Abstract
Mutations in the YMDD motif of the hepatitis B virus (HBV) DNA polymerase result in reduced susceptibility of HBV to inhibition by lamivudine, at a cost in replication fitness. The mechanisms underlying the effects of YMDD mutations on replication fitness were investigated using both a cell-based viral replication system and an in vitro enzyme assay to examine wild-type (wt) and YMDD-mutant polymerases. We calculated the affinities of wt and YMDD-mutant polymerases for each natural deoxyribonucleoside triphosphate (dNTP) and determined the intracellular concentrations of each dNTP in HepG2 cells under conditions that support HBV replication. In addition, inhibition constants for lamivudine triphosphate were determined for wt and YMDD-mutant polymerases. Relative to wt HBV polymerase, each of the YMDD-mutant polymerases showed increased apparent Km values for the natural dNTP substrates, indicating decreased affinities for these substrates, as well as increased Ki values for lamivudine triphosphate, indicating decreased affinity for the drug. The effect of the differences in apparent Km values between YMDD-mutant polymerase and wt HBV polymerase could be masked by high levels of dNTP substrates (>20 μM). However, assays using dNTP concentrations equivalent to those measured in HepG2 cells under physiological conditions showed decreased enzymatic activity of YMDD-mutant polymerases relative to wt polymerase. Therefore, the decrease in replication fitness of YMDD-mutant HBV strains results from the lower affinities (increased Km values) of the YMDD-mutant polymerases for the natural dNTP substrates and physiological intracellular concentrations of dNTPs that are limiting for the replication of YMDD-mutant HBV strains.
The YMDD motif (tyrosine, methionine, aspartate, aspartate) is a highly conserved amino acid sequence involved in deoxynucleoside triphosphate (dNTP) binding in the catalytic site of a number of RNA-dependent DNA polymerases, including hepatitis B virus (HBV) DNA polymerase (38). During lamivudine therapy for treatment of chronic HBV infection, diminished therapeutic responses may occur in some patients due to the emergence of mutant HBV species containing amino acid substitutions in the YMDD motif and in the proximal FLLAQ motif (phenylalanine, leucine, leucine, alanine, glutamine) of the viral polymerase (1, 9, 10, 19, 30, 46). The amino acid changes in the HBV polymerase that affect lamivudine sensitivity involve the methionine at position 552 (M552) within the YMDD motif and the proximal leucine at position 528 (L528) in the upstream FLLAQ motif. The substitutions observed in clinical isolates at M552 include the hydrophobic amino acid substitutions isoleucine (designated M552I or MI) and valine (designated M552V or MV). Although the M552I mutant has been observed in clinical isolates both as a single substitution (MI) and in combination with L528M (LMMI), the M552V substitution is observed in clinical isolates almost exclusively in combination with L528M (LMMV) (1, 10, 18, 25, 31, 46, 53, 54).
Changes in the YMDD motif alter viral sensitivity to lamivudine in cell culture and affect viral replication, even in the absence of lamivudine. In cell culture, YMDD-mutant strains show a wide range of sensitivities to lamivudine, with the decrease in sensitivity relative to wild-type (wt) polymerase varying with the specific amino acid change(s): LM < MV ≪ MI, LMMV, or LMMI (1, 16, 23, 24, 34). HBV strains with a single M552V or M552I substitution in the YMDD motif not only show decreased sensitivity to lamivudine but also display reduced replication competence, yielding lower levels of virus. A substitution of methionine at L528 in the upstream FLLAQ motif appears to modulate the impact of amino acid substitutions in the YMDD motif on both lamivudine sensitivity and viral fitness (15, 23, 24, 26, 28, 32, 35). The impact of the L528M substitution on both drug sensitivity and replication fitness differs depending on the specific substitution at M552 (i.e., M552V or M552I).
Similar substitutions of the methionine at position 184 in the YMDD motif of the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase are reported to produce viral species that exhibit both reduced sensitivity to lamivudine and lower replication efficiency in vitro (4, 5). These studies demonstrated that the replication defect in HIV-1 reverse transcriptase containing the M184V substitution was exacerbated at low cellular concentrations of dNTPs. Reduced replication efficiency was also reported for YMDD-mutant HBV polymerases in cells treated with hydroxyurea, which depletes intracellular dNTP levels (28).
Allen et al. (1) hypothesized that changes in the catalytic domain of HBV polymerase resulting from substitutions at M552 and L528 would alter the affinity of the enzyme for dNTP substrates, thus affecting replication efficiency. In this report, enzyme kinetic data were derived for both wt and YMDD-mutant polymerase and used to assess the affinity of each of the polymerases for the natural dNTPs. In addition, inhibition constants for lamivudine triphosphate were determined for wt and YMDD-mutant polymerases. The dNTP pools in cells that supported the replication of wt and YMDD-mutant HBV strains were also determined.
MATERIALS AND METHODS
Cell culture and reagents.
HepG2 cells were grown in medium consisting of Dulbecco's modified Eagle medium containing 4,500 mg of d-glucose per liter and 4 mg of pyridoxine hydrochloride per liter supplemented with 2 mM glutamine, 10 μmol of nonessential amino acids per liter and 10% fetal bovine serum (FBS) (HyClone, Logan, Utah). Cultures of 2.2.15 cells (a derivative line of HepG2 cells with stable integration of four tandem copies of the complete HBV genome [45]) were grown in RPMI 1640 medium supplemented with 2 mM glutamine and 10% heat-inactivated FBS. Cell cultures were grown in 75-cm2 flasks (catalogue no. 3376; Corning) and maintained at 37°C in a humidified incubator with 5% CO2.
Lamivudine triphosphate was obtained from Inspire Pharmaceuticals (Durham, N.C.). Cell media were obtained from Gibco (Grand Island, N.Y.) except as noted, and reagents and chemicals were obtained from Sigma (St. Louis, Mo.) unless noted otherwise.
Construction of recombinant plasmids.
The wt HBV genome was derived from pCMVhbv (14), which was kindly provided by Christoph Seeger (Fox Chase Cancer Center, Philadelphia, Pa.). Plasmids carrying the HBV genome with YMDD substitutions have been previously described (1). The recombinant shuttle vectors used to construct baculoviruses for delivery of HBV genomes to mammalian cells were created by subcloning a 3.6-kb SnaBI to NotI fragment containing the wt or YMDD-mutant HBV genome into the SnaBI and NotI sites of the pFastBacMam vector, creating pFBMamWT, pFBMamMV, pFBMamLM, pFBMamMI, pFBMamLMMI, and pFBMamLMMV (11).
The convention used for numbering nucleotides and amino acids is based on HBV genotype A (17, 33, 49, 51).
Construction and culture of recombinant baculovirus.
The recombinant baculoviruses were generated using the Bac-To-Bac system (Life Technologies, Grand Island, N.Y.). The viruses were amplified in Sf9 (Spodoptera frugiperda) cells grown in suspension in Grace's insect media (Life Technologies) supplemented with 10% FBS (HyClone), 0.1% Pluronic F-68, and 25 μg of gentamicin per ml. Virus titers were determined by plaque assay (36).
Transfection of HepG2 cells with recombinant plasmid and isolation of core particles.
HepG2 cells in 225-cm2 flasks (catalogue no. 3001; Corning) were transfected with wt and mutant HBV-producing plasmids as previously described (1). HBV core particles were isolated by a modification of a previously described procedure (12). Briefly, 5 days posttransfection the cells were harvested in situ by lysing the monolayer in a hypotonic buffer containing 0.25 mol of sucrose per liter, 3 mM dithiothreitol, and 0.5% NP-40 (Pierce, Rockford, Md.). The cell debris was pelleted, and the cell supernatant was digested with 0.005 μg of RNase A per ml and 6 U of DNase I per ml. The treated lysate was pelleted through a sucrose step gradient: (0.3, 0.6, and 0.9 mol of sucrose per liter) in 50 mM Tris-HCl (pH 7.5)-50 mM KCl-5 mM MgCl2 (TKM buffer). The resulting pellet containing HBV core particles was resuspended by sonication in TKM buffer with 0.1% NP-40 and stored at −80°C.
Transduction of HepG2 cells with recombinant baculovirus and isolation of core particles.
For transduction of HepG2 cells with the recombinant baculovirus, HepG2 cells were seeded at a density of ∼8 × 104 cells/cm2 in T-225 flasks and incubated for 24 h. The cell monolayer was washed twice with phosphate-buffered saline, and recombinant baculovirus was added at a multiplicity of infection of 10 to 50 PFU/cell in 10 ml of Sf9 medium. The cells were rocked gently for 1 h at room temperature, the inoculum was removed by aspiration, and 75 ml of HepG2 medium was added. The cells were incubated for 5 days, and core particles were harvested as described above for transfected cells.
Quantification of core particles by PCR.
Yields of extracellular viral particles from transfected HepG2 cells were quantified by a modification of a previously described PCR-based method (21). Intracellular core particles were quantified as previously described (18). Briefly, the HBV DNA within intracellular core particles was extracted, resuspended in 200 μl of 10 mM Tris-HCl buffer (pH 8.0), and quantified by PCR. With both procedures, the PCR primers used to amplify HBV DNA were EHBV372, 5′-TCGCTGGATGTGTCTGCGGCGTTTTAT-3′, complementary to the minus strand at nucleotides (nt) 372 to 398, and EHBV460, 5′-TAGAGGACAAACGGGCAACATACC-3′, complementary to the plus strand at nt 460 to 483. HBV DNA that has been replicated to a minimum of the first 1,457 nt (1829 to 372) of the minus strand is detected by this procedure.
Quantification of core particles from transduced cells by slot blot hybridization.
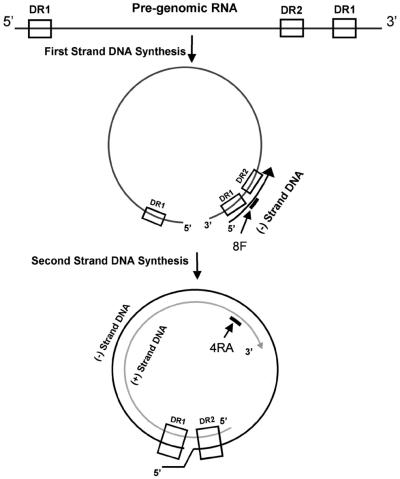
Plasmid pGEMX-HBV (18) was used as a control and diluted in Tris-EDTA to provide a series of seven samples ranging from 1,000 to 16 pg of HBV DNA/slot (1 pg of double-stranded HBV DNA is ∼3 × 105 genome equivalents). Ten microliters of control samples or core particle preparations were denatured in a final volume of 100 μl of 0.4 M NaOH, 0.01 M Na-EDTA at 100°C for 10 min. The samples were transferred to a Brightstar-Plus membrane (Ambion, Austin, Tex.) by vacuum blot using a Millipore MilliBlot-S system (Millipore Corp., Bedford, Mass.). The membrane was neutralized in 2× SSC (0.3 mM NaCl, 0.03 mM sodium citrate, pH 7.0), and the nucleic acids were covalently linked to the membrane by the UV-Stratalinker 2400 in auto-cross-link mode (Stratagene, La Jolla, Calif.). The membrane was hybridized for 1 h at 42°C in Rapid-Hyb buffer (Amersham Pharmacia Biotech, Piscataway, N.J.) containing 2 × 107 cpm of radiolabeled probe 8F or 4RA. The radiolabeled probe was prepared with the 3′-end-labeling kit (Amersham Pharmacia Biotech) with [33P]dATP (3,000 Ci/mmol; NEN Life Science Products, Boston, Mass.). Probe 8F (5′-ACGTTGCATGGAGACCAC-3′) is complementary to nt 1603 to 1620 of the HBV genome and therefore detects genomes that have replicated a minimum of 226 nt of the HBV minus strand (Fig. 1). Probe 4RA (5′-TGAGGCATAGCAGCAGGATGA-3′) hybridizes to nt 410 to 430 of the HBV plus strand. Probe 4RA detects genomes that have replicated the entire HBV minus strand and a minimum of 2,000 bases of the HBV plus strand (Fig. 1).
FIG. 1.
Schematic of HBV genome replication. Pregenomic RNA is shown as a linear molecule, with the locations of direct repeats 1 (DR1) and 2 (DR2) indicated. After initiation of replication at the 5′ end of the pregenomic RNA and translocation to the direct repeat 1 (DR1) at the 3′ end, elongation continues and produces the minus-strand DNA. Second-strand synthesis [producing the DNA plus strand] occurs after circularization of the minus strand. Probes 8F and 4RA are indicated by the heavy black lines.
After hybridization the membrane was washed with 0.5% sodium dodecyl sulfate in 1× SSPE (0.15 M NaCl, 0.01 M Na2HPO4 [pH 7.4], 0.001 M Na-EDTA) for one 20-min wash at 22°C, followed by two 15-min washes at 42°C. The membrane was exposed to a Molecular Dynamics Storage Phosphor Screen (Molecular Dynamics, Sunnyvale, Calif.) for 18 h. The exposed screen was scanned by a STORM 860 instrument (Molecular Dynamics), and densitometric analysis was performed with ImageQuant Software (Molecular Dynamics).
For comparison of the ratios of early-to-late replication complexes within each core particle preparation, the core particle preparations were diluted to equivalency with respect to hybridization with probe 8F.
Preparation of HBV core antibody capture plates.
Antibody capture plates were prepared by sequentially treating 96-well plates (catalogue no. 3632; Costar, Corning, N.Y.) at 22°C as follows: (i) 2 h in 0.15 ml of 0.1 mg of neutravidin (Pierce) per ml in 50 mM sodium carbonate (pH 9.6); (ii) 1 h in 0.15 ml of biotinylated goat anti-rabbit immunoglobulin G (Pierce) diluted to 0.0015 mg/ml in wash buffer (100 mM sodium phosphate [pH 7.2], 150 mM NaCl, 0.02% Tween 20 [Pierce], 0.02% sodium azide, and 0.01% bovine serum albumin); and (iii) 1 h in 0.15 ml of rabbit anti-HBV core antigen antibody (Zymed, San Francisco, Calif.) diluted 1:300 in wash buffer. After each incubation, the plates were washed three times with 0.25 ml of wash buffer. Wash buffer (0.25 ml) was then added to each well, and the plates were stored in a humidified chamber at 4°C until needed. Similar plates were prepared with 1.5 μg of biotinylated goat anti-mouse immunoglobulin G (Pierce) per ml and 0.67 μg of mouse anti-HBV core antigen monoclonal antibody (clone H3A4; Research Diagnostics, Inc., Flanders, N.J.) per ml in wash buffer.
HBV core polymerase assays.
The core polymerase assay was modified from a previously described protocol (12). The assay was performed in 96-well plates, and the reaction mixture consisted of 107 to 108 core particles in 33 mM Tris-HCl (pH 7.5), 230 mM KCl, 8 mM MgCl2, 5 mM dithiothreitol, 0.2% NP-40, 7 U of pyruvate kinase (Sigma) per ml, 2 mM phosphoenolpyruvate (Sigma), 35 μM unlabeled dNTPs (Amersham Pharmacia Biotech), and 0.001 to 4 μM α-33P-labeled dNTP (2,000 to 3,000 Ci/mmol; NEN) in a final volume of 10 μl. After termination with 90 μl of stop buffer (50 mM Tris-HCl [pH 7.4], 50 mM KCl, 5 mM MgCl2, 0.05% Tween 20, 2 mM sodium pyrophosphate, 0.02% sodium azide), the reaction mixtures were transferred to a 96-well antibody capture plate and incubated at room temperature with shaking for 2 h. The wells were then washed five times with 200 μl of wash buffer (100 mM sodium phosphate [pH 7.2], 150 mM NaCl, 0.05% Tween 20, 2 mM sodium pyrophosphate, 0.02% sodium azide). After the final wash, 170 μl of Optiphase Supermix (Wallac Inc., Gaithersburg, Md.) was added to each well and the radioactivity in the plates was determined by use of a Wallac 1450 Microbeta plus counter.
Extraction of dNTPs and calculation of intracellular concentrations in HepG2 and 2.2.15 cells.
For extractions of dNTPs, HepG2 or 2.2.15 cells were plated in triplicate in 25-cm2 flasks (catalogue no. 3056; Corning) and treated as described below. Cells from two flasks were used for dNTP extractions. Cells in the third flask were trypsinised, and the number of cells in the monolayer was determined by cell count.
dNTPs were extracted from HepG2 cells that had undergone mock transfections (transfections with plasmids containing no HBV sequences). To mimic conditions under which core particles or extracellular viruses were harvested, cells were mock-transfected and (for core particles) refed on day 1, with dNTPs extracted 4 days later, or (for extracellular viruses) refed on days 1, 3, and 5, with dNTPs extracted 2 days after each feeding (days 3, 5, and 7 after transfection). Nontransfected HepG2 cells were also plated at low density (1.6 × 104 cells/cm2) and refed on days 1, 3, 5, and 7, with dNTPs extracted 2 days after each feeding (days 3, 5, 7, and 9 after the initial plating). For dNTP extraction, the HBV-producing cell line 2.2.15 was plated at a density of 104 cells/cm2 in complete media containing 10% FBS and refed with the same media on day 4 and with complete media containing 1% FBS on days 7 and 11. Extractions of dNTPs from the 2.2.15 cells were on days 4, 7, 11, and 15 after plating. Cellular dNTPs were extracted as previously described (37).
The extracts were analyzed by a method using a modification of the polymerase assay previously described (47), in a final volume of 10 μl. The reaction was stopped by adding 5 μl of stop mix containing 250 mM Na-EDTA and 50 mM sodium pyrophosphate. Three microliters of the reaction mixture was spotted onto DE81 paper (Whatman Inc., Clifton, N.J.), dried briefly, washed five times for 10 min each in 5% Na2HPO4, once in distilled water, and once in 95% ethanol. The paper was dried and exposed to a Molecular Dynamics Storage Phosphor Screen, which was then scanned using a STORM 860 instrument. Densitometric analysis was performed with ImageQuant Software. The total amount of dNTP in each extract was calculated, and the intracellular concentration was determined, assuming a cell volume of 2.6 μl/106 cells for both HepG2 and 2.2.15 cells (37).
Calculation of kinetic constants.
Km values were calculated using the Michaelis-Menten equation. For calculating 50% inhibitory concentrations (IC50s), concentration-response curves were fit to the Hill equation. Ki values were calculated by using the equation Ki = IC50/(1 + [S]/Km), where IC50 is the lamivudine triphosphate concentration resulting in 50% enzyme inhibition and [S] is the concentration of [33P]dCTP. Vmax values for core polymerase activities in assays in which [33P]dNTP was kept at a constant concentration while concentrations of the other three dNTPs varied were calculated by using the following equation: v = (Vmax × S)/(Km + S) + Y, where v is the observed enzyme velocity, S is the cellular concentration of 33P-labeled dNTP, and Y is the amount of [33P]dNTP incorporated in the absence of the other three dNTPs. Data were fit to the above equations by using Robosage, a Microsoft EXCEL (Redmond, Wash.) add-in program developed at GlaxoSmithKline (M. W. Lutz, personal communication). The nonlinear optimization function in Robosage is based on the Marquardt-Levenberg algorithm. In all cases, unweighted nonlinear least-squares regression was used to perform the fit.
RESULTS
Viral yield following transfection or transduction with wt or YMDD-mutant HBV.
In an initial characterization of the YMDD-mutant HBV, the effect of amino acid substitutions in the YMDD region of the HBV polymerase on the production of viral particles in transfected or transduced cells was assessed. HepG2 cells were transfected with plasmids containing wt or YMDD-mutant HBV, and mature, enveloped viral particles were harvested at 7 days posttransfection and quantified by HBs-antibody capture and PCR detection. Cytoplasmic core particles containing replicative intermediates were also harvested, partially purified, and quantified. Under identical transfection and culture conditions, each of the YMDD-mutant polymerases produced lesser amounts of both immature intracellular viral particles and mature extracellular virus (Table 1). The relative yields obtained from the wt and YMDD-mutant HBV strains were similar for extracellular virus and for intracellular particles. While none of the YMDD mutants had a replication efficiency as high as that of wt, YMDD-mutant HBV polymerases containing the L528M substitution showed less reduction in replication efficiency than did YMDD-mutant polymerases without the substitution. Transductions of HepG2 cells with baculovirus containing identical wt and mutant HBV genome inserts gave similar results, except that absolute yields per transduction were ∼50- to 100-fold higher (data not shown).
TABLE 1.
Yields of extracellular and intracellular HBV DNA in transfected culturesa
| Viral construct | Extracellular HBV DNA
|
Intracellular HBV DNA
|
||
|---|---|---|---|---|
| DNA (pg/ml) | Fold decrease from wt | DNA (ng/flask) | Fold decrease from wt | |
| wt | 23.6 | NA | 27.9 | NA |
| LMMV | 4.8 | ∼5 | 8.0 | ∼3 |
| MI | 2.4 | ∼10 | 4.2 | ∼7 |
| LMMI | 6.8 | ∼3 | 12.6 | ∼2 |
| LM | 7.5 | ∼3 | 14.6 | ∼2 |
| MV | 2.9 | ∼8 | 3.0 | ∼9 |
Average of three to four assays; SE < 10 to 20%; abbreviations: NA, not applicable; LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I; LM, L528M; MV, M552V.
Kinetic parameters of wt and mutant HBV polymerases.
Enzyme kinetic parameters of wt and YMDD-mutant polymerases were calculated to determine the effects of the amino acid substitutions on polymerase activity. The wt and YMDD-mutant polymerases were assayed in their native state within subviral core particles, and the apparent Km of each of the four natural dNTP substrates was measured for each polymerase. As shown in Table 2, the apparent Km values of each of the four natural dNTP substrates for the YMDD-mutant polymerases were greater than the corresponding values for wt HBV polymerase, indicating that the YMDD-mutant polymerases require higher dNTP concentrations to reach maximum velocity.
TABLE 2.
Mean Km values for each dNTP determined by using wt and YMDD-mutant HBV polymerasesa
| HBV polymerase | Mean dNTP Km values (μM) ± SEM
|
|||
|---|---|---|---|---|
| dCTP | dATP | dGTP | TTP | |
| wt | 0.09 ± 0.01 | 0.18 ± 0.02 | 0.07 ± 0.01 | 0.08 ± 0.01 |
| LMMV | 0.16 ± 0.03 | 0.53 ± 0.1 | 0.16 ± 0.04 | 0.23 ± 0.06 |
| MI | 0.30 ± 0.06 | 0.79 ± 0.18 | 0.35 ± 0.10 | 0.42 ± 0.08 |
| LMMI | 0.24 ± 0.09 | 0.74 ± 0.07 | 0.18 ± 0.03 | 0.22 ± 0.02 |
| LM | 0.12 ± 0.01 | 0.40 ± 0.03 | 0.11 ± 0.03 | 0.19 ± 0.01 |
| MV | 0.32 ± 0.08 | 1.15 ± 0.18 | 0.29 ± 0.09 | 0.38 ± 0.06 |
Mean values are based on determinations from four to six core particle assays from transfected or transduced cells; abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I; LM, L528M; MV, M552V.
While the core polymerase assay indicated that the apparent Km values of the mutants deviated from those of the wild type, it was necessary to confirm that the assay could detect the differences between wt and mutant polymerases with respect to their interactions with lamivudine triphosphate, the active form of lamivudine. Therefore, a series of assays was undertaken to determine the Ki (inhibition constant) of lamivudine-triphosphate for wt and each mutant polymerase. Each core polymerase reaction contained [33P]dCTP at the apparent Km value calculated for that polymerase, the other three dNTPs at 35 μM (a concentration chosen to be 30-fold higher than the highest apparent Km value), and lamivudine triphosphate at concentrations ranging from 0.001 μM to 2 mM.
Ki values for lamivudine triphosphate calculated from the core polymerase assays were compared to previously reported IC50s for lamivudine obtained from cell-based assays in this laboratory (1). As expected, the YMDD-mutant polymerases were less sensitive to inhibition by lamivudine triphosphate than was the wt enzyme. Notably, the range of Ki values and the ranking of wt and YMDD-mutant polymerases were similar to the range and the ranking of the YMDD-mutant polymerase IC50s (Table 3). In both systems, the LM and MV mutant polymerases were much closer to wt polymerase in terms of sensitivity to lamivudine than were the mutants LMMV, MI, and LMMI, each of which showed a decrease in sensitivity of ∼1,000-fold for the Ki of lamivudine triphosphate and ∼10,000-fold for the IC50 of lamivudine.
TABLE 3.
Comparison of wt and YMDD-mutant polymerases based on IC50s for lamivudine and on Ki values for lamivudine triphosphatea
| Polymerase | Lamivudineb
|
Lamivudine Triphosphatec
|
||
|---|---|---|---|---|
| IC50 (μM) | Fold increase relative to wt | Ki (μM) | Fold increase relative to wt | |
| wt | 0.049 | NA | 0.032 | NA |
| LMMV | >200 | >10,000 | 37 | 1,100 |
| MI | >200 | >10,000 | 31 | 970 |
| LMMI | >200 | >10,000 | 43 | 1,300 |
| LM | 0.9 | 18 | 0.255 | 8 |
| MV | 7.5 | 153 | 1.8 | 55 |
Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I; LM, L528M; MV, M552V; NA, not applicable.
Data from Allen et al. (1).
Data shown are averages of three assays. Standard error of the mean is <10%.
The determination of Km or Ki is not dependent on the amount of enzyme present, but enzyme concentration is necessary to calculate kcat, the catalytic efficiency or turnover rate, which is the amount of substrate converted per unit of time per molecule of enzyme. Since the polymerase enzyme is covalently bound to the primer template during replication (42), measuring the number of molecules of minus-strand DNA with a minimum length of 200 nt was taken as a surrogate measure of active HBV polymerase in a polymerase reaction. Since the minus-strand DNA molecules that have replicated to a minimum length of 200 nt may not all remain enzymatically active, this calculation may result in an underestimation of the actual rate of incorporation. The amount of newly replicated minus strand was quantitated by hybridization of radiolabeled oligonucleotide probe 8F (Fig. 1). The turnover rate, kcat, was then calculated as the number of molecules of a radiolabeled dNTP that were incorporated per minute in a polymerase reaction (determined from the specific activity of radioisotope in the reaction) when all four dNTPs were at saturating concentrations, divided by the number of HBV polymerase molecules (calculated by a surrogate determination based on hybridization to the 8F probe). For a given dNTP substrate, there were no appreciable differences between any of the HBV polymerases in the kcat values (Table 4). For each of the polymerases assayed, the average incorporation of dNTPs at Vmax (saturating conditions for all dNTPs) was approximately 5 ± 0.4 molecules of dNTPs per min (mean ± standard deviation), calculated as the sum of the incorporation rates of all four dNTPs. This result indicates that there are no appreciable differences in the catalytic efficiency of the wt and YMDD-mutant polymerases at nonlimiting dNTP levels. If this method overestimates the number of active polymerase molecules, the absolute rates may be higher, but the relative rates at high dNTP concentrations will be unaffected. By comparison, the incorporation rate of the HIV-1 reverse transcriptase has been calculated to be between 1 and 3 molecules of dNTPs per s (8, 13), or 60 to 180 molecules per min.
TABLE 4.
Estimated kcat values of wt and YMDD-mutant HBV polymerasesa
| Polymeraseb | Rate of incorporation of each dNTP (no. of molecules/min/enzyme)
|
Rate of total incorporation (no. of molecules of all dNTPs/min) | |||
|---|---|---|---|---|---|
| dATP | dGTP | dCTP | TTP | ||
| wt | 1.2 | 0.9 | 1.0 | 0.8 | 3.9 |
| LMMV | 1.6 | 1.1 | 1.2 | 0.9 | 4.9 |
| MI | 1.2 | 0.8 | 1.1 | 0.8 | 3.9 |
| LMMI | 2.0 | 1.5 | 1.7 | 1.3 | 6.5 |
| LM | 2.1 | 1.2 | 1.1 | 1.4 | 5.8 |
| MV | 1.9 | 0.9 | 1.2 | 0.9 | 4.9 |
Values based on the assumption that the number of active DNA polymerase molecules is represented by the number of (−) strands that have completed synthesis to at least 200 nt.
Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I; LM, L528M; MV, M552V.
Whereas kcat is determined under saturating conditions, the catalytic specificity, kcat/Km, is a measurement of enzyme activity at limiting substrate concentrations. Specifically it measures the turnover rate of the enzyme at a concentration of substrate sufficient for the enzyme to proceed at only 50% of the maximum rate. When catalytic specificity was calculated by using the apparent Km and kcat values previously determined, the YMDD-mutant polymerases showed a characteristic decrease associated with the presence of specific amino acid substitutions, indicating that they are not as active as wt polymerase at relatively low substrate concentrations (Table 5). The relative differences in activity between the polymerases were calculated by normalizing the value for wt polymerase to 100% and expressing the activities of the YMDD-mutant polymerases as a percentage of wt activity (Table 6). Taken together, these kinetic results indicate that the enzymatic activities of YMDD-mutant polymerases decrease relative to the activity of wt polymerase as the concentrations of normal dNTPs decrease.
TABLE 5.
Catalytic specificities (kcat/Km) for wt and YMDD-mutant polymerases, determined by core polymerase assays using one labeled dNTP at Km and unlabeled dNTPs at 35 μM
| HBV polymerasea |
kcat/Km valuesb for each dNTP (turnover rate/μM)
|
|||
|---|---|---|---|---|
| dATP | dGTP | dCTP | TTP | |
| wt | 6.6 | 13.3 | 10.9 | 9.8 |
| LMMV | 3.0 | 7.0 | 7.7 | 4.1 |
| MI | 1.5 | 2.3 | 3.8 | 1.9 |
| LMMI | 2.6 | 8.5 | 6.9 | 6.0 |
| LM | 5.4 | 10.5 | 9.1 | 7.3 |
| MV | 1.6 | 3.2 | 3.7 | 2.4 |
Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I; LM, L528M; MV, M552V.
Values based on the assumption that the number of active DNA polymerase molecules is represented by the number of (−) strands that have completed synthesis to at least 200 nt.
TABLE 6.
Enzymatic activities of YMDD-mutant polymerases relative to wt polymerase, calculated at low substrate concentrations
| HBV polymerasea | % Enzyme activity relative to wt polymerase
|
|||
|---|---|---|---|---|
| dATP | dGTP | dCTP | TTP | |
| wt | 100 | 100 | 100 | 100 |
| LMMV | 46 | 53 | 71 | 42 |
| MI | 23 | 17 | 35 | 19 |
| LMMI | 40 | 64 | 63 | 62 |
| LM | 82 | 79 | 83 | 75 |
| MV | 25 | 24 | 34 | 25 |
Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I; LM, L528M; MV, M552V
Intracellular dNTP pool sizes.
To investigate whether the concentration of intracellular dNTPs might influence the replication fitness of YMDD-mutant polymerases, the concentrations of dNTP pools within HepG2 and 2.2.15 cells under various growth conditions were determined. The conditions chosen for dNTP extraction included both normal growing conditions for HepG2 cells and growing conditions used to produce virus and core particles from transfected HepG2 cells or virus from 2.2.15 cells. Control experiments indicated that the cellular extracts were not inhibitory to the activity of the Klenow polymerase under the assay conditions used (data not shown).
With the exception of [TTP] (12.4 μM) in normal HepG2 cells grown at the lowest cell density, dNTP concentrations ranged from <0.1 to ∼4 μM (Table 7). Although individual dNTP concentrations differed slightly between the various extracts, the results from the other extracts were similar (data not shown). Cultures with higher cell densities (i.e., confluent cultures) tended to have lower concentrations of dNTPs. These observations are consistent with results from other studies, in which it was shown that rapidly dividing cells have higher concentrations of dNTPs than confluent or slower growing cells (6, 7, 39, 40).
TABLE 7.
dNTP pools in nontransfected HepG2 cells, by day harvested
| Day harvested | dATP (μM) | dGTP (μM) | dCTP (μM) | TTP (μM) | Cell density (105/cm2) |
|---|---|---|---|---|---|
| 3 | 3.2 | 0.6 | 1.2 | 12.4 | 0.9 |
| 5 | 1.6 | 0.5 | 0.7 | 4.8 | 2.6 |
| 7 | 0.74 | 0.15 | 0.41 | 1.8 | 7.1 |
| 9 | 0.51 | 0.07 | 0.23 | 1.1 | 7.3 |
Core polymerase activity at limiting concentrations of dNTPs.
Both the Km and kcat constants were determined for wt and YMDD-mutant polymerases by measuring incorporation of a single radiolabeled dNTP in the presence of nonlimiting concentrations of the other three dNTPs. However, the catalytic specificity values (kcat/Km) predicted that at limiting substrate levels there should be easily measurable differences in polymerase activity between wt and YMDD-mutant polymerases. Based on measured apparent Km values, at 35 μM the nonlimiting concentration of dNTPs represents an excess over the Km value ranging from 30-fold for the highest Km calculated (MV, dATP, 1.15 μM) to >500-fold for the lowest Km (wt, dGTP, 0.07 μM). These concentrations ranged from 8-fold higher (TTP at 4 μM) to >350-fold higher (dGTP at <0.1 μM) than the intracellular concentrations of the corresponding dNTPs in HepG2 cells grown at high density. In order to examine polymerase activity under conditions that were closer to the conditions encountered in cultured liver-derived cells, assays were carried out over a range of dNTP concentrations bracketing the concentrations observed in HepG2 cells.
The midrange concentrations and relative amounts of dNTPs chosen were the highest levels found in HepG2 cells grown under the conditions used for harvesting core particles: dATP, 4.1 μM; dGTP, 2.0 μM; dCTP, 4.4 μM; and TTP, 4.6 μM. For one of the comparison assays, the concentration of [33P]dCTP was held at a constant 4.4 μM (>10 times the apparent Km for each polymerase) and the concentrations of the three unlabeled dNTPs were altered while their relative ratios were maintained (Table 8). The results indicated that wt polymerase is near its maximum velocity at the onefold concentrations, while the mutant polymerases require higher levels of dNTPs to achieve their maximum velocities. Moreover, the apparent activity differences between wt and YMDD-mutant polymerases increase as the levels of dNTPs decrease. In a slightly different assay the labeled dNTP was held constant at the apparent Km for each polymerase, thus normalizing the labeled dNTP concentration to kinetic equivalency. Therefore any kinetic differences between wt and mutant polymerases would be due solely to the levels of the other three substrates, as shown in Table 9 (note that the maximum rate calculated in this assay is still only one-half that of the true Vmax). Again it is apparent that even when the polymerases are kinetically normalized with respect to one substrate, the YMDD-mutant polymerases are not as efficient as wt polymerase when the concentrations of the other dNTPs approach levels seen in confluent HepG2 cells.
TABLE 8.
Activities of wt and YMDD-mutant HBV polymerases at 4.4 μM dCTP and limiting concentrations of dGTP, dATP, and TTPa
| Concn of dATP, dGTP, and TTP | % of HBV polymerase activity
|
||||
|---|---|---|---|---|---|
| Actual (μM) | Fold (relative to measured cellular dNTP levels) | wt | LMMV | MI | LMMI |
| 16.4, 8.0, 18.4 | 4 | 101.9 | 91.5 | 80.6 | 93.1 |
| 4.1, 2.0, 4.6 | 1 | 93.3 | 66.0 | 46.0 | 67.7 |
| 2.0, 1.0, 2.3 | 0.5 | 85.6 | 49.2 | 30.9 | 50.3 |
| 0.41, 0.2, 0.46 | 0.1 | 62.0 | 23.4 | 12.7 | 23.6 |
Enzymatic activities were determined as the mean values of three measurements and expressed relative to calculated Vmax values at nonlimiting dNTP concentrations. Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I.
TABLE 9.
Activities of wt and YMDD-mutant polymerases at low dNTP concentrations
| [dCTP] at Km (μM) | % Activity of calculated Vmax valuesa
|
|||
|---|---|---|---|---|
| wt | LMMV | MI | LMMI | |
| 10 | 102.7 | 93.2 | 85.6 | 92.9 |
| 3.16 | 96.1 | 76.0 | 60.7 | 76.0 |
| 1 | 87.6 | 54.6 | 41.0 | 56.0 |
| 0.316 | 71.0 | 35.1 | 24.0 | 35.0 |
Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I.
Indirect measurement of polymerization velocity by wt and YMDD-mutant HBV polymerases within subviral particles.
To assess relative velocities of wt and the three clinically significant YMDD-mutant polymerases included in this study, partially purified core particles of wt, LMMI, LMMV, or MI were isolated from HepG2 cells that had been transduced with baculovirus constructs and grown and harvested under conditions identical to those described above. Within each core particle preparation, the number of DNA molecules hybridizing to probe 8F (Fig. 1) was quantified as a direct measurement of molecules that had replicated a minimum of 200 nt. The various core particle preparations were diluted to equivalence with respect to the number of molecules hybridizing to probe 8F, and equivalent amounts of core particle preparations were hybridized to probe 4RA, as a measure of the number of replication complexes that had completed replication of the entire minus strand (∼3,000 bases) and approximately 2,000 bases of the HBV plus strand (Fig. 1). As shown in Table 10, a greater percentage of wt core particles have reached the later stage of replication, indicating that under identical conditions of replication the YMDD-mutant polymerases are unable to extend the replicating DNA to the same extent as wt polymerase. Notably, the rank order of the differences is the same as that predicted by the kinetic data: wt > LMMV; LMMI > MI.
TABLE 10.
Measurement of polymerization velocity of wt and YMDD-mutant polymerases, using probes specific for early and late HBV genomic replication
| HBV polymerasea | (−) strand probe 8F (107 molecules) | (+) strand probe 4RA (106 molecules) | % of polymerase HBV particles hybridizing to (+) strand probe 4RA | % Replication relative to wt polymerase |
|---|---|---|---|---|
| wt | 5.5 | 9.9 | 18 | 100 |
| LMMV | 5.6 | 4.9 | 9 | 50 |
| MI | 5.5 | 3.4 | 6 | 33 |
| LMMI | 4.9 | 4.6 | 10 | 55 |
Abbreviations: LMMV, L528M M552V; MI, M552I; LMMI, L528M M552I.
DISCUSSION
Variant species of HBV with reduced susceptibility to lamivudine emerge in chronically infected patients on long-term lamivudine therapy (1, 9, 10, 19, 30, 46). In these HBV species, the affinity of the viral polymerase for lamivudine triphosphate is decreased by the substitution of either valine or isoleucine for the methionine at position 552 in the conserved YMDD region of the polymerase. In many clinical isolates, an upstream methionine substitution at L528 also emerges, presumably as a compensatory mutation (15, 23, 24, 26, 28, 35). While these substitutions result in decreased viral susceptibility to lamivudine, HBV appears to pay a replicative price, as previous studies have indicated that HBV strains with YMDD-mutant polymerase replicate with lower efficiency than wt strains (1, 16, 23, 24, 34). Based on a computer model of the HBV polymerase, these amino acid changes were shown to be in the catalytic domain of the enzyme and could possibly influence interactions with the normal dNTP substrates as well as interactions with lamivudine triphosphate (1). One study indicated that not only were amino acid changes in the polymerase involved in decreased replication efficiency but also the concentrations of dNTPs within HBV-producing cells may play a role in diminishing replication efficiency (28). To examine the influence of dNTP concentrations on replication fitness of YMDD-mutant HBV strains, the levels and relative amounts of dNTPs were determined for cells that both supported HBV replication and exhibited differences in replication efficiency between wt and YMDD-mutants. The enzyme kinetics of the HBV polymerase reaction was measured in order to investigate the molecular interactions between the dNTPs and HBV polymerase, both wt and mutant.
The system chosen for kinetic analysis was one in which the HBV polymerase activity measured is in the native state. During replication the polymerase is enclosed within the HBV core particle, using the encapsidated HBV pregenomic RNA as a template for the RNA-directed DNA polymerase activity and the minus-strand DNA as a template for the DNA-directed DNA polymerase activity. We assayed the polymerase activity found within partially replicated subviral core particles isolated from liver-derived cells which support HBV replication. This assay system is in contrast to other systems which have used recombinant HBV polymerases to obtain kinetic data (44, 52). In highlighting the fact that the Ki values reported in their assay did not match the values that had been reported in a study using a cell-based system, Xiong et al. (52) noted that the enzymes studied were partially purified recombinant protein preparations isolated from insect cells and that the primer template was activated calf thymus DNA, rather than the normal HBV template (K. P. Fischer, Letter, Hepatology 29:996, 1999; X. Xiong, Letter, Hepatology 29:996, 1999). While the Ki values determined from the core polymerase assay show as much as a 1,000-fold difference between wt and YMDD-mutant polymerases (Table 3), the assay using isolated polymerase preparations derived from insect cells showed a maximum difference of only 20-fold between the polymerases (52). The core polymerase assay also discriminates between the two single M552 substitutions by demonstrating that MV is much less sensitive to lamivudine triphosphate than is MI, consistent with results previously reported in cell-based systems (1, 26, 34). However, the opposite conclusion was reached with the recombinant polymerase assay, suggesting that recombinant HBV polymerases produced in heterologous systems cannot discriminate between natural substrates or lamivudine triphosphate as well as core polymerases in the native state. Moreover, the kinetic data were derived by using linear regression analysis (44, 52), which is not as precise as the nonlinear regression analysis used in this study (described at http://www.curvefit.com/avoid_linearizing.htm).
Determination of enzyme kinetic constants for an enzymatic reaction gives insight into the nature of the reaction, especially the interaction between substrate and enzyme. In a simple enzymatic reaction with one substrate, the Km value is the concentration of substrate at which 50% of the enzyme is bound to the substrate (43). When the substrate concentration is equal to the Km value, the rate of the enzymatic reaction is 50% of the maximal rate, Vmax. During replication the HBV polymerase utilizes five substrates: four dNTPs and a template primer. The steady-state velocities for wt and mutant polymerases are the result of the complex interplay of all reactants and depend on the Km values of each dNTP, the concentrations of each dNTP, and the nucleotide sequence of the template primer. Consequently, the kinetic equation describing the velocity of the polymerase reaction is quite complex. To simplify the interpretation of the data presented here, an apparent Km value for each naturally occurring dNTP substrate has been defined as the concentration of the selected dNTP yielding 50% of the Vmax value when the other three dNTPs and template primer are at saturating concentrations.
The higher apparent Km values for the mutant polymerases determined by the core polymerase assay indicate that higher concentrations of dNTPs are required for activity equivalent to that of wt polymerase. One indication that the Km differences are involved in replication efficiency is the fact that the differences in apparent Kms between wt and mutant polymerases correlated with the change in yields and not the changes in lamivudine sensitivity. Because of the complexity of the actual polymerase reaction, as noted above, both the actual concentration of each dNTP and their relative amounts will have a profound effect on the observed velocity. Thus, it was important to determine the levels and relative amounts of dNTPs in cells that both supported HBV replication and exhibited differences in replication efficiency between wt and mutants.
dNTP pools were determined in HepG2 cells that had been grown under a variety of conditions: (i) mock production of extracellular virus, (ii) mock production of intracellular particles, and (iii) normal passage. Pools of dNTPs were also measured in the HepG2-derived 2.2.15 producer cell line under conditions that induce overexpression of extracellular virus. For the most part, maximum levels of dNTP concentrations were in the low micromolar range (<5 μM), with the highest concentrations observed at very low cell density or under conditions used for mock intracellular harvest. In most cases at high cell densities the concentrations of individual dNTPs were ≤1 μM. The levels of dCTP in HepG2 cells reported in this paper are similar to those reported recently (22). A review of the literature indicates that similarly low concentrations of dNTPs are found in liver or liver-derived cells from the rat or mouse (41, 48). Nakamura et al. (29) measured dNTP levels in mouse fetal liver and in mouse adult liver; the dNTP levels for mouse fetal liver reported by these investigators are similar in ratio and concentration to those measured in low-density, rapidly growing HepG2 cells after 3 days of growth (Table 7). By contrast, dNTP levels in the liver of the adult dam were lower (<0.4 μM) than those found in dense HepG2 monolayers. These data indicate that as HepG2 cells increase in density, intracellular dNTP levels decrease to levels that may approximate those in normal adult liver (2, 20, 27, 29, 41, 48, 50). One caveat to these calculations is that assessment of the dNTP pools was based upon a cellular average and does not reflect possible intracellular compartmentation. If there are two distinct dNTP pools, with cytoplasmic dNTP concentrations lower than those in the nucleus (3), then actual concentrations available for HBV replication may be lower than our estimates indicate.
A comparison of the catalytic specificities, kcat/Km, of wt and mutant polymerases indicated that at low levels of dNTPs the relative velocity of HBV polymerase would be lower for the mutants than for the wild type (Table 5). Polymerase assays were performed to see whether the levels of dNTPs measured in HepG2 cells were sufficiently low to detect differences between wt and mutant polymerases. As predicted by the enzyme kinetic data, there were marked differences between wt and mutant polymerases at low levels of dNTPs (Tables 8 and 9). The results were similar whether the nonlabeled dNTPs were equimolar, with the labeled dNTP at Km, or whether the concentrations of the nonlabeled dNTPs varied and the labeled dNTP was present in high concentration. The differences in polymerase activity between wt and mutant polymerases based on kcat/Km calculations are almost identical to the differences observed in polymerase assays performed at low dNTP levels.
The results of the kinetic experiments and polymerase assays at limiting levels of dNTP substrates indicated that the elongation rate of polymerases containing YMDD mutations would be slower than the elongation rate of wt polymerase in HepG2 cells. In order to test this hypothesis, the extent of DNA polymerization within core particles of wt and YMDD-mutant HBV grown in HepG2 cells was measured. The results showed that in a population of core particles that had polymerized at least the first 200 nt of DNA, a much greater number of wt core particles than of YMDD-mutant core particles had extended polymerization by at least 5,000 nt, which is twice as many wt core particles as LMMV or LMMI core particles and three times as many wt core particles as MI core particles.
In summary, the results presented in this paper confirm the lower replication efficiency of YMDD-mutant HBV strains and show that they have reduced fitness. The YMDD-mutant polymerases have a lowered affinity for the natural dNTP substrates used for viral DNA replication, and cells capable of supporting HBV replication have intracellular concentrations of dNTPs that are low enough to influence the differences in the enzymatic rates between YMDD-mutant polymerases and wt polymerase.
Acknowledgments
We thank Michael Lutz for advice on statistical analysis, David Porter and Misty Burnette for useful discussions, and Barbara J. Rutledge for editing assistance.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, and L. D. Condreay. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Arezzo, F. 1987. Determination of ribonucleoside triphosphates and deoxyribonucleoside triphosphates in Novikoff hepatoma cells by high-performance liquid chromatography. Anal. Biochem. 160:57-64. [DOI] [PubMed] [Google Scholar]
- 3.Aw, T. 2000. Intracellular compartmentation of organelles and gradients of low molecular weight species. Int. Rev. Cytol. 192:223-253. [DOI] [PubMed] [Google Scholar]
- 4.Back, N. K., and B. Berkhout. 1997. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 41:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi, V. 1998. Regulation of deoxynucleotide pools by substrate cycles. Adv. Exp. Med. Biol. 431:501-506. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi, V., S. Borella, C. Rampazzo, P. Ferraro, F. Calderazzo, L. C. Bianchi, S. Skog, and P. Reichard. 1997. Cell cycle-dependent metabolism of pyrimidine deoxynucleoside triphosphates in CEM cells. J. Biol. Chem. 272:16118-16124. [DOI] [PubMed] [Google Scholar]
- 8.Borroto-Esoda, K., and L. R. Boone. 1994. Development of a human immunodeficiency virus-1 in vitro DNA synthesis system to study reverse transcriptase inhibitors. Antivir. Res. 23:235-249. [DOI] [PubMed] [Google Scholar]
- 9.Buti, M., R. Jardi, M. Cotrina, F. Rodriguezfrias, R. Esteban, and J. Guardia. 1998. Transient emergence of hepatitis B variants in a patient with chronic hepatitis B resistant to lamivudine. J. Hepatol. 28:510-513. [DOI] [PubMed] [Google Scholar]
- 10.Chayama, K., Y. Suzuki, M. Kobayashi, M. Kobayashi, A. Tsubota, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, N. Murashima, K. Ikeda, and H. Kumada. 1998. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild-type after cessation of therapy. Hepatology 27:1711-1716. [DOI] [PubMed] [Google Scholar]
- 11.Condreay, J. P., S. M. Witherspoon, W. C. Clay, and T. A. Kost. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 96:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, M. G., J. E. Wilson, N. A. Vandraanen, W. H. Miller, G. A. Freeman, S. M. Daluge, F. L. Boyd, A. E. Aulabaugh, G. R. Painter, and L. R. Boone. 1996. DNA polymerase activity of hepatitis B virus particles—differential inhibition by L-enantiomers of nucleotide analogs. Antivir. Res. 30:133-145. [DOI] [PubMed] [Google Scholar]
- 13.Debyser, Z., A. M. Vandamme, R. Pauwels, M. Baba, J. Desmyter, and E. De Clercq. 1992. Kinetics of inhibition of endogenous human immunodeficiency virus type 1 reverse transcription by 2′,3′-dideoxynucleoside 5′-triphosphate, tetrahydroimidazo-[4,5,1-jk][1,4]-benzodiazepin-2(1H)-thione, and 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine derivatives. J. Biol. Chem. 267:11769-11776. [PubMed] [Google Scholar]
- 14.Fallows, D. A., and S. P. Goff. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, L., and Y. C. Cheng. 1998. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(-)SddC (3TC) resistance. Biochem. Pharmacol. 55:1567-1572. [DOI] [PubMed] [Google Scholar]
- 16.Fu, L., S. H. Liu, and Y. C. Cheng. 1999. Sensitivity of L-(-)2,3-dideoxythiacytidine resistant hepatitis B virus to other antiviral nucleoside analogues. Biochem. Pharmacol. 57:1351-1359. [DOI] [PubMed] [Google Scholar]
- 17.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 18.Gauthier, J., E. J. Bourne, M. W. Lutz, L. M. Crowther, J. L. Dienstag, N. A. Brown, and L. D. Condreay. 1999. Quantitation of hepatitis B viremia and emergence of YMDD variants in patients with chronic hepatitis B treated with lamivudine. J. Infect. Dis. 180:1757-1762. [DOI] [PubMed] [Google Scholar]
- 19.Honkoop, P., H. G. M. Niesters, R. A. M. de Man, A. D. M. E. Osterhaus, and S. W. Schalm. 1997. Lamivudine resistance in immunocompetent chronic hepatitis B. J. Hepatol. 26:1393-1395. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, R. C., M. S. Lui, T. J. Boritzki, H. P. Morris, and G. Weber. 1980. Purine and pyrimidine nucleotide patterns of normal, differentiating, and regenerating liver and of hepatomas in rats. Cancer Res. 40:1286-1291. [PubMed] [Google Scholar]
- 21.Jansen, R. W., L. C. Johnson, and D. R. Averett. 1993. High-capacity in vitro assessment of anti-hepatitis B virus compound selectivity by a virion-specific polymerase chain reaction assay. Antimicrob. Agents Chemother. 37:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kewn, S., P. Hoggard, S. Sales, M. Johnson, and D. Back. 2000. The intracellular activation of lamivudine (3TC) and determination of 2′-deoxycytidine-5′-triphosphate (dCTP) pools in the presence and absence of various drugs in HepG2 cells. Br. J. Clin. Pharmacol. 50:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladner, S. K., T. J. Miller, and R. W. King. 1998. The M539V polymerase variant of human hepatitis B virus demonstrates resistance to 2′-deoxy-3′-thiacytidine and a reduced ability to synthesize viral DNA. Antimicrob. Agents Chemother. 42:2128-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladner, S. K., T. J. Miller, M. J. Otto, and R. W. King. 1998. The hepatitis B virus M539V polymerase variation responsible for 3TC resistance also confers cross-resistance to other nucleoside analogues. Antivir. Chem. Chemother. 9:65-72. [PubMed] [Google Scholar]
- 25.Liaw, Y. F., R. N. Chien, C. T. Yeh, S. L. Tsai, and C. M. Chu. 1999. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology 30:567-572. [DOI] [PubMed] [Google Scholar]
- 26.Ling, R., and T. J. Harrison. 1999. Functional analysis of mutations conferring lamivudine resistance on hepatitis B virus. J. Gen. Virol. 80:601-606. [DOI] [PubMed] [Google Scholar]
- 27.Maybaum, J., F. K. Klein, and W. Sadee. 1980. Determination of pyrimidine ribotide and deoxyribotide pools in cultured cells and mouse liver by high-performance liquid chromatography. J. Chromatogr. 188:149-158. [DOI] [PubMed] [Google Scholar]
- 28.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology 27:628-633. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, M., T. Kakutani, K. Samejima, Y. Wataya, and H. Hayatsu. 1998. Deoxyribonucleoside triphosphates in mouse fetal liver cells assayed by high-pressure liquid chromatography. Toxicol. Methods 8:11-16. [Google Scholar]
- 30.Niesters, H. G., P. Honkoop, E. B. Haagsma, R. A. de Man, S. W. Schalm, and A. D. Osterhaus. 1998. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J. Infect. Dis. 177:1382-1385. [DOI] [PubMed] [Google Scholar]
- 31.Ogata, N., K. Fujii, S. Takigawa, M. Nomoto, T. Ichida, and H. Asakura. 1999. Novel patterns of amino acid mutations in the hepatitis B virus polymerase in association with resistance to lamivudine therapy in Japanese patients with chronic hepatitis B. J. Med. Virol. 59:270-276. [PubMed] [Google Scholar]
- 32.Ono, S. K., N. Kato, Y. Shiratori, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Investig. 107:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono, Y., H. Onda, R. Sasada, K. Igarashi, Y. Sugino, and K. Nishioka. 1983. The complete nucleotide sequences of the cloned hepatitis B virus DNA subtype adr and adw. Nucleic Acids Res. 11:1747-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono-Nita, S. K., N. Kato, Y. Shiratori, K. H. Lan, H. Yoshida, F. J. Carrilho, and M. Omata. 1999. Susceptibility of lamivudine-resistant hepatitis B virus to other reverse transcriptase inhibitors. J. Clin. Investig. 103:1635-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono-Nita, S. K., N. Kato, Y. Shiratori, T. Masaki, K. H. Lan, F. J. Carrilho, and M. Omata. 1999. YMDD motif in hepatitis B virus DNA polymerase influences on replication and lamivudine resistance: a study by in vitro full-length viral DNA transfection. Hepatology 29:939-945. [DOI] [PubMed] [Google Scholar]
- 36.O'Reilly, D. R. 1997. Use of baculovirus expression vectors. Methods Mol. Biol. 62:235-246. [DOI] [PubMed] [Google Scholar]
- 37.Paff, M. T., D. R. Averett, K. L. Prus, W. H. Miller, and D. J. Nelson. 1994. Intracellular metabolism of (−)- and (+)-cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]cytosine in HepG2 derivative 2.2.15 (subclone P5A) cells. Antimicrob. Agents Chemother. 38:1230-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reichard, P. 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57:349-374. [DOI] [PubMed] [Google Scholar]
- 40.Reichard, P. 1985. Ribonucleotide reductase and deoxyribonucleotide pools. Basic Life Sci. 31:33-45. [DOI] [PubMed] [Google Scholar]
- 41.Rottgen, V., and H. M. Rabes. 1989. Deoxyribonucleoside triphosphate pools in regenerating rat liver: effect of hydroxyurea and exogenous deoxypyrimidines. Biochim. Biophys. Acta 992:349-354. [DOI] [PubMed] [Google Scholar]
- 42.Seeger, C., and W. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segel, I. H. 1993. Enzyme kinetics. John Wiley and Sons, New York, N.Y.
- 44.Seifer, M., R. K. Hamatake, R. J. Colonno, and D. N. Standring. 1998. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob. Agents Chemother. 42:3200-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sells, M. A., M.-L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 84:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seta, T., O. Yokosuka, F. Imazeki, M. Tagawa, and H. Saisho. 2000. Emergence of YMDD motif mutants of hepatitis B virus during lamivudine treatment of immunocompetent type B hepatitis patients. J. Med. Virol. 60:8-16. [DOI] [PubMed] [Google Scholar]
- 47.Sherman, P. A., and J. A. Fyfe. 1989. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal. Biochem. 180:222-226. [DOI] [PubMed] [Google Scholar]
- 48.Soderhall, S. S., A. Larsson, and K. L. Skoog. 1973. Deoxyribonucleotide pools during liver regeneration. Eur. J. Biochem. 33:36-39. [DOI] [PubMed] [Google Scholar]
- 49.Stuyver, L. J., S. A. Locarnini, A. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33:751-757. [DOI] [PubMed] [Google Scholar]
- 50.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 51.Valenzuela, P., P. Gray, M. Quiroga, J. Zaldivar, H. M. Goodman, and W. J. Rutter. 1979. Nucleotide sequence of the gene coding for the major protein of hepatitis B virus surface antigen. Nature 280:815-819. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, X., C. Flores, H. Yang, J. J. Toole, and C. S. Gibbs. 1998. Mutations in hepatitis B DNA polymerase associated with resistance to lamivudine do not confer resistance to adefovir in vitro. Hepatology 28:1669-1673. [DOI] [PubMed] [Google Scholar]
- 53.Yeh, C. T., R. N. Chien, C. M. Chu, and Y. F. Liaw. 2000. Clearance of the original hepatitis B virus YMDD-motif mutants with emergence of distinct lamivudine-resistant mutants during prolonged lamivudine therapy. Hepatology 31:1318-1326. [DOI] [PubMed] [Google Scholar]
- 54.Zollner, B., A. Stoehr, A. Plettenberg, H. H. Feucht, M. Schroter, P. Schafer, and R. Laufs. 2000. In vivo dynamics and pathogenicity of wild-type and resistant hepatitis B virus during long-term lamivudine monotherapy—a clinical note. J. Clin. Virol. 17:183-188. [DOI] [PubMed] [Google Scholar]