Abstract
Ectopic expression of ebp1, a member of the PA2G4 family, inhibits the proliferation and induces the differentiation of human breast and prostate cancer cell lines. Ebp1 inhibits transcription of E2F1 and androgen receptor regulated genes such as prostate specific antigen (PSA) through its interactions with histone deacetylases (HDACs). To further understand Ebp1's interactions with other components of the transcriptional repression machinery, we examined the association of Ebp1 with the corepressor Sin3A. Ebp1 interacted with Sin3A both in vitro and in vivo as demonstrated by glutathione S-transferase (GST) pull-down and coimmunoprecipitation analysis. The C-terminal domain of Ebp1, responsible for its ability to repress transcription and arrest cell growth, was necessary and sufficient for binding Sin3A. The C-terminal domain of Sin3A, containing the paired amphipathic domain 4 and the HDAC interacting domain, bound Ebp1. Recombinant Sin3A bound Ebp1 directly, but recombinant HDAC2 failed to bind Ebp1. Chromatin immunoprecipitation (ChIP) and DNA affinity precipitation analysis demonstrated that Ebp1 and Sin3A associate at the PSA and E2F1 promoters. Functionally, Sin3A enhanced the ability of Ebp1 to repress transcription of androgen receptor (AR) and E2F1 regulated genes. These results demonstrate that Ebp1 participates in transcriptional regulation via its interaction with the Sin3–HDAC.
INTRODUCTION
Ebp1, a member of the PA2G4 family of proliferation regulated proteins, was isolated as an ErbB3 binding protein in our laboratory (1). The ectopic expression of Ebp1 inhibits the growth of human breast and prostate cancer cells and induces cellular differentiation (2). Ebp1 represses transcription of both E2F1 (3) and androgen receptor (AR) (4,5) regulated genes. Ebp1 binds to the E2F1 promoter in a complex with the E2F1 transcription factor and Rb and Ebp1 activity on the E2F1 promoter is regulated by the ErbB3 ligand heregulin (6). Ebp1 contains an autonomous C-terminal transcriptional repression domain that binds histone deacetylase (HDAC) activity and HDAC2. The ability of Ebp1 to repress transcription is partially reversed by HDAC inhibitors (3).
Histone acetylation has emerged as a major mechanism of the control of gene expression. Hyperacetylation of histones H3 and H4 is generally associated with transcriptionally active chromatin, while the chromatin of inactive regions is enriched in deacetylated histones. The acetylation status of histones at specific DNA regulatory sequences depends on the recruitment of histone acetyltransferases or histone deacetylase activities (7). HDACs are frequently recruited to specific DNA sites by association with corepressor molecules as part of multimolecular repressor complexes. HDAC1 and 2 exist in at least three multiprotein complexes: the Sin3, the NuRD/NRD/M12 and the CoRest complexes (8). The Sin3 complex contains the Sin3A and Sin3B corepressors, class I HDAC1/2 and additional associated proteins such as RbAp46 and 48, SAP18 and SAP30 (9,10). The abundance and relative stability of both Sin3 and HDAC proteins have led to the proposal that the core Sin3 represssor complexes are pre-assembled and available for recruitment by transient association with gene specific transcription factors. To date, several transcriptional repressors have been shown to associate with the Sin3 complex to mediate repression of target genes. This is exemplified by MAD proteins whose transcriptional repressing activity has been correlated to the recruitment of HDACs together with the Sin3A and Sin3B (11). Other examples of transcriptional repressors that bind Sin3A include MECp2, Alien, Ikaros, p53 and PLZF (12). Certain unliganded nuclear receptors exert transcriptional regulatory effects via their ability to recruit the corepressor SMRT which in turn associates with both Sin3A and HDACs (13,14).
The purpose of the present study was to further understand the basis of the ability of Ebp1 to repress transcription of E2F1 and AR regulated promoters by examining its interactions with other proteins involved in transcriptional regulation. Sin3A was identified as a candidate interacting partner initially because of its interactions with HDAC2. Second, Sin3A has been demonstrated to be associated with both AR (15) and Rb (16) regulated promoters and to contribute to repression of the activities of such promoters. Therefore, we reasoned that Sin3A might contribute to the activity of Ebp1 for both AR and E2F1 regulated genes. We therefore investigated the involvement of Sin3A in Ebp1-mediated gene repression. We determined that Ebp1 could bind Sin3A and mapped their interaction domains. We also assessed if Ebp1 and Sin3A could interact on endogenous promoters and if Sin3A could enhance the ability of Ebp1 to repress transcriptional activity. We found that Sin3A is a functional corepressor for Ebp1, supporting the model that Ebp1 mediates the repression of AR and E2F1 regulated promoters by the recruitment of a corepressor complex containing HDAC2 and Sin3A.
MATERIALS AND METHODS
Cell culture
All cell lines were obtained from the American Type Culture Collection (Manassas, Va) and maintained at 37°C in a humidified atmosphere of 5% CO2 in air. Cell lines were routinely cultured in RPMI 1640 media supplemented with 10% FBS.
Plasmids
The E2F1 reporter plasmid contains a 225 bp fragment of the E2F1 promoter upstream of the luciferase reporter gene (17). Glutathione S-transferase (GST)–Sin3A PAH1, 2 and PAH2, 3 were from Dr Maureen Murphy (18) and GST–Sin3A 545–1157 was received from Dr M. Privalsky (19). The HDAC2 expression plasmid was from Dr A. Otte.
The bacterial expression vectors encoding full-length and truncated GST–Ebp1 fusion proteins and a mammalian expression vector (pcDNA3) encoding full length ebp1 were described previously (20). The ebp1-LXXAA mutant pcDNA expression plasmid was described previously (4).The GFP–Ebp1 plasmid was constructed by cloning full-length Ebp1 into the EcoR1/BamH1 restriction sites of the EGFP-C1 vector (Clontech, Palo Alto, CA).
GST pull-down assays
In vitro expression and purification of recombinant GST–Ebp1 fusion proteins were performed essentially as described (20). For pull-down assays, cells were rinsed with phosphate-buffered saline (PBS) and lysed in buffer consisting of 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10% glycerol, 2 µg/ml each of aprotinin and leupeptin and 1 mM phenylmethlysulfonyl fluoride (PMSF). Cell lysates (1 mg of protein) were mixed with equal amounts of GST or wild-type or GST–Ebp1 mutants loaded onto glutathione–Sepharose beads and incubated overnight at 4°C with gentle rotation. An aliquot of bound GST or GST–Ebp1 fusion constructs was also analyzed by Coomassie blue staining of SDS–PAGE gels to confirm equal loading of fusion proteins. The pelleted beads were then washed in lysis buffer, mixed with SDS sample buffer, boiled and proteins separated on SDS gels. After electrophoresis, the proteins were transferred to Immobilon-P membranes, and immunoblotted as described (21). The blots were probed with Sin3A polyclonal antibodies (either K-20 or AK-11 Santa Cruz, CA) and anti-horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (BioRad, Richmond, CA). Proteins were detected using an ECL kit (Pierce, Rockford, IL).
Immunoprecipitation and western blot analysis
Cell lysates were immunoprecipitated where indicated as described previously (20). Briefly, total cell extracts from cells transfected with a myc-tagged Sin3A expression construct and Flag-tagged Ebp1 were prepared by direct lysis of cells with buffer containing 50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 250 mM NaCl, 1% Triton X-100, 0.5 mM DTT and 1 mM PMSF. Proteins concentrations were measured using a detergent compatible kit (BioRad, Hercules, CA). Cell lysates were pre-cleared with Protein A/Protein G agarose and immunoprecipitated for 4 h at 4°C with 2 µg of a polyclonal antibody directed against the myc-epitope (A-14, Santa Cruz) and 20 µl packed Protein A/Protein G agarose beads. The immunoprecipitates were washed and resuspended in Laemmli sample buffer. Proteins were resolved by SDS–PAGE and analyzed by western blotting as described (21) using the M2 antibody to the FLAG epitope (Sigma, St Louis, MO). In reciprocal experiments, MCF-7 cells stably transfected with a FLAG-tagged Ebp1 vector were immunoprecipitated with the M2 antibody and blots stained with a mouse monoclonal antibody to Sin3A or a rabbit polyclonal anti Ebp1 antibody (Upstate, Lake Placid, NY).
In vitro binding assays
Sin3A and HDAC2 were labeled and synthesized in the presence of [35S]methionine (Amersham) using the TnT coupled reticulocyte lysate system (Promega) as per manufacturer's instructions. GST fusion proteins purified from bacteria were incubated with in vitro-translated proteins with glutathione–Sepharose 4B beads in buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10% glycerol, 2 µg/ml each aprotinin and leupeptin and 1 mM PMSF. GST pull-down assays were performed as described above and the resulting proteins associated with the beads eluted with sample buffer and samples resolved by SDS–PAGE. Gels were fixed and soaked in AMPLIFY (Amersham) as per manufacturer's instructions. Gels were dried and analyzed by fluorography.
Chromatin immunoprecipitation (ChIP) assays
The method of Shang et al. (22) was used. Briefly, LNCaP cells were grown in RPMI 1640 medium supplemented with 5% charcoal stripped fetal bovine serum (FBS) (Sigma). After 3 days of culture, cells were treated with 5 µM bicalutamide for 1 h, washed with PBS and centrifuged at 14 000 g for 5 min. The pellets were then resuspended in 0.3 ml of lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1) and 1× protease inhibitor cocktail (Roche, Indianapolis, IN)]. DNA was sheared on ice to the appropriate lengths (∼500 bp) and diluted in NET-N buffer [20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 10 mM EDTA, 0.5% NP-40, 1.5 mM MgCl2, 10% glycerol and protease inhibitor cocktail] for a final volume of 1.5 ml. A portion of the diluted cell supernatant (1%) was kept to have crosslinks reversed and quantitate the amount of DNA present in samples for the PCR protocol. After pre-clearing with salmon sperm DNA/Protein A agarose slurry for 30 min at 4°C, immunoprecipitation was performed overnight on a rotary shaker at 4°C with specific antibodies or pre-immune IgG as a control. The samples were then mixed with sonicated salmon sperm DNA (100 µg/ml) and Protein A/Protein G agarose (Oncogene Research Products, San Diego, CA) for another 6 h incubation. Agarose beads were washed sequentially in low salt, high salt, LiCl and TE buffer provided with a kit from Upstate and extracted two times with freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3). Eluates were pooled and incubated at 65°C for 6 h to reverse the formaldehyde cross-links. DNA was purified by phenol/chloroform extraction and precipitated in the presence of 0.3 M sodium acetate, 20 µg tRNA in 2 vol of ethanol at −20°C overnight. The DNA pellets were dissolved in 50 µl of water. Nested PCR amplification of a 210 bp prostate specific antigen (PSA) promoter fragment (−250 to −39) was carried out using a 5′ primer 5′-TCTGCCTTTGTCCCCTAGAT-3′ and a 3′ primer 5′-AACCTTCATTCCCCAGGACT-3′. The PCR products were resolved on 2.5% agarose gels and visualized with ethidium bromide.
DNA affinity precipitation
The method of Alliston et al. (23) was used as described. Briefly, MCF-7 cells, transfected with an EGFP-C1 control vector or EGFP–Ebp1 and selected for 1 week in G418 (500 µg/ml) were lysed with NET-N buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10% glycerol and protease and phosphatase inhibitors. An oligonucleotide derived from the human E2F1 promoter (−35 to +1) (GGCTCTTTCGCGGCAAAAAGGATTTGGCGCGTAAAA and containing two overlapping E2F consensus sites (indicated in bold) was modified by the addition of biotin to the 5′ end of the coding strand. Biotinylated oligonucleotides were annealed with non-biotinylated complementary oligonucleotides. Annealed oligonucleotides were coupled to streptavidin magnetic beads (Promega), washed in NET-N buffer and added to 700 µg of cell lysates and 10 µg of salmon sperm DNA and incubated for 2 h at 4°C. The precipitates were collected on a magnetic stand and washed once with NET-N buffer. Precipitated proteins were analyzed by SDS–PAGE and western blotting using antibodies to GFP (Clontech), Ebp1 (Upstate) and Sin3A (AK-11).
Luciferase reporter assays
For the E2F1 reporter assays, MCF-7 cells (5 × 104) were plated in 12-well plates in complete media. When cells reached 50–60% confluence, they were transfected with 0.5 µg of an E2F1 reporter plasmid and 5 ng of the Renilla-TK control plasmid and transfected using the Fugene-6 reagent (Roche). Cells lysates were collected 24 h later and luciferase activity assessed using a Promega Dual-Luciferase Assay kit (Madison, WI). All transfection experiments were carried out in triplicate wells and repeated three times. The activities of Renilla luciferase were used to normalize any variations in transfection efficiency.
For the androgen regulated gene promoter assays, LNCaP cells (5 × 104) were plated in 12-well plates in complete media. When cells reached 50–60% confluence, they were transfected using the Fugene-6 Reagent (Roche, Indianapolis, IN) with 0.5 µg of the MMTV-luc reporter and 5 ng of the Renilla-TK plasmid (Promega, Madison, WI) as an internal control. Complete medium was replaced 24 h after transfection with phenol red free RPMI 1640 with 5% CSS with or without R1881 (10−8 M) (24). Luciferase activity was determined using the Promega Dual luciferase assay kit as described by the manufacturer. The levels of luciferase activity were normalized using the Renilla luciferase as an internal control. The ratio of luciferase activity to the Renilla control derived from cells that were transfected with vector alone and not treated with R1881 was given a relative luciferase activity value of 1. All values presented in the individual Figures were derived by comparison to this ratio observed in control cells.
Statistical analysis
Results were analyzed using a two-tailed Student's t-test. Significance was established at P < 0.05.
RESULTS
Ebp1 associates with Sin3A in vitro and in vivo
To test whether Ebp1 can interact with Sin3A, we performed GST pull-down assays using HeLa cell lysates. Western blot analysis demonstrated that Sin3A specifically associated with GST–Ebp1, but not GST alone (Figure 1A).
Figure 1.
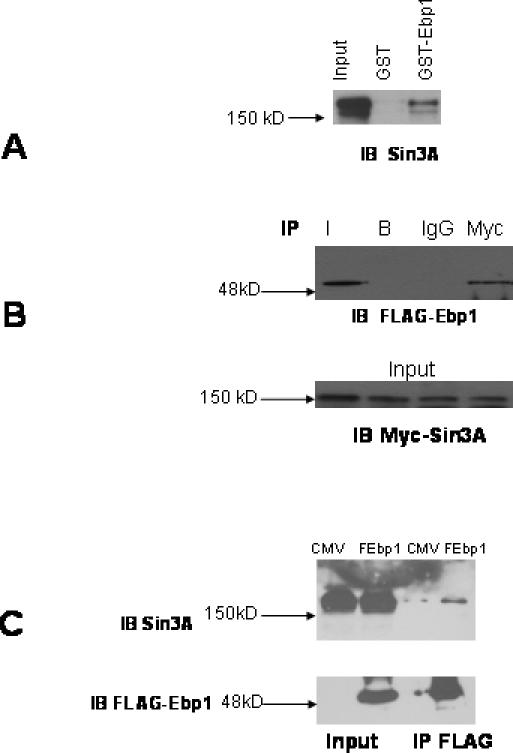
Ebp1 interacts with Sin3A in vitro and in vivo. (A) Equal amounts of GST–Ebp1 or GST alone were prepared and incubated with lysates of logarithmically growing HeLa cells. Ebp1 associated proteins were analyzed by western blotting using a Sin3A antibody. Aliquots of cell lysates (5% of input) were also loaded directly onto the gels and analyzed by western blotting (Input, Lane 1). (B) COS-7 cells were transfected with expression plasmids for a Myc-tagged Sin3A and a Flag-tagged Ebp1. Cellular proteins were prepared and aliquots (1 mg) immunoprecipitated with a polyclonal antibody to c-Myc (A-14), non-specific rabbit IgG, or Protein A/Protein G beads (B). Immunocomplexes were separated by SDS–PAGE and proteins were transferred to PVDF membranes. Blots were probed with the mouse monoclonal M-2 Flag antibody (upper panel). (C) MCF-7 cells stably transfected with a FLAG-tagged ebp1 expression construct (FEbp1) or a control vector (CMV) were lysed. Cell lysates were immunoprecipitated with the M2 monoclonal antibody to FLAG. Immunocomplexes were separated by SDS–PAGE and proteins were transferred to PVDF membranes. Blots were probed with a monoclonal antibody to Sin3A and then stripped and reprobed with an antibody to FLAG. Input = 10% of the volume used in the immunoprecipitation assays.
The interactions between Ebp1 and Sin3A observed in vitro were confirmed in mammalian cells by coimmunoprecipitation analysis. COS-7 cells were transfected with a myc-tagged Sin3A expression construct and a FLAG-tagged ebp1 expression construct. Cell lysates were immunoprecipitated with a polyclonal antibody to the myc-epitope tag and probed with a mouse monoclonal antibody to the FLAG-tag. Results indicated that Ebp1 was found in myc, but not isotype control, immunoprecipitates (Figure 1B). This finding suggests that Ebp1 and Sin3A can bind in vivo. Reciprocal experiments were performed using MCF-7 cells that had been stably transfected with a Flag-tagged ebp1 construct or a vector control. Lysates were immunoprecipitated with the Flag antibody and probed with a mouse antibody to Sin3A. The anti-FLAG antibody did not precipitate proteins from lysates of cells that had been transfected with the control vector. The FLAG antibody did precipitate FLAG–Ebp1 protein from FLAG–Ebp1 transfectants. Endogenous Sin3A was found in FLAG immunoprecipitates from the FLAG–Ebp1, but not vector control, transfectants (Figure 1C). Incubation of cell lysates with beads alone or control IgG failed to immunoprecipitate either Flag reactive or Sin3A proteins (data not shown). These results indicated that Ebp1 associated with endogenous Sin3A in vivo.
Sin3A interacts with a domain in Ebp1 required for transcriptional repression
To determine the Sin3A-binding domain of Ebp1, a series of GST–Ebp1 truncated fusion proteins was prepared (Figure 2A and B). Equal amounts of the fusion proteins or GST alone (Figure 2C) were incubated with HeLa cell lysates. The results showed that the last 79 C-terminal amino acids of Ebp1 (amino acid 293–372) were both necessary and sufficient to bind Sin3A (Figure 2D). This is of interest as the interacting region of Ebp1 for HDAC2 also maps to amino acids 293–372 (3). Mutation of the LXXLL motif to LXXAA also abrogated the ability of Ebp1 to bind to Sin3A.
Figure 2.
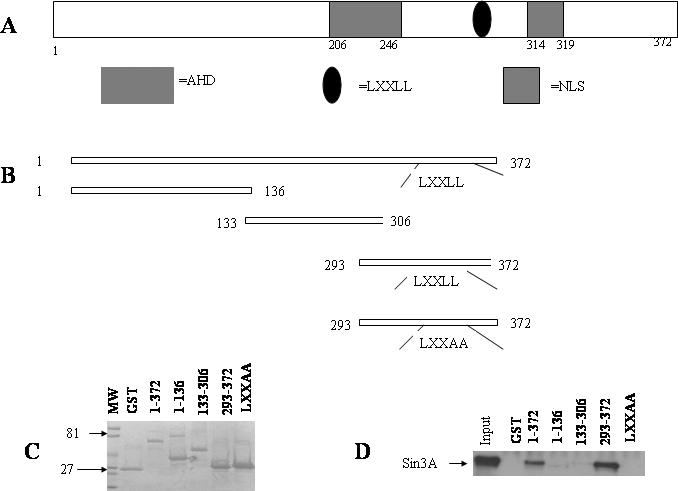
The C-terminal end of Ebp1 is necessary and sufficient to bind Sin3A. (A) Schematic of the Ebp1 protein indicating the location of the amphipathic helical domain (AHD), LXXLL and putative nuclear localization signal (NLS). (B) Diagram of Ebp1 truncation mutants. (C) Equal amounts of the fusion proteins used in the pull-down assays were loaded onto parallel SDS–PAGE gels and stained with Coomassie blue. (D) Equal amounts of GST–Ebp1 fusion proteins (shown in A) or GST alone were prepared and incubated with lysates of logarithmically growing HeLa cells. Ebp1 associated proteins were analyzed by western blotting using a Sin3A antibody (the arrow indicates the position of Sin3A). Aliquots of the cell lysates (5% of the input) were also loaded directly onto the gels and analyzed by western blotting (Input, Lane 1).
Ebp1 interacts with the C-terminal domain of Sin3A
The Sin3A protein is highly modular and contains four PAH domains that mediate protein–protein interactions with distinct transcription factors and HDAC proteins to modify chromatin structure. Three of the domains (PAH1, 3) are clustered at the amino terminus of the protein (Figure 3A). To determine which regions of Sin3A are required for interaction with Ebp1, we performed pulldown assays using GST–Sin3A fusion proteins and HeLa cell nuclear lysates. A fragment containing PAH4 and most of the C-terminal domain of Sin3A (545–1157) pulled down endogenous Ebp1. No interaction was observed with GST proteins fused to amino acids 127–187, 314–381 or 314–532 containing the PAH1, 2 and 2, 3 domains, respectively (Figure 3B, top panel). Similarly we showed that domain 545–1157 bound to HDAC2 (Figure 3B, bottom panel) in keeping with previous reports (11) which demonstrated that a region between PAH3 and PAH4 was sufficient to bind HDAC2.
Figure 3.
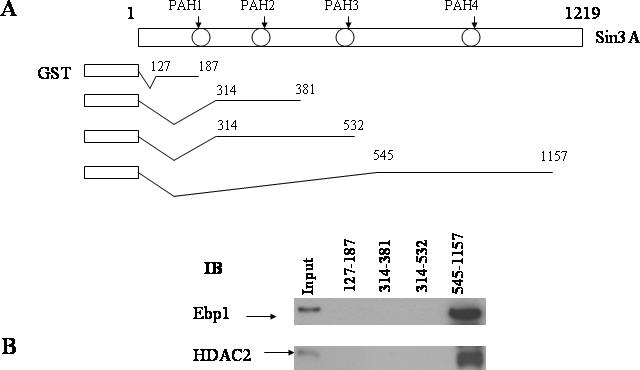
The C-terminal domain of Sin3A binds both Ebp1 and HDAC. (A) Equal amounts of GST–Sin3A fusion proteins (shown in A) or GST alone were prepared and incubated with lysates of logarithmically growing HeLa cells. (B) Sin3A associated proteins were analyzed by western blotting using Ebp1 or HDAC2 antibodies as indicated. Aliquots of the cell lysates (5% of the input) were also loaded directly onto the gels and analyzed by western blotting (Input, Lane 1).
Ebp1 directly interacts with Sin3A
Our previous studies showed that Ebp1 can bind HDAC2 and HDAC enzymatic activity (3). As the interacting region of Ebp1 for both Sin3A and HDAC2 mapped to amino acids 293–372, we tested if the Ebp1–HDAC2 interaction was mediated through the Sin3A protein. We first examined the ability of in vitro synthesized Sin3A and HDAC2 added individually to bind to Ebp1 using in vitro protein pull-down experiments. Full-length in vitro translated Sin3A protein alone was added to GST–Ebp1 beads. GST–Ebp1, but not GST, bound full-length in vitro translated Sin3A protein (Figure 4A). Full-length in vitro translated HDAC2 was added alone to GST–Ebp1 beads. In contrast to results with Sin3A, no binding was detected between GST–Ebp1 and in vitro-translated HDAC2, suggesting the binding of HDAC2, derived from cell lysates to Ebp1 was indirect. Although HDAC1 has been reported to bind to Sin3A in in vitro translation assays (19), the binding of HDAC2 to Sin3A has not been reported in such systems. To verify that HDAC2 was intact and that it could bind to Sin3A, we incubated [35S] HDAC2 to GST–Sin3A. HDAC2 was able to bind to Sin3A directly (Figure 4B). We also tested the ability of GST–Ebp1 fusion proteins to interact with in vitro translated Sin3A. As shown in Figure 4C, Sin3A specifically bound to full-length and Ebp1 293–372 as observed in assays using cell lysates. This study demonstrates that the same region that binds Sin3A as part of a protein complex from cell lysates also binds Sin3A directly.
Figure 4.
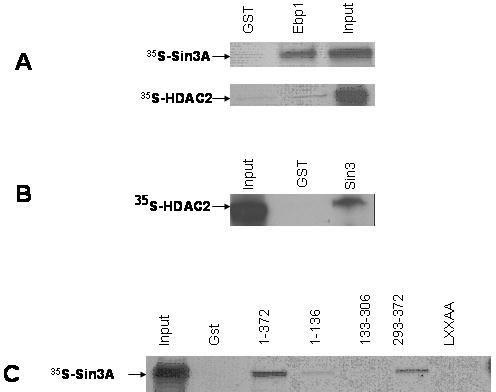
Sin3A, but not HDAC2, interacts directly with Ebp1. (A) In vitro translated 35S-labeled full length Sin3A or HDAC2 were incubated with equal amounts of GST or GST–Ebp1. Ebp1 associated proteins were resolved by SDS–PAGE and visualized by fluorography. Input = 10% of the incubation mixture. (B) In vitro translated 35S-labeled full length HDAC2 was incubated with equal amounts of GST or GST–Sin3A (545–1157). Sin3A associated proteins were resolved by SDS–PAGE and visualized by fluorography. Input = 10% of the incubation mixture. (C) In vitro translated 35S-labeled Sin3A was incubated with equal amounts of GST or GST–Ebp1 fusion proteins. Ebp1 associated proteins were resolved on SDS–PAGE gels and visualized by fluorography.
HDAC2, Ebp1 and Sin3A interact with the PSA and E2F1 promoters
Our previous studies demonstrated that Ebp1 is a DNA-binding protein as are other members of the PA2G4 gene family (6). To test the hypothesis that Ebp1, Sin3A and HDAC could assemble on the endogenous AR regulated PSA promoter, we conducted ChIP experiments. Three AREs have been identified in the PSA gene: ARE I and ARE II reside in the 630 bp promoter region, whereas ARE III resides in the enhancer region located 4 kb upstream of the PSA transcription start site (22). The corepressors NCoR and SMRT are associated with ARE I only. LNCaP cells were serum starved and then treated with the androgen antagonist bicalutamide for 1 h. Ebp1 was not associated with the ARE I response element of the PSA promoter in the absence of bicalutamide, but was recruited to the PSA promoter after bicalutamide exposure (Figure 5B) as demonstrated previously (5). In contrast, Ebp1 was not found on the ARE II response element of the PSA promoter (Figure 5C) indicating that the association of Ebp1 with ARE I was specific. Sin3A was found on the ARE I response element of the PSA promoter under growth arrested conditions in the absence of bicalutamide, and its presence was only slightly increased after treatment with bicalutamide. HDAC2 was associated with the PSA promoter before bicalutamide treatment, but its association was increased after exposure to bicalutamide as reported previously (22). Thus all three proteins assembled at the PSA promoter after treatment with an androgen antagonist.
Figure 5.
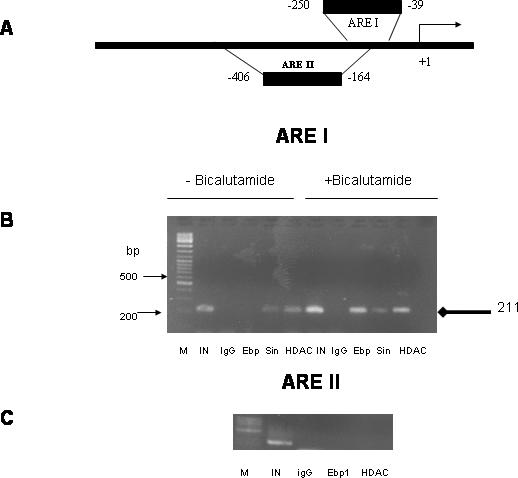
Assembly of Ebp1, Sin3A and HDAC2 complexes at the promoter of the PSA gene in response to bicalutamide treatment. (A) Schematic diagram of the PSA gene regulatory region. (B and C) ChIP assays of Ebp1 involvment in the PSA gene regulatory region. Log phase LNCaP cells, in the absence or presence of bicalutamide as indicated, were fixed with formaldehyde and chromatin lysates immunoprecipitated with pre-immune IgG or antibodies to Ebp1, Sin3A or HDAC2 as indicated. Samples were processed as described in the Materials and Methods and PCR products visualized by ethidium bromide staining. Immunoprecipitated DNA was amplified by using primers specific for the human PSA promoter ARE I or ARE II as illustrated in (A). IN = Input which represents 1% of the total amount of chromatin added to each immunoprecipitation reaction. M = MW markers. Diamond headed arrow indicates the position of the 211 bp PCR product. In (C) cells were treated with bicalutamide.
We next used DNA affinity precipitation assays to determine if Sin3A associates with Ebp1 at an E2F1 consensus element. MCF-7 cells were transfected with an EGFP vector control or an EGFP–Ebp1 expression plasmid TA. Cell lysates were incubated with the biotinylated E2F1 promoter consensus oligonucleotide and bound proteins determined by western blotting. GFP–Ebp1 was found associated with the E2F1 oligonucleotide. No GFP reactive proteins from lysates of cells that had not been transfected or cells that were transfected with the GFP vector alone (Figure 6B, upper panel) were found associated with the E2F1 oligo. Endogenous Ebp1 was also associated with the oligonucleotide, although binding was decreased in the GFP only transfected cells (Figure 6B, middle panel). Sin3A was found associated with the E2F1 consensus oligonucleotide in all three cell lysates. However, overexpression of Ebp1 increased the binding of Sin3A to the consensus oligonucleotide suggesting binding of Sin3A to the E2F1 promoter may be via its association with Ebp1 (Figure 6B, bottom panel). As reported previously (6) Ebp1 from MCF-7 cell lysates was unable to bind to an E2F1 oligo in which the two E2F1 consensus sites were mutated (data not shown).
Figure 6.
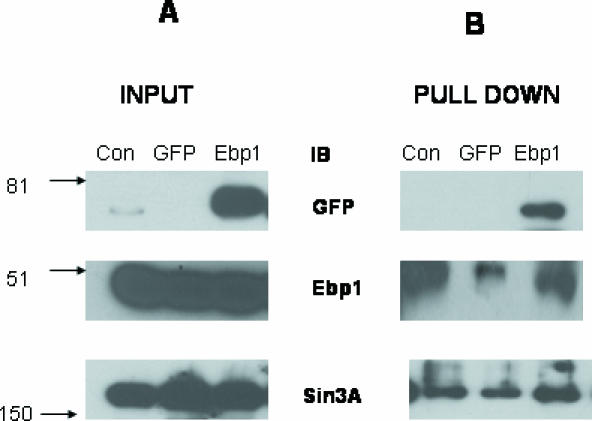
Endogenous Ebp1 and Sin3A bind to an E2F1 oligonucleotide MCF-7 cells were transfected with a control EGFP vector (GFP) or an EGFP–Ebp1 (Ebp1) vector. Transfected cells were selected in 500 µg/ml of G-418 for 1 week and collected as described. Untransfected logarithmically growing MCF-7 cells were also lysed (Con). Cell lysates were incubated with a biotinylated double-stranded E2F1 consensus oligonucleotide. DNA bound proteins were precipitated by magnetic streptavidin beads. Western blotting was used to detect the presence of bound proteins. Input is illustrated in A and proteins associated with the E2F1 oligo in B. The GFP antibody detected the presence of exogenous Ebp1 (upper panel); a polyclonal antibody to Ebp1 detected the presence of endogenous Ebp1 (middle panel); a mouse monoclonal antibody to Sin3A detected endogenous Sin3A (lower panel).
Cooperation of Ebp1, Sin3A and HDAC to repress the transcription
To determine if Sin3A and Ebp1 could cooperate to repress transcriptional activity, we first analyzed the effect of Ebp1 on the E2F1 promoter in the presence and absence of ectopically expressed Sin3A and HDAC2 in MCF-7 cells. To observe potential cooperation, pcDNA3-ebp1 was transfected under conditions that resulted in a partial repression of the promoter. Ectopically expressed Sin3A alone did not affect E2F1 transcription and did not increase the ability of Ebp1 to repress transcription. In contrast, transfection of HDAC2 repressed the E2F1 promoter. However, a stronger repression was observed after cotransfection of Ebp1 with HDAC and Sin3A than that observed with either plasmid alone or with any two plasmids in combination (Figure 7A). These findings suggests that these proteins can cooperate in mediating repression of an E2F1 regulated promoter. To further support the hypothesis that direct interactions of Ebp1 with Sin3A and HDAC2 were important for these effects, we also transfected ebp1 in which the LXXLL domain had been mutated to LXXAA to prevent Ebp1–Sin3A interactions. The LXXAA mutant did not inhibit E2F1 signaling. The lack of activity of the mutant was not changed by transfection of Sin3A, HDAC2 or the combination.
Figure 7.
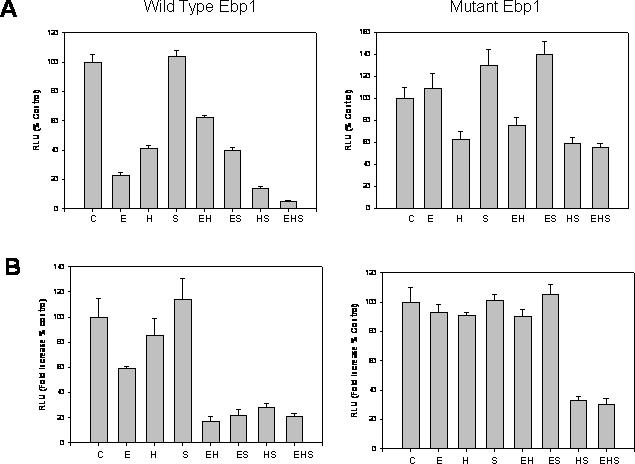
Sin3A and HDAC cooperate with Ebp1 for repression of promoter activity. (A) MCF-7 cells were transfected with an E2F1 luciferase reporter plasmid (500 ng) in the absence (C) or presence of either wild-type or mutant (LXXAA) Ebp1 (200 ng) (E), HDAC2 (100 ng) (H) or Sin3A (200 ng) (S) expression vectors as indicated. Cells were also cotransfected with a Renilla-TK control plasmid. Promoter activity was determined 36 h later as indicated in the Materials and Methods. (B) LNCaP cells were transfected with the MMTV-luciferase reporter plasmid (500 ng) in the absence (C) or presence of either wild-type or mutated (LXXAA) Ebp1 (200 ng) (E), HDAC2 (100 ng) (H) or Sin3A (200 ng) (S) expression vectors as indicated. Cells were also cotransfected with a Renilla-TK control plasmid. After 24 h, the cells were switched to RPMI 1640 phenol-red free media with 5% CSS and R1881. Cell lysates were assayed for luciferase activity 16 h later. The relative luciferase activity in the absence of R1881 was set at 1 for cells that received the vector control. The fold change in the presence of R1881 was calculated after transfection with empty vector and set at 100%. None of the expression vectors affected basal luciferase activity in the absence of R1881. Each point represents mean ±S.E. of triplicate wells. Representative of three experiments.
We also examined the ability of the three proteins to repress the androgen regulated transcription of an AR regulated promoter. Cells were transfected with Ebp1, HDAC, Sin3A expression plasmids or the combination and an AR regulated MMTV-luciferase reporter plasmid. Cells were then stimulated with R1881. None of the constructs alone or in combination affected the low level of transcription observed in the absence of R1881 (data not shown). However, in the presence of R1881, Ebp1 at limiting concentrations repressed AR transcription ∼40%. Sin3A and HDAC alone at the concentrations tested had no effect on activation of AR by R1881. Unlike in the case of the E2F1 promoter, Sin3A alone was able to enhance the ability of Ebp1 to repress AR regulated transcription. Similarly, HDAC alone potentiated the ability of Ebp1 to repress R1881 induced transcription. All three constructs did not further increase the transcriptional repression observed (Figure 7B). As described above, we also transfected the LXXAA mutant which was reported previously to be unable to affect transcription (4). The LXAAA mutant did not affect transcription by itself and had no effects on the ability of HDAC2 and Sin3A in combination to repress transcription.
DISCUSSION
Ebp1 has been previously demonstrated to repress transcription of endogenous and exogenous AR and E2F1 regulated genes via its association with HDACs (3). In this study, we investigated whether the known corepressor Sin3A, a crucial component of many inducible repression systems involving HDACs, was involved in Ebp1-mediated transcriptional repression. We found that Sin3A interacted with the C-terminal domain of Ebp1 that is required to repress transcription. The fact that Ebp1 associated with Sin3A further supports the role of Ebp1 in transcriptional repression and provides a mechanistic basis for its ability to inhibit cell growth. These findings also demonstrate that a protein such as Ebp1, that can be activated via ErbB signaling, is involved in Sin3A-mediated transcriptional repression.
GST pull-down assays mapped the sites of interaction on the two proteins. Amino acids 293–372 of Ebp1 were necessary and sufficient for interaction with Sin3A. This region has been previously shown to be responsible both for the transcriptional repression and growth inhibitory activity of Ebp1 (3,20). These C-terminal 79 amino acids also bind HDAC2 (3). Thus, HDAC2 and Sin3A are likely to be found as a complex in vivo. To further clarify interactions between Ebp1, Sin3A and HDAC2, we performed pull-down assays using in vitro-translated proteins. Results using in vitro-translated Sin3A indicated that Ebp1 bound Sin3A directly. In contrast, in vitro-translated HDAC2, which binds Ebp1 as a component of cell lysates, failed to interact with Ebp1. Thus, we postulate that Sin3A acts as a bridge between Ebp1 and HDAC2 in vivo. Our results also indicated that mutation of the LXXLL motif to LXXAA abrogated the ability of Ebp1 to bind to Sin3A. Although LXXLL motifs were originally identified as components of nuclear hormone receptor binding proteins (25), recent studies have demonstrated that there are numerous examples of other types of protein–protein interactions involving LXXLL motifs (26). Indeed, transcription factors such as Bcd use hydrophobic surfaces to interact with Sin3A (27). Therefore, disruption of this motif may result in a change of conformation that leads to failure to bind Sin3A.
The Ebp1 interacting domain of Sin3A mapped to amino acid 545–1147 encoding the PAH4 and the C-terminal domain. This is of interest as HDAC2, derived from whole cell lysates, was demonstrated to bind to amino acid 480–680 of Sin3A (11,19). Thus, Ebp1 may be bound in tandem with HDAC2 or in a complex at the same site of Sin3A. Similarly, the corepressor SMRT binds to Sin3A through amino acid 57–215 and 533–724 domains. The C-terminal domain outside of PAH4 (HCR domain) interacts with other transcriptional repressors (12) including the nuclear receptor corepressor Alien (28). The binding of Ebp1 to the C-terminal region of Sin3A stands in contrast to its binding to the repressors MAD (11) which binds PAH2 and REST (29) which binds the N-terminal region of Sin3A. The N-terminal domain of Sin3A also interacts with TGIF, another repressor of AR-mediated transcription (15). It is possible that both TGIF and Ebp1 may be recruited to Sin3A under conditions such as androgen antagonist treatment which results in transcriptional inactivation of the androgen receptor.
ChIP assays showed that Ebp1 was absent from the PSA promoter under steroid-reduced conditions, but was recruited to the PSA promoter in the presence of the androgen antagonist bicalumtamide. Ebp1 was not recruited to ARE II indicating a specific association with genomic DNA. Similarly, other AR repressors have been mapped to ARE I (22,30), but their presence on ARE II has not been reported. HDAC was associated at the ARE I of the PSA promoter under serum-starved conditions. Similarly, Gaughan et al. (31) reported previously that HDACs were associated with the ARE I region of the PSA promoter under serum-starved conditions. However, HDAC2 was also recruited to the PSA promoter by bicalutamide. In contrast, the amount of Sin3A detected by ChIP analysis appeared to be little changed. Sin3A may be on the PSA promoter in the absence of bicalutamide because cells are in a resting state. Sin3A may serve as a platform onto which other cofactors are recruited in response to different stimuli. Although Ebp1 and Sin3A appear not be recruited in tandem, the addition of Ebp1 to the HDAC–Sin3A complex may enhance the ability of these molecules to repress transcription. Our ChIP studies support the functional role of Ebp1, Sin3A and HDAC binding in transcriptional repression mediated by bicalutamide.
DNA affinity precipitation assays also indicated that Ebp1 and Sin3A bound an E2F1 promoter consensus oligonucleotide. DNA affinity precipitation assays previously indicated that Rb and HDAC were also recruited to an E2F1 consensus oligonucleotide with Ebp1 (6). Similarly, the retinoblastoma binding protein RBP1 was demonstrated to recruit Sin3A complexes containing HDAC and Rb to E2F promoters (16). Although Ebp1 can bind Rb (20), we do not know if Ebp1 is part of a multiprotein complex containing RBP1. However, the ability of Ebp1 to bind transcription factors such as E2F1 and AR suggest that Ebp1 may recruit Sin3A to DNA in the presence of sequence-specific DNA-binding factors to repress transcription.
To test the functional importance of Ebp1–Sin3A–HDAC complexes, we examined the ability of these proteins to inhibit transcriptional activity. Luciferase reporter assays indicated that Sin3A alone did not enhance the ability of Ebp1 to repress E2F1-mediated transcription. However, HDAC2 and Sin3A together enhanced the transcriptional repression observed using low concentrations of Ebp1 plasmid suggesting the importance of a multiprotein complex. An Ebp1 LXXAA mutant, which does not bind Sin3A, was unable to repress transcription by itself or to lead to transcriptional repression in the presence of Sin3A. This finding suggests that interactions of Ebp1 and Sin3A are important for the observed synergistic effects. Sin3A or HDAC2 alone were able to enhance the ability of wild-type, but not mutant, Ebp1 to repress AR-mediated transcription. Other studies also indicate that Sin3A–HDAC is important for silencing of AR (32–34). Sharma and Sun (15) demonstated that the transcriptional repressor TGIF could inhibit AR transcriptional activity in the presence of androgens via its association with Sin3A and HDAC1. It is of interest that TGIF is a member of the TGF-β signal transduction pathway whose activity is modulated by TGF-β. Similarly, Ebp1 is a member of a HRG activated ErbB3 pathway and its ability to modulate AR activity is dependent on HRG concentration (35). These studies indicate that AR may be regulated by multiple physiological signals originating from different signal transduction pathways.
In conclusion, the results presented here indicate that Ebp1 can bind the transcriptional repressor Sin3A both in vivo and in vitro. The C-terminal domain of Ebp1 that is required for its ability to repress transcription and to inhibit cell growth interacts with Sin3A. Ebp1, Sin3A and HDAC can associate on promoters and interact to repress transcription. These studies provide a model of Ebp1-mediated silencing through recruitment of Sin3A and subsequently HDACs.
Acknowledgments
This work was supported in part by NIH grants R01 CA76047 and R21 088882-01 and a grant from the Department of Pathology (to A.W.H.). We thank Dr W.D. Cress for the E2F1 plasmid, Drs Maureen Murphy and Martin Privalsky for the GST–Sin3A plasmids, Dr Robert Eisenman for the Sin3A mammalian expression plasmid and Dr A. Otte for the HDAC2 plasmid. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yoo J.Y., Wang X.W., Rishi A.K., Lessor T., Xia X.M., Gustafson T.A., Hamburger A.W. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer. 2000;82:683–690. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lessor T.J., Yoo J.Y., Xia X., Woodford N., Hamburger A.W. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J. Cell. Physiol. 2000;183:321–329. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y.X., Woodford N., Xia X.M., Hamburger A.W. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Res. 2003;31:2168–2177. doi: 10.1093/nar/gkg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y.X., Fondell J.D., Wang Q.B., Xia X.M., Cheng A.W., Lu M.L., Hamburger A.W. Repression of androgen receptor mediated transcription by the ErbB-3 binding protein, Ebp1. Oncogene. 2002;21:5609–5618. doi: 10.1038/sj.onc.1205638. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Wang X.W., Jelovac D., Nakanishi T., Yu M.H., Akinmade D., Goloubeva O., Ross D.D., Brodie A., Hamburger A.W. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc. Natl Acad. Sci. USA. 2005;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Hamburger A.W. Heregulin regulates the ability of the ErbB3-binding protein Ebp1 to bind E2F promoter elements and repress E2F-mediated transcription. J. Biol. Chem. 2004;279:26126–26133. doi: 10.1074/jbc.M314305200. [DOI] [PubMed] [Google Scholar]
- 7.Lehrmann H., Pritchard L.L., Harel-Bellan A. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 8.Sengupta N., Seto E. Regulation of histone deacetylase activities. J. Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Sun Z.W., Iratni R., Erdjument-Bromage H., Tempst P., Hampsey M., Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 11.Laherty C.D., Yang W.M., Sun J.M., Davie J.R., Seto E., Eisenman R.N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 12.Silverstein R.A., Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 13.Heinzel T., Lavinsky R.M., Mullen T.M., Soderstrom M., Laherty C.D., Torchia J., Yang W.M., Brard G., Ngo S.D., Davie J.R., et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 14.Nagy L., Kao H.Y., Chakravarti D., Lin R.J., Hassig C.A., Ayer D.E., Schreiber S.L., Evans R.M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 15.Sharma M., Sun Z. 5′-TG-3′ interacting factor interacts with Sin3A and represses AR-mediated transcription. Mol. Endocrinol. 2001;15:1918–1928. doi: 10.1210/mend.15.11.0732. [DOI] [PubMed] [Google Scholar]
- 16.Lai A., Kennedy B.K., Barbie D.A., Bertos N.R., Yang X.J., Theberge M.C., Tsai S.C., Seto E., Zhang Y., Kuzmichev A., et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cress W.D., Nevins J.R. A role for a bent DNA structure in E2F-mediated transcription activation. Mol. Cell Biol. 1996;16:2119–2127. doi: 10.1128/mcb.16.5.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zilfou J.T., Hoffman W.H., Sank M., George D.L., Murphy M. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell Biol. 2001;21:3974–3985. doi: 10.1128/MCB.21.12.3974-3985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong C.W., Privalsky M.L. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol. Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia X., Cheng A., Lessor T., Zhang Y., Hamburger A.W. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell. Physiol. 2001;187:209–217. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- 21.Xia X., Lessor T.J., Zhang Y., Woodford N., Hamburger A.W. Analysis of the expression pattern of Ebp1, an ErbB-3-binding protein. Biochem. Biophys. Res. Commun. 2001;289:240–244. doi: 10.1006/bbrc.2001.5942. [DOI] [PubMed] [Google Scholar]
- 22.Shang Y., Myers M., Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 23.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadar M.D., Gleave M.E. Ligand-independent activation of the androgen receptor by the differentiation agent butyrate in human prostate cancer cells. Cancer Res. 2000;60:5825–5831. [PubMed] [Google Scholar]
- 25.Heery D.M., Hoare S., Hussain S., Parker M.G., Sheppard H.M. Core LXXLL motif sequences in CBP, SRC1 and RIP140 define affinity and selectivity for steroid and retinoid receptors. J. Biol. Chem. 2000;276:6695–6702. doi: 10.1074/jbc.M009404200. [DOI] [PubMed] [Google Scholar]
- 26.Plevin M.J., Mills M.M., Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 2005;30:66–69. doi: 10.1016/j.tibs.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhao C., Fu D., Dave V., Ma J. A composite motif of the Drosophila morphogenetic protein bicoid critical to transcription control. J. Biol. Chem. 2003;278:43901–43909. doi: 10.1074/jbc.M302714200. [DOI] [PubMed] [Google Scholar]
- 28.Moehren U., Dressel U., Reeb C.A., Vaisanen S., Dunlop T.W., Carlberg C., Baniahmad A. The highly conserved region of the co-repressor Sin3A functionally interacts with the co-repressor Alien. Nucleic Acids Res. 2004;32:2995–3004. doi: 10.1093/nar/gkh621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimes J.A., Nielsen S.J., Battaglioli E., Miska E.A., Speh J.C., Berry D.L., Atouf F., Holdener B.C., Mandel G., Kouzarides T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 30.Hodgson M.C., Astapova I., Cheng S., Lee L.J., Verhoeven M.C., Choi E., Balk S.P., Hollenberg A.N. The androgen receptor recruits nuclear receptor corepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J. Biol. Chem. 2005;280:6511–6519. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- 31.Gaughan L., Logan I.R., Neal D.E., Robson C.N. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.List H.J., Smith C.L., Rodriguez O., Danielsen M., Riegel A.T. Inhibition of histone deacetylation augments dihydrotestosterone induction of androgen receptor levels: an explanation for trichostatin A effects on androgen-induced chromatin remodeling and transcription of the mouse mammary tumor virus promoter. Exp. Cell Res. 1999;252:471–478. doi: 10.1006/excr.1999.4638. [DOI] [PubMed] [Google Scholar]
- 33.Petre C.E., Wetherill Y.B., Danielsen M., Knudsen K.E. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J. Biol. Chem. 2002;277:2207–2215. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 34.Niki T., Takahashi-Niki K., Taira T., Iguchi-Ariga S.M., Ariga H. DJBP: a novel DJ-1-binding protein, negatively regulates the androgen receptor by recruiting histone deacetylase complex, and DJ-1 antagonizes this inhibition by abrogation of this complex. Mol. Cancer Res. 2003;1:247–261. [PubMed] [Google Scholar]
- 35.Zhang Y., Hamburger A.W. Specificity and heregulin regulation of Ebp1 (ErbB3 binding protein 1) mediated repression of androgen receptor signalling. Br. J. Cancer. 2005;92:140–146. doi: 10.1038/sj.bjc.6602257. [DOI] [PMC free article] [PubMed] [Google Scholar]


