Abstract
We determined the sequence of the entire marRAB operon in Enterobacter aerogenes. It is functionally and structurally analogous to the Escherichia coli operon. The overexpression of E. aerogenes MarA induces a multidrug resistance phenotype in a susceptible strain, demonstrated by a noticeable resistance to various antibiotics, a decrease in immunodetected porins, and active efflux of norfloxacin.
The mar regulon identified in Escherichia coli (mar-Eco) plays a key role in the expression of a multidrug resistance phenotype, and specific mutations located in marR have been identified in resistant strains (2, 14, 19). The regulatory function associated with the marA locus simultaneously induces a decrease in antibiotic uptake by altering the porin content of the outer membrane and an increase of antibiotic ejection by activating efflux mechanisms (1). This response supports an efficient resistance to a range of commonly used antibiotics.
The emergence of clinical strains resistant to several structurally unrelated antibiotics is a contributing factor in bacterial dissemination, outbreaks, and changes in patients' flora. Among the emerging resistant bacteria, Enterobacter aerogenes is now the third leading cause of nosocomial respiratory tract infections (6, 10). It generally exhibits high resistance to broad-spectrum antibiotics. Such high resistance is the result of enzymatic responses, mutations in the antibiotic target, and modifications in envelope permeability, including porin alteration and induction of drug efflux (7, 16, 21). A rapid modulation of porin synthesis is observed during the course of patient therapy. With broad-spectrum β-lactams, a decrease in porin expression is associated with a resistant phenotype, and, after a pause in antibiotic therapy, a restoration of pretreatment levels of bacterial porin coincides with de novo antibiotic susceptibility. This observation suggests that in this case, resistance is mediated by a regulation cascade rather than a mutation (5, 8). A concomitant decrease in porin synthesis and a quinolone efflux in several clinical isolates of E. aerogenes have been reported (16). These characteristics suggest that a mar operon may have a role in the antibiotic resistance strategy. In the present study, we identified a homologue of the mar operon in E. aerogenes (mar-Ea) and demonstrated the involvement of the marA gene in the cascade that governs outer membrane impermeability and the ejection of intracellular antibiotics in E. aerogenes.
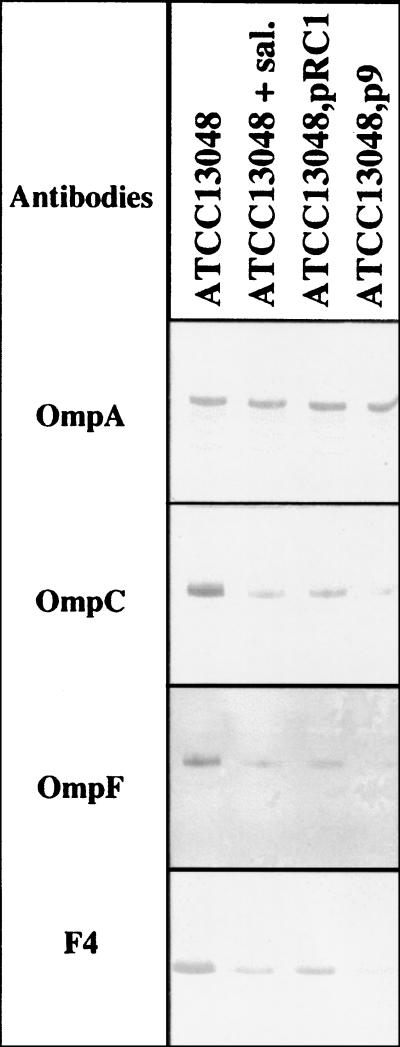
As a first step we investigated whether a MarA-inducible resistance system exists in E. aerogenes. To test this, the plasmid p9, containing the marA-Eco gene, was introduced into the antibiotic-susceptible E. aerogenes type strain ATCC 13048 (12). MICs were determined by using E-test strips (AB Biodisk) on Mueller-Hinton medium or Mueller-Hinton medium supplemented with 5 mM sodium salicylate. The gene marA-Eco efficiently conferred resistance to various antibiotics (Table 1). Overexpression of MarA-Eco in a sensitive E. aerogenes strain generated a multidrug resistance (MDR) phenotype, characterized by an alteration in porin expression, as revealed by immunodetection (Fig. 1). These observations suggest the existence of a system homologous to MarA in E. aerogenes.
TABLE 1.
Susceptibility of E. aerogenes ATCC 13048 and E. coli JM109 to various antibiotics
| Antibiotic | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
|
E. aerogenes ATCC 13048
|
E. coli JM109
|
|||||
| LB
|
LB + salicylatea | Without plasmid | With marA-Eac | |||
| Without plasmid | With marA-Ecob | With marA-Eac | ||||
| Imipenem | 0.038 | 8 | 4 | 2 | 0.047 | 0.128 |
| Norfloxacin | 0.05 | 2 | 2 | 1 | 0.19 | 0.75 |
| Cefepime | 0.032 | 0.19 | 0.064 | 0.064 | 0.016 | 0.094 |
| Tetracycline | 2 | 32 | 16 | 16 | 0.5 | 3 |
| Chloramphenicol | 4 | 32 | 32 | 8 | 4 | 32 |
5 mM sodium salicylate.
Plasmid p9.
Plasmid pRC1.
FIG. 1.
Immunodetection of E. aerogenes outer membrane proteins. The proteins were electrotransferred to membranes and immunodetected with polyclonal antibodies directed against denatured OmpA, OmpC, and OmpF porins and F4 antibodies against peptide from the internal L3 loop domain, which determines pore permeability properties (11). sal., salicylate.
In a previous study, Southern analysis using mar-Eco as a DNA probe provided evidence for a homologue of the mar operon in Klebsiella pneumoniae but not in E. aerogenes (9). Alignment of the mar-Eco operon with the genome of K. pneumoniae (http://genome.wustl.edu/gsc/) allowed us to confirm the presence of a similar operon sequence. Since E. aerogenes and K. pneumoniae are closely related, we used PCR primers constructed from the putative K. pneumoniae marRAB sequence (mar-Kp) except for M12, which is specific for the E. aerogenes sequence (Table 2). The hexadecyltrimethylammonium bromide method was used to obtain DNA (3). PCR amplifications were performed in a GeneAmp PCR system 2400 thermocycler (Perkin Elmer), programmed for an initial 5-min denaturation at 94°C followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C and a 7-min extension at 72°C. The sequences of the PCR products were determined with an ABI Prism 377 DNA sequencer with dye fluorescent terminators and the primers used in the initial PCR amplification.
TABLE 2.
Selective primers used to obtain marRAB-Ea PCR products
| Primer | Sequence | Size (bp) and nature of PCR product obtained with primer combination
|
||
|---|---|---|---|---|
| M4 | M7 | M18 | ||
| M3 | 5′-GATCGCCTGCTCAATGACTAC-3′ | 353 (′marR′) | 985 (marRAB′) | |
| M4 | 5′-GCAGGACCTTCTTGAGCA-3′ | |||
| M5 | 5′-CCTGTTTATTACGCTCGGCGT-3′ | 819 (marOR′) | 1,451 (marORAB′) | 1,695 (marORAB) |
| M7 | 5′-TATGATTGAAATCAAACGGCG-3′ | |||
| M12 | 5′-CAG ACGAAGTGGTCATGCTTG-3′ | 632 (marAB′) | 900 (marAB) | |
| M18 | 5′-TGGCGC GCTGTCGCTGCT-3′ | |||
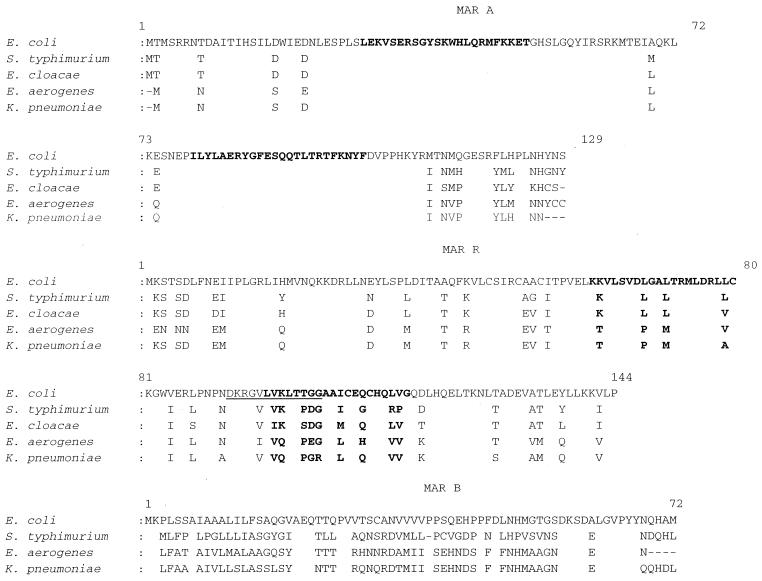
The marORAB-Ea locus is mapped into an operon of 1,335 bp. The predicted amino acid sequence alignments of MarA, MarR, and MarB from E. aerogenes with the known homologues in E. coli, Salmonella enterica serovar Typhimurium, Enterobacter cloacae (except MarB), and K. pneumoniae are presented in Fig. 2. The various MarA proteins exhibit a high interspecific conservation. MarA-Ea showed greater amino acid similarity to MarA-Kp (96.1%) than MarA-Eco (88.4%), and several amino acid substitutions or deletions were observed in the COOH-terminal region. The amino acid sequences varied in length: 125 amino acids for MarA-Kp, 128 for MarA-Ecl, and 129 for MarA-Eco and MarA-St. The length of the putative sequence of the MarR-Ea protein is comparable to the length of the known MarR proteins. Amino acid similarities of 91% between MarR-Ea and MarR-Kp and 81.2% between MarR-Ea and MarR-Eco were observed. The differences are scattered throughout the protein and are located in the two helix-turn-helix regions and the conserved motif comprising amino acids 92 to 104, but in particular at position 103, which showed a high interspecies variability (neutral and acidic residue transition between the five species). Moreover, four of the first amino acids in the NH2 portion of MarR were specific to E. aerogenes. As regards MarB, important differences in size and in sequence were observed. Searches of protein databases with the amino acid residues that are conserved between species gave no significant results. These differences are consistent with the lack of a discernible role of MarB.
FIG. 2.
Amino acid sequence alignment of MarA, MarR, and MarB from E. coli (AG100) (12), S. enterica serovar Typhimurium (X3181) (23), E. cloacae (DSM 3264) (accession no. 302680 and 302681), E. aerogenes (ATCC 13048), and K. pneumoniae (MGH 78578). Boldface indicates helix-turn-helix binding motifs, and underlining indicates the conserved motif in MarR at amino acids 92 to 104.
Plasmid pRC1, a multicopy plasmid containing the cloned marA-Ea gene, was constructed by cloning the 632-bp PCR product (marA and most of marB) obtained with the M12 and M7 primers in pGEM-T (Promega) and the Kanr gene derived from plasmid pUC4K digested with HincII was introduced into the Ampr gene of pGEM-T following ScaI digestion. For the transformed strains, selection was made after overnight incubation on prewarmed Luria-Bertani agar (LB) containing tetracycline (5 μg/ml) for p9 and kanamycin (30 μg/ml) for pRC1. To characterize the role of MarA-Ea in the MDR phenotype in E. aerogenes, we determined the susceptibility to unrelated antibiotics by E-tests (Table 1). The introduction of pRC1 into E. aerogenes ATCC 13048 or E. coli JM109 caused a 2- to 100-fold increase in MIC of the antibiotics tested. Similar modifications were obtained when ATCC 13048 was grown in the presence of sodium salicylate. Effectively, in E. coli, salicylate binds to the MarR repressor at the binding site and allows constitutive expression of the mar operon, and for comparative purposes, the antibiotic resistance phenotype induced by salicylate via the induction of MarA was investigated in E. aerogenes (18).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and porin immunodetection were investigated (15). Polyclonal antibodies directed against porin monomers OmpC, OmpF, and OmpA and the antipeptide F4, directed against the internal porin L3 loop (described elsewhere), were used to determine if plasmid-borne marA-Ea has an effect on the immunorelated porin expression in E. aerogenes (8, 11). As a control, sodium salicylate was used to induce the MDR phenotype. Overexpression of MarA-Ea in the strain transformed by pRC1, as in E. aerogenes grown in the presence of salicylate, induced a strong diminution of porins (Fig. 1). The amount of OmpA, an outer membrane protein that plays a role in envelope architecture, was not significantly modified, irrespective of the culture conditions used or the plasmids present. These results indicate the absence of a general membrane modification that would alter the overall profile of outer membrane proteins. This low level of porin synthesis suggests that salicylate and MarA-Eco or MarA-Ea have an efficient repressor activity in the genetic cascade of porin regulation in E. aerogenes cells. Finally, no modification of the F4 epitopes from the L3 loop domain, which determine the pore properties, was observed.
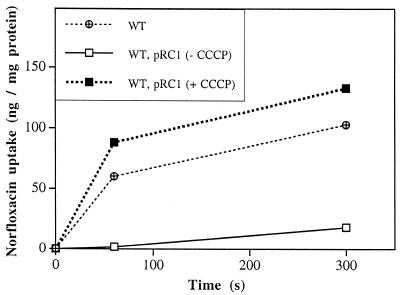
The accumulation of radiolabeled norfloxacin was measured in the presence of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP), which inhibits the energy-dependent efflux of antibiotic as described (16). Since the increase in the MICs of quinolones and tetracycline for the strain containing pRC1 indicates the activation of an efflux mechanism (Table 1), we determined the intracellular accumulation of radiolabeled norfloxacin in this strain. The results showed a severe reduction in the intracellular accumulation of norfloxacin in the presence of pRC1, with a reduction of about sixfold compared to the wild-type strain (Fig. 3). CCCP blocks the efflux pump and restores a level of intracellular norfloxacin similar to that in the wild-type strain. These results suggest that MarA-Ea expression from pRC1 is able per se to induce a complete expression of the active efflux pump.
FIG. 3.
Accumulation of [14C]norfloxacin in the E. aerogenes reference strain 13048 (wild type [WT]) and the transformed strain (carrying pRC1) in the absence and presence of CCCP.
No functional or genetic information is available concerning the regulation of the MDR phenotype in E. aerogenes, although some data suggest similarities with the E. coli cascade (16). In other Enterobacteriaceae, the presence of a mar operon was confirmed, and it was shown to be structurally identical in the genus Salmonella and in Shigella flexneri (4, 13). A mar operon has been partially identified in Klebsiella oxytoca, E. cloacae, and Enterobacter agglomerans by Southern blot hybridization with an E. coli marRAB probe (9). Here, we describe a marRAB locus in E. aerogenes that is structurally and functionally almost identical to that identified in marRAB-Eco and marRAB-St (23).
The crystal structure of MarA-Eco indicates the existence of two comparable subdomains containing a helix-turn-helix DNA binding motif. The differences that we have observed in the amino acid sequence of the protein MarA-Ea do not map to the two conserved helix-turn-helix regions involved in the regulatory function. The COOH-terminal region, which does not interact in the MarA-DNA complex, is the most variable (22). The similarity between MarA-Ea and MarA-Kp was greater, and this is certainly due to their genetic proximity.
Three regions of MarR repressor are important for its activity: two helix-turn-helix DNA binding domains and the first 31 amino acids, which are involved in the dimerization process (2). Alignment of the different MarR repressors shows that there are some variations in the two helix-turn-helix motifs. Except for three cases (N3→S, E52→A, and E103→D103→G), substitutions do not modify the charge of the residue. The first eight amino acids of the COOH-terminal helix-turn-helix MarR-sequence belong to the conserved motif, corresponding to amino acids 92 to 104, which plays a critical role in repressor function and constitutes the DNA binding site (2). Interestingly, major interspecies substitutions are found at residues 102 and 103. Moreover, some of these interspecific amino acid differences are located at mutation sites (positions 3, 53, 96, 103, and 137) identified in MarR-Eco in multidrug-resistant E. coli strains (2, 17, 19, 20). These five positions are particularly susceptible to amino acid variations among Enterobacteriaceae and are also responsible for marA overexpression. It is worth mentioning the substitutions G103→E and Y137→Q, found in E. aerogenes in particular, which introduce a local negative and positive charge, respectively. Mutations which allow a similar charge transition, G103→S and Y137→H, have previously been identified in several clinical E. coli isolates that exhibit an antibiotic efflux mechanism (24). The N-terminal helix-turn-helix amino acid composition is more conserved, probably because it is fundamental to the function of DNA binding and determines its specificity. The MarR sequence is not particularly conserved, perhaps because it is specifically active on marO and differences do not influence its function.
In this study, we identified the E. aerogenes mar operon and characterized the role of MarA in the induction of an MDR phenotype. Clinical E. aerogenes isolates that have a decreased susceptibility to cephalosporins associated with a loss of the major porins and an active efflux are frequently found. A MarA-mediated response could support the in vivo selection of such resistant E. aerogenes strains. Further studies are now necessary to evaluate the distribution of MarA expression levels in clinical strains and characterize the role of potential inducers in the regulation of the mar cascade.
Nucleotide sequence accession number.
The nucleotide sequence of the marORAB-Ea locus has been submitted to the EMBL database and has been given accession no. AJ404624.
Acknowledgments
We thank E. Pradel for helpful advice concerning the molecular characterization of the mar operon. We thank P. F. Miller for generously providing plasmid p9 and D. Parzy and M. Torentino (Parasitologie, Institut de Médecine Tropicale du Service de Santé des Armées) for help in sequencing.
This work was supported by the Université de la Méditerranée, the Institut National de la Santé et de la Recherche Médicale, and the Assistance Publique à Marseille (AORC 1998).
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. E. Brent, D. D. Kingston, J. G. Moore, J. A. Seidman, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornet, C., A. Davin-Regli, C. Bosi, J. M. Pagès, and C. Bollet. 2000. Fast regulation of outer membrane permeability synthesis originated imipenem resistance of Enterobacter aerogenes prevalent type strains in France. J. Clin. Microbiol. 38:1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosi, C., A. Davin-Regli, C. Bornet, M. Malléa, J. M. Pagès, and C. Bollet. 1999. Most Enterobacter aerogenes strains in France belong to a prevalent clone. J. Clin. Microbiol. 37:2165-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charrel, R. N., J. M. Pagès, P. De Micco, and M. Malléa. 1996. Prevalence of outer membrane porin alteration in β-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob. Agents Chemother. 40:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevalier, J., J. M. Pagès, and M. Malléa. 1999. In vivo modification of porin activity conferring antibiotic resistance to Enterobacter aerogenes. Biochem. Biophys. Res. Commun. 266:248-251. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, S. P., Y. William, and S. B. Levy. 1993. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J. Infect. Dis. 168:484-488. [DOI] [PubMed] [Google Scholar]
- 10.Davin-Regli, A., D. Monnet, P. Saux, C. Bosi, R. N. Charrel, A. Barthelemy, and C. Bollet. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J. Clin. Microbiol. 34:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dé, E., A. Basle, M. Jaquinod, N. Saint, M. Mallea, G. Molle, and J.-M. Pagès. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 12.Gambino, L., S. J. Gracheck, and P. F. Miller. 1993. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 175:2888-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunonga, N. I., R. J. Sobieski, and S. S. Crupper. 2000. Prevalence of the multiple antibiotic resistance operon (marRAB) in the genus Salmonella. FEMS Microbiol. Lett. 187:155-160. [DOI] [PubMed] [Google Scholar]
- 14.Linde, H. J., F. Notka, M. Metz, B. Kochanowski, P. Heisig, and N. Lehn. 2000. In vivo increase in resistance to ciprofloxacin in Escherichia coli associated with deletion of the C-terminal part of MarR. Antimicrob. Agents Chemother. 44:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malléa, M., V. Simonet, L. Eun-Hee, E. Collatz, R. Gervier, L. Gutmann, and J. M. Pagès. 1995. Biological and immunological comparisons of Enterobacter cloacae and Escherichia coli porins. FEMS Microbiol. Lett. 129:273-280. [DOI] [PubMed] [Google Scholar]
- 16.Malléa, M., J. Chevalier, C. Bornet, A. Eyraud, A. Davin-Regli, C. Bollet, and J. M. Pagès. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003-3009. [DOI] [PubMed] [Google Scholar]
- 17.Maneewannakul, K., and S. B. Levy. 1996. Identification of mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, R. G., and J. L. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oethinger, M., L. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oethinger, M., W. V. Kern, A. S. Jellen-Ritter, L. M. McMurry, and S. B. Levy. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitout, J. D. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, P. Coudron, and C. C. Sanders. 1998. Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob. Agents Chemother. 42:596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davis. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulavik, M. C., M. Dazer, and P. F. Miller. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179:1857-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webber, M. A., and L. J. V. Piddock. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]