracycline efflux protein TetA and were expected to have low levels of tetracycline uptake. Results showed that these cells did have the lowest levels of accumulation, about 5-fold lower than that of cells carrying pATJ1 and 12-fold lower than that of E. coli TOP10 cells. It also appears that tetracycline efflux in cells harboring pATJ1 was not as efficient as in cells carrying pCT71. Cells containing pATJ1 did not achieve very low levels of steady-state accumulation compared to cells with pCT71.
FIG. 3.
Accumulation of tetracycline by susceptible E. coli TOP10 cells and resistant clones. [3H]tetracycline was added to cells at time zero. (A) Accumulation in E. coli carrying no plasmid (▪), pATJ1 (•), or pCT71 (▴); (B) effect of CCCP (0.2 mM) on accumulation in E. coli with no plasmid (▪) or pATJ1 (•). The graphs are typical of at least two independent experiments.
Active efflux systems are driven by the energy provided by an electrochemical proton gradient (15). An energy inhibitor like CCCP should be able to block the transport process. When CCCP was added to E. coli TOP10 cells, a drastic decrease in tetracycline uptake was observed, indicating that in susceptible cells, the process of drug accumulation is one of active uptake (19) and not a consequence of increased membrane permeability which could allow the drug to enter cells. The addition of CCCP to cells with pATJ1 caused an increase in uptake, with levels of accumulation similar to that in E. coli TOP10 cells. These data point to a PMF driving an energy-dependent drug export process in E. coli cells carrying pATJ1. Results of the accumulation studies show that the DNA insert present in pATJ1 is capable of encoding the tetracycline efflux function, and sequence analysis of pATJ1 shows that the tet35 ORF is most likely to mediate the active efflux.
DISCUSSION
In this study, we report the isolation of two tetracycline resistance determinants from V. harveyi M3.4L which enabled susceptible E. coli cells to grow in the presence of tetracycline. A recombinant E. coli clone carrying pCT71 was found to possess tetA and tetR gene sequences identical to Tn10 of Shigella flexneri. It is well known that tetA and tetR in Tn10 confer high levels of inducible tetracycline resistance (2). TetA functions as a metal-tetracycline antiporter, while tetR encodes a repressor protein that negatively regulates transcription of both the tetA and tetR genes (15, 38). Therefore, the tetA sequence found in V. harveyi M3.4L probably functions in a similar manner and is responsible for the high level of tetracycline resistance. This is supported by the observation that high levels of tetracycline efflux are achieved in E. coli hosts carrying pCT71 and tetracycline resistance is lost when the tetA ORF is disrupted by transposon mutagenesis. The inability to obtain flanking regions of the pCT71 insert by PCR amplification suggests that V. harveyi M3.4L may not harbor the entire Tn10. This is interesting, as Tn10 has been reported to retain its physical integrity through multiple cycles of transposition (2). Analysis of the genetic sequences surrounding tetA may reveal the kind of transposition events that have taken place and how it may have been acquired by V. harveyi.
The tetracycline resistance determinant of Tn10, as identified by DNA hybridization, is found widely in enteric bacteria like E. coli, Klebsiella, Proteus, Pseudomonas, Vibrio spp., and Salmonella (2, 18). Tn10 is usually a plasmid-associated element, and it is also possible that V. harveyi M3.4L acquired the transposon by conjugative transfer of resistance plasmids. In strain M3.4L, a single 500-kb band hybridized to the insert fragment present in pCT71, but we have yet to determine if the fragment is derived from the chromosome or is part of a large, hitherto-undetected plasmid. Conjugation studies have shown that the transmission range of tetA is narrow and will transfer to a V. harveyi recipient but not to other gram-negative bacteria. It is possible that tetA in M3.4L is present on a narrow-host-range plasmid, thus limiting its transfer to V. harveyi strains.
The second resistance determinant is encoded by the insert fragment carried in pATJ1, which has two ORFs, tet35 and txr. Transposon mutations generated in each ORF suggest that both ORFs are important for tetracycline resistance in E. coli, as mutants with disruptions in either ORF were unable to grow on media containing tetracycline. Sequence homology suggests that Tet35 is an integral membrane protein with nine TMS and has low homology to Na+/H+ antiporters. Homology searches suggest that the second ORF, txr, may encode a putative transcriptional regulator. Hence, transcriptional analysis by RT-PCR was performed to determine if txr could influence the expression of tet35. Results of RT-PCR from E. coli cells carrying pJKM1159 (disrupted txr) show that txr does not act as a transcriptional activator of tet35, because even when txr is mutated, tet35 transcripts could still be produced. This led us to propose that while txr does not act as a transcriptional regulator of tet35, Txr may be required to interact with Tet35 to enable the proper functioning of Tet35. For example, in the doxorubicin efflux pump of Streptomyces peucetius, DrrB, the peripheral membrane protein requires another protein, DrrA, in order to be stably maintained, and biochemical coupling between the two proteins has been demonstrated (14).
Uptake studies with [3H]tetracycline indicate that the determinant carried on pATJ1 could mediate the active efflux of tetracycline from cells through an energy-dependent process that was likely to be driven by the PMF. This is substantiated by the fact that cells with pATJ1 accumulated tetracycline to a lower extent than susceptible E. coli cells this and drug export was inhibited when the proton uncoupler CCCP was added to the assay. However, Tet35 may not function as efficiently as TetA, because cells carrying pCT71 tend to have lower levels of tetracycline accumulation than cells with pATJ1. Also, MIC data show that E. coli cells carrying pCT71 had a twofold-greater resistance to tetracycline than cells with pATJ1.
The majority of bacterial or drug-specific pumps use PMF as the driving force for efflux and belong to the MFS, SMR, or RND family of transporters (25). Prototype tetracycline efflux proteins have 12 TMS, although many of its MF family members have 14. Multidrug efflux transporters of the SMR family are small (100-amino-acid) proteins that span the cell membrane four times (23), but the RND family are large (1,000-amino-acid) proteins which have 12 TMS. No sequence similarity to members of the MFS, SMR, or RND family was found for either Tet35 or Txr. Neither signature sequences or consensus motifs typical of any class (26) were detected on the putative nine-TMS Tet35. MFS and SMR transporters tend to have a narrower substrate range than RND transporters. For example, the Bacillus subtilis transporter Bmr transports only organic cations and fluoroquinones (22). On the other hand, RND transporters pump out a wide range of substrates, including almost all lipophilic and amphiphilic antibiotics, dyes, detergents, solvents, and chemotherapeutic agents (23). When the drug substrate range of pATJ1 was tested, the plasmid was found to encode resistance to tetracyclines but not to structurally different antimicrobial agents, such as cationic dyes, aminoglycosides, quinolones, and chloramphenicol. Hence, pATJ1 does not carry multidrug resistance determinants, and Tet35 appears to be a novel efflux pump that lacks homology to any other antibiotic efflux pumps.
Hybridization studies using the tetA probe showed the presence of at least two copies in the genome of V. harveyi M3.4L (data not shown), while tet35 exists as a single chromosomal copy in the V. harveyi genome, and expression levels may not be sufficiently high to produce the tetracycline resistance phenotype. The tetracycline resistance observed in E. coli may be due to the high copy number of the pUC18 cloning vector, which enables tet35 to be present in multiple copies in E. coli cells. It is likely that the high levels of tetracycline resistance in strain M3.4L can be attributed to the presence of tetA rather than tet35. Screening of six other V. harveyi strains by Southern blot hybridization with tet35 also showed that it is present as a chromosomal gene. However, the MIC of tetracycline for these strains was considerably lower than that for strain M3.4L, which carries both tet35 and tetA.
It has been suggested that multidrug transporters may have evolved to protect bacteria from diverse environmental toxins or to transport physiological compounds and that the ability to expel drugs is only a fortuitous side effect (21, 26). Convincing evidence for this comes from Tet(A)L of B. subtilis, a multifunctional antiporter that catalyzes metal-tetracycline/H+ antiport as well as Na+/H+ and K+/H+ antiport. It plays a physiological role in Na+ resistance and pH homeostasis in addition to conferring tetracycline resistance on its host (6). A similar role may be suggested for Tet35: its primary function may not be to transport tetracycline, but rather, tetracycline may be an incidental substrate. This is reflected in tetracycline accumulation studies which show that the protein encoded by pATJ1 is less effective than that encoded by pCT71 in pumping out tetracycline. The low sequence homology of Tet35 to the Na+/H+ antiporter suggests that the primary role of Tet35 is a physiological one involving Na+/H+ antiport.
Acknowledgments
This work was supported by National University of Singapore Academic Research grant RP182000039213 to C. L. Poh.
We thank Yeo Chew Chieng for critical reading of the manuscript and A. Suwanto for the gift of V. harveyi strain M3.4L.
REFERENCES
- 1.Bambeke, F. V., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 2.Berg, D. E., and M. M. Howe. 1989. Mobile DNA. American Society for Microbiology, Washington, D.C.
- 3.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdett, V. 1986. Streptococcal tetracycline resistance mediated at the level of protein biosynthesis. J. Bacteriol. 165:564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalmers, R., P. Crellin, S. Sewitz, and K. Liplow. 2000. Complete nucleotide sequence of transposon Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J. B., A. A. Guffanti, W. Wang, T. A. Krulwich, and D. H. Bechhofer. 1996. Chromosomal tetA(L) gene of Bacillus subtilis: regulation of expression and physiology of a tetA(L) deletion strain. J. Bacteriol. 178:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dantley, K. A., H. K. Dannely, and V. Burdett. 1998. Binding interaction between Tet(M) and the ribosome: requirements for binding. J. Bacteriol. 180:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar, R., and E. Bibi. 1997. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179:2274-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grinius, L., G. Dreguniene, E. B. Goldberg, C. H. Liao, and S. J. Projan. 1992. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid 27:119-129. [DOI] [PubMed] [Google Scholar]
- 10.Hillen, W., and K. Schollmeier. 1983. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 11:525-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillen, W., C. Gatz, L. Altschmied, K. Schollermeier, and I. Meier. 1983. Control of expression of the Tn10-encoded tetracycline resistance genes. I. Equilibrium and kinetic investigation of the regulatory reactions. J. Mol. Biol. 169:707-721. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen, R. A., and W. S. Reznikoff. 1979. Organization of structural and regulatory genes that mediate tetracycline resistance in transposon Tn10. J. Bacteriol. 138:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karunasagar, I. R., R. Pai, and G. R. Malathi. 1994. Mass mortality of Penaeus monodon larvae due to antibiotic-resistant Vibrio harveyi infection. Aquaculture 128:203-209. [Google Scholar]
- 14.Kaur, P., and J. Russell. 1998. Biochemical coupling between the DrrA and DrrB proteins of the doxorubicin efflux pump of Streptomyces peucetius. J. Biol. Chem. 273:17933-17939. [DOI] [PubMed] [Google Scholar]
- 15.Levy, S. B. 1992. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 36:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Heart. 1993. Molecular cloning and characterization of arcA and arcE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsutani, S. 1991. Multiple copies of IS10 in the Enterobacter cloacae MD36 chromosome. J. Bacteriol. 173:7802-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurry, L., R. E. Petrucci, Jr., and S. B. Levy. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:3974-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 1997. Performance standards for the antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6. National Committee for the Clinical Laboratory Standards, Wayne, Pa.
- 21.Neyfakh, A. A. 1997. Natural functions of bacterial multidrug transporters. Trends Microbiol. 8:309-313. [DOI] [PubMed] [Google Scholar]
- 22.Neyfakh, A. A., V. E. Bidnenko, and L. B. Chen. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA 88:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 24.Pasquier, C., and S. J. Hamodrakas. 1999. An hierarchical artificial neural network system for the classification of transmembrane proteins. Protein Eng. 12:631-634. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 28.Ruangpan, L. 1998. Luminous bacteria associated with shrimp mortality, p. 206-211. In T. W. Flegel (ed.), Advances in shrimp biotechnology. National Center for Genetic Engineering and Biotechnology, Bangkok, Thailand.
- 29.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Speer, B. S., and A. A. Salyers. 1989. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J. Bacteriol. 171:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speer, B. S., and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suwanto, A., M. Yuhana, E. Herawaty, and S. L. Angka. 1998. Genetic diversity of luminous Vibrio isolated from shrimp larvae, p. 217-224. In T. W. Flegel (ed.), Advances in shrimp biotechnology. National Center for Genetic Engineering and Biotechnology, Bangkok, Thailand.
- 33.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teo, J. W., A. Suwanto, and C. L. Poh. 2000. Novel beta-lactamase genes from two environmental isolates of Vibrio harveyi. Antimicrob. Agents Chemother. 44:1309-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjahjadi, M. R., S. L. Angka, and A. Suwanto. 1994. Isolation and evaluation of marine bacteria for biocontrol of luminous bacterial disease in tiger shrimp larvae (Penaeus monodon, Fab.). Asia Pac. J. Mol. Biol. Biotechnol. 2:347-352. [Google Scholar]
- 36.Xiong, X. F., N. de la Cruz, and W. S. Reznikoff. 1991. Downstream deletion analysis of the lac promoter. J. Bacteriol. 173:4570-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi, A., T. Udagawa, and T. Sawai. 1990. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J. Biol. Chem. 265:4809-4813. [PubMed] [Google Scholar]
- 38.Yamaguchi, A., Y. Iwasaki-Ohba, N. Ono, M. Kaneko-Ohdera, and T. Sawai. 1991. Stoichiometry of metal-tetracycline/H+ antiport mediated by transposon Tn10-encoded tetracycline resistance protein in E. coli. FEBS Lett. 282:415-418. [DOI] [PubMed] [Google Scholar]
- 39.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-109. [DOI] [PubMed] [Google Scholar]