Abstract
In order to track the evolution of primary protease inhibitor (PI) resistance mutations in human immunodeficiency virus type 1 (HIV-1) isolates, baseline and follow-up protease sequences were obtained from patients undergoing salvage PI therapy who presented initially with isolates containing a single primary PI resistance mutation. Among 78 patients meeting study selection criteria, baseline primary PI resistance mutations included L90M (42% of patients), V82A/F/T (27%), D30N (21%), G48V (6%), and I84V (4%). Despite the switching of treatment to a new PI, primary PI resistance mutations present at the baseline persisted in 66 of 78 (85%) patients. D30N persisted less frequently than L90M (50% versus 100%, respectively; P < 0.001) and V82A/F/T (50% versus 81%, respectively; P = 0.05). HIV-1 isolates from 38 (49%) patients failing PI salvage therapy developed new primary PI resistance mutations including L90M, I84V, V82A, and G48V. Common combinations of primary and secondary PI resistance mutations after salvage therapy included mutations at amino acid positions 10, 82, and 46 and/or 54 in 16 patients; 10, 90, and 71 and/or 73 in 14 patients; 10, 73, 84, 90, and 46 and/or 54 in 5 patients; 10, 48, and 82 in 5 patients; and 30, 88 and 90 in 5 patients. In summary, during salvage PI therapy, most HIV-1 isolates with a single primary PI resistance mutation maintained their original mutations, and 49% developed additional primary PI resistance mutations. The persistence of L90M, V82A/F/T, G48V, and I84V during salvage therapy suggests that these mutations play a role in clinical resistance to multiple PIs.
Mutations at more than 20 positions in human immunodeficiency virus type 1 (HIV-1) protease have been associated with resistance to present protease inhibitors (PIs). Several PI resistance mutations are of particular importance because they occur in the substrate cleft or independently reduce drug susceptibility. Six of these mutations (D30N, G48V, I50V, V82A/F/S/T, I84V, and L90M) have been designated primary resistance mutations in recent published guidelines for antiretroviral drug resistance testing (10).
Much of what is known about PI resistance mutations is based on preclinical studies of laboratory isolates and early clinical studies in patients given PI monotherapy. These isolates usually contain a single or just a few protease mutations. As a consequence, there are major gaps in what is known about the development of PI resistance in patients receiving PIs in combination or in sequence. Specifically, the extent to which protease mutations confer selective resistance to some PIs but not others is not completely known. Knowing the extent of cross-resistance conferred by each of the protease mutations is essential for choosing antiretroviral drugs when genotypic testing is done in patients failing PI-containing regimens as well as for designing new non-cross-resistant PIs.
The primary PI resistance mutations do not occur as natural polymorphisms in HIV-1 isolates from untreated persons. Each of these mutations has also been shown to impair HIV-1 replication in vitro (3, 14-17). Mutations selected during treatment with one PI that persist during treatment with a second PI are therefore likely to contribute resistance to the second PI because, in the absence of selective drug pressure, these mutations would be expected to have been replaced with wild-type residues (5, 24).
Genetic sequences of HIV-1 isolates from patients failing antiretroviral therapy demonstrate which mutations are most significant in vivo. To elucidate the extent of cross-resistance conferred by each of the primary PI resistance mutations, we looked at how HIV-1 isolates containing only one primary PI resistance mutation evolved in patients undergoing changes in PI therapy.
MATERIALS AND METHODS
Patients.
The patients included individuals who had HIV-1 isolates sent to Stanford University Hospital (SUH) for protease and reverse transcriptase (RT) sequencing between 1 August 1997 and 1 April 2001. All patients were from northern California and had failed a PI-containing regimen (plasma HIV-1 RNA level, >500 to 1,000 copies/ml) at the time sequencing was done. Detailed treatment histories were obtained, and sequence analyses were performed on isolates from those patients who had been treated with PIs and had had an original sequence containing one primary PI mutation and whose HIV-1 isolates were resequenced after a change in PI therapy.
HIV-1 protease sequencing.
Sequencing was performed by using a previously described method (21). Briefly, RNA was extracted from 0.2 ml of plasma by using the guanidine-thiocyanate lysis reagent contained within the AMPLICOR HIV Monitor test kit (Roche Diagnostic Systems, Branchburg, N.J.). Reverse-strand cDNA was generated from viral RNA, and first-round PCR was performed by using Superscript One-Step RT-PCR (Life Technologies, Rockville, Md.). A 1.3-kb product encompassing the protease gene and the first 300 residues of the RT gene was then amplified with nested PCR primers. Direct PCR (population-based) cycle sequencing was performed by using AmpliTaq DNA FS polymerase and dRhodamine terminators (Applied Biosystems, Foster City, Calif.). Electropherograms were created with Sequence Analysis software, version 3.0 (Applied Biosystems), and the sequences were assembled with the manufacturer's FACTURA and AUTOASSEMBLER sequence analysis software. Each amino acid sequence was compared to the subtype B consensus reference sequence, and those sequences with differences from the reference sequence were considered mutations.
HIV-1 protease mutations.
D30N, G48V, I50V, V82A/F/S/T (but not V82I), I84V, and L90M were defined as primary PI resistance mutations in agreement with recent International AIDS Society-USA Panel drug resistance testing guidelines (10). Secondary PI resistance mutations were defined as those with any change(s) from the consensus B reference sequence and included the following: L10I/V/F, K20R/M/I, L24I, V32I, M36I/L/V, M46I/L, I47V, F53L, I54V/L, A71V/T/I, G73C/S/T/A, and N88D/S/T. Mutations at positions 63, 77, and 93 were not categorized as drug resistance mutations because, although mutations at these positions contribute to resistance, these positions are highly polymorphic within individuals. Positions containing mixtures of wild-type and mutant residues were classified as mutant.
Sequence analyses.
Nucleotide distances were calculated between the presenting and follow-up sequences of each study patient. Synonymous and nonsynonymous nucleotide substitution rates were calculated by using the method of Nei and Gojobori (18) using the Synonymous-Nonsynonymous Analysis Program (http://hiv-web.lanl.gov) (12). PS represents the number of observed synonymous substitutions (causing no amino acid change) divided by the number of possible synonymous substitutions in a sequence. PN represents the number of observed nonsynonymous substitutions (causing an amino acid change) divided by the number of possible nonsynonymous substitutions in a sequence. The Jukes-Cantor correction adjusts for multiple substitutions at the same codon and was used to calculate DS (synonymous nucleotide distance) and DN (nonsynonymous nucleotide distance) from PS and PN.
Phylogenetic trees of paired sequences from each of the study patients confirmed the absence of laboratory cross-contamination. Subtypes were determined by comparing the protease and RT sequences of HIV-1 isolates to reference sequences, and all sequences were found to belong to subtype B (8). The GenBank accession numbers for the 156 sequences are AY030416, AY030418, AY030452, AY030476, AY030496, AY030529, AY030572, AY030592, AY030597, AY030601, AY030603, AY030618, AY030625, AY030628, AY030636, AY030652, AY030666, AY030689, AY030721, AY030723, AY030726, AY030733, AY030741, AY030773, AY030818, AY030854, AY030875, AY030903, AY030906, AY030940, AY030953, AY031074, AY031176, AY031179, AY031231, AY031259, AY031262, AY031275, AY031280, AY031353, AY031758, AY032084, and AY047367 to AY047480.
Statistical analysis.
Comparisons were made between groups of patients with different primary PI resistance mutations. Chi-square and Kruskal-Wallis rank sum tests were used to compare proportions and medians, respectively, between multiple patient groups. Fisher exact and Mann-Whitney tests were used to compare proportions and medians, respectively, between two patient groups.
RESULTS
Prevalence of primary PI resistance mutations.
Between 1 August 1997 and 1 April 2001, 670 persons had more than one sequence sent to SUH for HIV-1 sequencing. Treatment histories were available for 430 of these patients. Of these 430 patients, 166 had received at least one PI and had a sequence containing exactly one primary PI resistance mutation. Of these 166 patients, 78 changed to a new PI and thus met study selection criteria; 88 remained on their initial PI (54 patients) or discontinued PI therapy (34 patients).
Among the 78 patients in this study, isolates from 33 contained HIV-1 with the protease mutation L90M (42%), isolates from 21 contained V82A/F/T (27%), isolates from 16 contained D30N (21%), isolates from 5 contained G48V (6%), isolates from 3 contained I84V (4%), and no isolates contained I50V.
PI treatment regimens.
The PIs received by the 78 study patients prior to protease sequencing are shown in the third column of Table 1. with L90M, the most common previous PIs included indinavir (IDV) (20 patients), saquinavir (SQV) (19 patients), and nelfinavir (NFV) (18 patients). Among the 21 patients with V82A/F/T, 19 had received IDV. Among the 16 patients with D30N, all had received NFV. Among the five patients with G48V, all had received SQV. Among the three patients with I84V, all had received IDV. The median duration of PI therapy before presentation was 68 weeks (range, 8 to 164 weeks). Forty patients had had previous treatment with more than one PI regimen. The median duration of previous PI therapy and the proportion of patients receiving more than one PI regimen did not differ among the different patient groups.
TABLE 1.
Evolution of primary and secondary PI mutations during salvage therapy in 78 patients presenting with a single PI resistance mutation
| Amino acid no. of presenting mutation(s) | Previous therapy
|
Salvage therapy
|
Amino acid no. of follow-up mutation(s)
|
||||
|---|---|---|---|---|---|---|---|
| Primary | Secondary | Protease inhibitor(s) | Duration (wk) | Protease inhibitor(s) | Duration (wk) | Primary | Secondary |
| 30 | 36 | NFV | 12 | RTV-SQV | 44 | 30, 90 | 20, 36, 54, 71, 88 |
| 30 | 36 | SQV, IDV, NFV | 51 | RTV-SQV | 19 | 30 | 36 |
| 30 | 36 | SQV, NFV | 107 | IDV | 78 | 36, 46, 71, 88 | |
| 30 | 20, 36 | SQV, NFV | 53 | RTV-SQV | 8 | 84, 90 | 20, 36, 71 |
| 30 | 20, 36 | NFV | 20 | RTV-SQV | 9 | 84, 90 | 20, 36, 71, 73 |
| 30 | 71 | IDV, NFV | 27 | RTV-SQV | 6 | 90 | 20, 32, 36, 71, 73 |
| 30 | 36 | IDV, NFV | 68 | RTV-SQV | 71 | 30, 84, 90 | 36, 88 |
| 30 | 10 | SQV, IDV, NFV | 113 | RTV-SQV | 38 | 30 | 71, 88 |
| 30 | 71, 88 | NFV | 57 | IDV | 76 | 30, 90 | 20, 46, 71, 88 |
| 30 | 10, 20, 71, 88 | NFV | 49 | RTV-SQV | 41 | 30, 90 | 10, 20, 46, 53, 54, 71, 88 |
| 30 | 36, 71, 88 | IDV, NFV | 88 | APV | 50 | 30 | 36, 46, 71, 88 |
| 30 | 36 | RTV, NFV | 108 | IDV | 85 | 82 | 36, 46, 71, 88 |
| 30 | NFV | 73 | RTV-SQV, IDV | 35 | 46, 73 | ||
| 30 | 10, 88 | IDV, RTV, NFV | 157 | RTV-SQV, RTV-APV, RTV-IDV | 41 | 90 | 10, 54, 88 |
| 30 | 88 | SQV, NFV | 52 | SQV | 40 | 90 | 36 |
| 30 | 10, 36 | NFV | 16 | RTV-SQV | 44 | 30, 90 | 20, 36, 54, 88 |
| 48 | SQV | 24 | IDV | 24 | 48, 82 | 10, 71 | |
| 48 | 71 | SQV | 24 | IDV | 24 | 48, 82 | 10, 54, 71 |
| 48 | 71 | SQV | 26 | IDV, RTV-SQV | 46 | 48, 82 | 10, 54, 71 |
| 48 | 10, 36, 54 | SQV | 40 | NFV, RTV-IDV | 100 | 48, 82 | 10, 36, 54 |
| 48 | 10 | SQV | 24 | IDV | 24 | 48, 82 | 10 |
| 82 | 10, 46, 54, 88 | SQV, IDV | 59 | RTV-SQV | 18 | 90 | 10, 20, 71, 73 |
| 82 | 10, 46, 71, 88 | IDV | 84 | NFV-SQV, RTV-SQV | 120 | 84, 90 | 10, 46, 71, 88 |
| 82 | 10, 46, 71 | IDV | 53 | RTV-SQV | 43 | 82, 90 | 10, 32, 46, 53, 71 |
| 82 | 10, 24, 32, 46, 71, 73 | IDV | 68 | RTV-SQV | 75 | 84 | 10, 24, 46, 73 |
| 82 | 71 | SQV, IDV | 78 | RTV-SQV | 21 | 82 | 71 |
| 82 | 10, 36, 54, 73 | IDV | 44 | RTV-SQV | 76 | 48, 82 | 10, 20, 36, 54, 73 |
| 82 | 32, 46, 47 | IDV | 93 | SQV, NFV-SQV | 46 | 82, 90 | 10, 46, 54, 71 |
| 82 | IDV | 68 | NFV-SQV, NFV | 61 | 90 | ||
| 82 | 54 | RTV | 30 | NFV | 71 | 82, 90 | 10, 54, 71 |
| 82 | 10, 20, 24, 36, 71, 73 | IDV, NFV-SQV | 89 | APV | 49 | 82 | 10, 20, 24, 36, 46, 54, 71, 73 |
| 82 | 10, 54 | IDV, NFV | 20 | RTV-SQV | 22 | 48, 82, 84 | 10, 53, 54, 71 |
| 82 | 32, 36, 46, 71 | IDV | 59 | RTV-SQV | 90 | 82 | 10, 24, 36, 54, 71 |
| 82 | 10, 24, 46 | IDV, SQV, NFV | 136 | APV | 56 | 82 | 10, 24, 46 |
| 82 | 10, 24, 46 | SQV, IDV | 52 | NFV | 43 | 82 | 10, 24, 46 |
| 82 | 10, 24, 46 | NFV, IDV | 83 | NFV, NFV-SQV | 26 | 82, 84 | 10, 24, 46 |
| 82 | 46 | IDV | 124 | NFV | 44 | 82 | 10, 46 |
| 82 | 46 | IDV | 96 | APV | 24 | 82 | 46 |
| 82 | 20, 46, 71 | IDV | 54 | NFV | 16 | 82 | 20, 46, 71 |
| 82 | 10, 20, 24, 36, 46 | IDV | 9 | RTV-SQV | 44 | 82 | 10, 20, 24, 36, 46, 54 |
| 82 | 10, 46, 53, 54, 71 | IDV | 58 | RTV-SQV | 16 | 82, 90 | 10, 53, 54, 71 |
| 82 | 10, 36, 54 | NFV | 123 | RTV-SQV | 15 | 48, 82, 90 | 10, 36, 46, 54 |
| 84 | 10, 36, 71 | IDV, NFV | 68 | RTV-SQV | 35 | 84 | 10, 36, 71 |
| 84 | 46, 71 | IDV | 57 | RTV-SQV | 81 | 48, 84 | 10, 46, 71 |
| 84 | 10, 46, 54, 71 | IDV, SQV | 65 | APV | 13 | 84 | 10, 46, 54, 71 |
| 90 | 10, 36 | SQV | 52 | IDV | 11 | 90 | 10, 20, 36, 71 |
| 90 | 10, 54 | IDV | 43 | RTV-SQV | 28 | 84, 90 | 10, 46, 73 |
| 90 | 10, 46, 53, 71, 73 | SQV, IDV, NFV | 116 | RTV-SQV, IDV | 36 | 90 | 10, 53, 71, 73 |
| 90 | 71, 73 | SQV | 48 | RTV-SQV | 149 | 84, 90 | 10, 71, 73 |
| 90 | 71 | IDV | 48 | RTV-SQV | 41 | 90 | 71 |
| 90 | 10, 53, 71, 73 | SQV | 40 | NFV, IDV, RTV-SQV | 69 | 90 | 10, 24, 46, 53, 71, 73 |
| 90 | 71 | SQV, NFV | 104 | IDV | 124 | 90 | 46, 73 |
| 90 | IDV | 67 | RTV-SQV | 17 | 84, 90 | 73 | |
| 90 | 10, 20, 36, 73 | SQV, IDV, RTV-SQV, NFV-SQV | 135 | IDV | 28 | 90 | 10, 20, 36, 73 |
| 90 | 20, 71, 73 | IDV | 98 | RTV-SQV | 29 | 90 | 20, 71, 73 |
| 90 | 71, 88 | SQV, NFV | 73 | RTV-SQV | 14 | 90 | 10, 71, 88 |
| 90 | 10, 20, 71 | SQV, NFV | 55 | IDV | 37 | 90 | 10, 71, 73 |
| 90 | 10, 46, 73 | IDV | 66 | NFV | 59 | 90 | 10, 73 |
| 90 | 10, 71, 73 | SQV, IDV, NFV | 94 | RTV-IDV, RTV-APV-SQV | 71 | 84, 90 | 10, 54, 71, 73 |
| 90 | 20, 36, 71 | IDV, NFV | 79 | RTV-SQV | 65 | 90 | 20, 36, 54, 71/PICK>{tt} |
| 90 | 20, 46, 73 | NFV | 56 | RTV-IDV | 72 | 90 | 20, 73 |
| 90 | 10, 46, 73 | IDV, NFV | 112 | RTV-SQV | 35 | 90 | 10, 73 |
| 90 | 46, 71, 73 | IDV | 6 | NFV, IDV-SQV, RTV-SQV, APV-NFV | 154 | 84, 90 | 10, 54, 71, 73 |
| 90 | SQV, SQV-APV | 36 | NFV, RTV, RTV-IDV | 169 | 84, 90 | 10, 20, 36, 71 | |
| 90 | 10, 46, 73 | IDV, NFV-SQV | 108 | NFV-SQV, RTV-SQV | 76 | 90 | 10, 46, 73 |
| 90 | 10, 54, 71, 73 | IDV, IDV-SQV, NFV-SQV | 124 | NFV, RTV-APV | 118 | 90 | 10, 36, 71, 73 |
| 90 | 10, 20, 36, 46, 73 | NFV | 68 | RTV-SQV | 28 | 84, 90 | 10, 20, 36, 46, 73 |
| 90 | 71, 73 | IDV, NFV, SQV | 83 | RTV-APV, LPV | 75 | 84, 90 | 10, 54, 71, 73 |
| 90 | 10, 46 | IDV, NFV-SQV, SQV | 125 | APV | 47 | 90 | 10 |
| 90 | 46, 71 | NFV | 137 | APV | 46 | 90 | 10, 71 |
| 90 | 32, 46, 47 | IDV | 58 | NFV, SQV, RTV-NFV | 83 | 90 | 10 |
| 90 | 10, 71, 73 | SQV | 35 | NFV | 17 | 90 | 10, 71, 73 |
| 90 | 20, 46 | IDV, SQV | 136 | APV | 35 | 90 | 10, 20 |
| 90 | 10, 46, 73 | IDV, NFV-SQV, IDV | 89 | APV | 33 | 90 | 73 |
| 90 | 20, 36 | NFV | 72 | APV | 51 | 90 | 10, 73 |
| 90 | 46 | NFV, NFV-SQV | 96 | APV, RTV-SQV | 62 | 90 | |
| 90 | 10, 46, 53, 73 | SQV, IDV, RTV-SQV, NFV | 164 | RTV-IDV | 20 | 84, 90 | 10, 53, 73 |
| 90 | 10, 20, 36, 46, 71 | IDV | 106 | APV | 37 | 90 | 10, 20, 36, 71 |
The fifth column of Table 1 shows the PIs that were used as salvage therapy for each of the 78 study patients. The most commonly used follow-up PI regimens included ritonavir (RTV)-SQV (45 patients), IDV (21 patients), NFV (17 patients), and amprenavir (APV) (17 patients). RTV-SQV was used as salvage therapy for about two-thirds of the patients with D30N, V82A/F/T, or L90M. IDV was used as salvage therapy for all of the patients with G48V and for about one-third of the patients with D30N or L90M. NFV was used as salvage therapy for about one-third of the patients with V82A/F/T or L90M. APV was used as salvage therapy for about one-third of the patients with L90M. The median duration of salvage therapy was 48 weeks (range, 8 to 169 weeks). Eighteen patients had follow-up treatment with more than one PI regimen before their second sequences. The median duration of salvage therapy and the proportion of patients with more than one follow-up PI regimen did not differ between the different patient groups.
Evolution of primary PI resistance mutations.
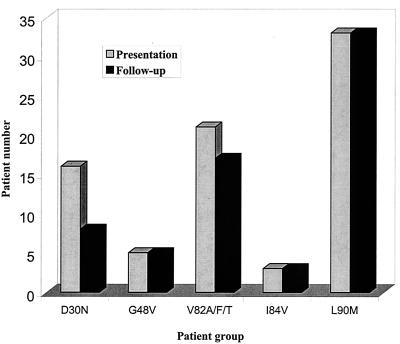
Following salvage therapy, each of the patients with L90M, 17 of 21 patients with V82A/F/T, 8 of 16 patients with D30N, and each of the patients with G48V and I84V maintained a virus population in which the initial primary PI resistance mutation continued to predominate (Fig. 1). D30N persisted less often than L90M (P < 0.001), V82A/F/T (P = 0.05), or G48V (P = 0.06). New primary PI resistance mutations developed in 9 of 33 (27%) patients starting with L90M, in 12 of 21 (57%) starting with V82A/F/T, in 11 of 16 (69%) starting with D30N, in 5 of 5 (100%) starting with G48V, and in 1 of 3 (33%) starting with I84V. New primary PI resistance mutations were significantly less likely to develop in patients starting with L90M than in those starting with D30N (P = 0.007) or V82A/F/T (P = 0.03).
FIG. 1.
Bar graph indicating the extent to which primary protease inhibitor resistance mutations present at baseline (gray bars) were also present following salvage therapy (black bars).
Following salvage therapy, additional primary PI resistance mutations that were acquired included L90M in 18 patients, I84V in 16 patients, V82A in 6 patients, and G48V in 4 patients. No patients acquired D30N or I50V during salvage therapy. L90M developed in 10 of 16 patients who started with D30N and in 8 of 21 who started with V82A/F/T. I84V developed in 9 of 33 patients who started with L90M, in 4 of 21 who started with V82A/F/T, and in 3 of 16 who started D30N. V82A developed in all patients who started with G48V and in 1 of 16 who started with D30N. G48V developed in 3 of 21 patients who started with V82A/F/T and in 1 of 3 who started with I84V.
More than one primary PI resistance mutation was observed at follow-up in 31 of 78 (40%) patients. The most common combinations of primary PI resistance mutations that developed during follow-up included mutations at positions 84 and 90 in 12 patients, 48 and 82 in 6 patients, 82 and 90 in 4 patients, and 30 and 90 in 4 patients. Mutations at positions 48 and 84; 82 and 84; 30, 84, and 90; 48, 82, and 84; and 48, 82, and 90 occurred in one patient each.
Secondary PI resistance mutations.
The mean number of secondary PI resistance mutations per patient at baseline was 2.5 for patients presenting with L90M, 2.7 for patients with V82A/F/T, 1.6 for patients with D30N, 1.2 for patients with G48V, and 3.0 for patients with I84V (Table 1). Common secondary drug resistance mutations before salvage therapy included mutations at positions 10 (17 of 33 patients), 46 (15 of 33), and 73 (17 of 33) in patients with L90M; mutations at positions 10 (13 of 21 patients), 46 (14 of 21), and 54 (6 of 21) in patients with V82A/F/T; and mutations at positions 36 (9 of 16 patients) and 88 (5 of 16) in patients with D30N.
The most commonly acquired secondary PI resistance mutations occurred at positions 10 (17 patients), 54 (14 patients), 71 (13 patients), 46 (11 patients), 73 (9 patients), 20 (8 patients), and 88 (6 patients). New secondary PI resistance mutations were more likely to develop in patients presenting with D30N (15 of 16 patients [94%]) than in those presenting with L90M (17 of 33 [52%]; P = 0.003) or V82A/F/T (10 of 21 [48%]; P = 0.003).
Common combinations of primary and secondary PI resistance mutations after salvage therapy included mutations at positions 10, 82, and 46 and/or 54 in 16 patients; 10, 90, and 71 and/or 73 in 14 patients; 10, 73, 84, 90, and 46 and/or 54 in 5 patients; 10, 48, and 82 in 5 patients; and 30, 88, and 90 in 5 patients.
Plasma HIV-1 RNA levels.
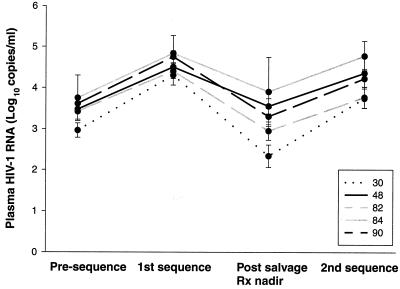
Plasma HIV-1 RNA levels (in log10 copies per milliliter) were available from 68 of the 78 patients, including 29 of 33 presenting with L90M, 19 of 21 presenting with V82A/F/T, 13 of 16 presenting with D30N, 4 of 5 presenting with G48V, and 3 of 3 presenting with I84V (Fig. 2). The mean plasma HIV-1 RNA levels (± standard error) of each patient group at four times are shown in Fig. 2 as follows: (i) prior to the first sequence, (ii) at the time of the first sequence, (iii) at the time with the lowest RNA level between the first and second sequences, and (iv) at the time of the second sequence. Plasma HIV-1 RNA levels increased 1.1 log10 copies/ml between times i and ii, indicating that the patients were on average experiencing virologic rebound at the time of the first sequence. Plasma HIV-1 RNA levels decreased 1.4 log10 copies/ml between times ii and iii after salvage therapy was begun. Plasma HIV-1 RNA levels then increased 1.0 log10 copies/ml by the time of the second sequence (iv), indicating virologic rebound on salvage therapy.
FIG. 2.
Plasma HIV-1 RNA levels (log10 copies per milliliter, means ± standard errors) from 68 of 78 patients, including 29 of 33 presenting with L90M, 19 of 21 presenting with V82A/F/T, 13 of 16 presenting with D30N, 4 of 5 presenting with G48V, and 3 of 3 presenting with I84V. “Presequence” indicates the lowest plasma RNA levels in the year (median, 6 months) before the first sequence. “1st sequence” indicates the RNA levels at the time of the first sequence (i.e., that containing a single primary drug resistance mutation). “Post salvage Rx nadir” indicates the lowest plasma HIV-1 RNA levels between the first and second sequences (median, 6 months after the first sequence). “2nd sequence” indicates the plasma HIV-1 RNA levels at the time of the second sequence.
Patients presenting with isolates containing D30N or V82A/F/T consistently had lower plasma HIV-1 RNA levels than those presenting with G48V, I84V, and L90M. The patients with D30N had the greatest response to salvage therapy (P = 0.03, analysis of variance test). However, this group of patients also had the greatest increase in plasma HIV-1 RNA levels by the time of the second sequence.
Nucleotide distances.
Table 2 characterizes the nucleotide differences between the first and second sequences in each of the 78 study patients. Nucleotide differences were categorized as either synonymous or nonsynonymous, and nonsynonymous changes were further categorized as to whether they occurred at positions associated with drug resistance.
TABLE 2.
Nucleotide distances between baseline and follow-up sequences in the 78 patients presenting with a single primary PI resistance mutationa
| Patient group | No. of patients | Time (mo) between sequences | No. of nucleotide changes/patientb | No. of nonsynonymous changesc
|
No. of synonymous changes | DS/DN ratio | |
|---|---|---|---|---|---|---|---|
| Drug resistance positions | Other positions | ||||||
| D30N | 16 | 10 | 8.5 | 4 | 2 | 2 | 1.2 |
| G48V | 5 | 22 | 6 | 1 | 2 | 2 | 2.4 |
| V82A/F/T | 21 | 10 | 7 | 3 | 3 | 1 | 1.1 |
| I84V | 3 | 9 | 1 | 0.5 | 0.5 | 0.5 | 1.3 |
| L90M | 33 | 12 | 5 | 2 | 2 | 2 | 0.9 |
Numbers in columns 2 to 7 indicate median values; DS/DN ratios indicate average values.
The number of nucleotide changes during salvage therapy for patients starting therapy with L90M was significantly lower than the number of nucleotide changes during salvage therapy for patients starting therapy with D30N (P = 0.02, Wilcoxon rank sum test).
The number of changes at drug resistance positions for patients starting therapy with L90M was significantly lower than the number of changes at drug resistance positions during salvage therapy for patients starting therapy with D30N (P = 0.003, Wilcoxon rank sum test).
Patients presenting with D30N had the most nucleotide changes between their first and second sequences (median, 8.5 changes per patient), and this was significantly greater than the median number of changes between the two sequences from patients presenting with L90M (median, 5 changes per patient). For each of the five groups of patients, most nucleotide changes were nonsynonymous (range, 64% to 72%), and this was reflected by the low DS/DN ratios for each patient group (range, 0.9 to 2.4).
DISCUSSION
Among 78 patients presenting with HIV-1 isolates containing a single primary PI resistance mutation, the most common mutations included L90M (42%), V82A/F/T (27%), and D30N (21%). G48V and I84V occurred in 6 and 4% of patients, respectively. I50V did not occur in this cohort. Despite switching therapy to a regimen containing a new PI, the original primary PI resistance mutations persisted in 85% (66 of 78) of patients. The only mutation that did not persist in the majority of patients was D30N. The differences in persistence between D30N and the other two most common mutations, V82A/F/T (50% versus 81%, respectively; P = 0.05) and L90M (50% versus 100%, respectively; P < 0.001), were statistically significant.
Our findings support the results of previously reported studies of PI salvage therapy. In patients changing from one PI-containing regimen to a second PI-containing regimen, successful salvage therapy occurs most often in patients with HIV-1 isolates containing D30N and no other primary PI resistance mutations (2, 23, 27) and in patients receiving dual PIs in conjunction with a nonnucleoside RT inhibitor (11, 13). The fact that D30N was the only mutation in our study that often reverted to the wild type corroborates the clinical data that this mutation confers less cross-resistance to other PIs than the other primary PI resistance mutations.
Most HIV-1 isolates from patients failing the follow-up PI-containing regimen developed new primary and secondary PI resistance mutations. Although classified as primary PI resistance mutations (i.e., those that appear first in the course of development of resistance mutations), L90M, I84V, V82A/F/T, and G48V mutations often developed during salvage therapy. D30N and I50V did not occur during salvage despite the fact that NFV and/or APV was used for salvage in 31 patients. This accumulation of primary and secondary mutations was not random, and several combinations of drug resistance mutations occurred in a high proportion of patients. These patterns provide important insight into how HIV-1 develops multidrug resistance, which might help in the development of new non-cross-resistant PIs. In 18 patients, the additional mutations could not be linked to a specific drug regimen, because more than one regimen was received between the two sequences.
We studied the evolution of mutations in patients with virologic failure associated with a single primary PI resistance mutation because in such patients, the primary PI resistance mutation is by itself likely to be directly responsible for drug resistance. Isolates with a single primary resistance mutation may also be less likely than isolates with multiple primary resistance mutations to be restricted in their evolution. Isolates with multiple resistance mutations that are already resistant to multiple drugs may be less likely to gain mutations during salvage therapy. Isolates with multiple resistance mutations may also be less likely to lose mutations due to significant dependencies between multiple primary and secondary mutations (1).
This study is the largest to date designed to examine the evolution of genotypic PI resistance in sequential HIV-1 isolates from patients receiving more than one course of PI therapy. There have been two large cross-sectional studies of genotypic PI resistance in clinical settings (9, 26) and several small studies containing longitudinal genotypic data from patients receiving consecutive PI-containing regimens (4, 6, 7, 13, 19, 20, 22, 25). The longitudinal studies contain data from 53 patients, of whom 35 had a single primary PI resistance mutation at the time salvage therapy was begun (L90M in 18 patients, V82A in 13 patients, and G48V in 4 patients). HIV-1 isolates from each of these 35 maintained their original primary PI resistance mutation during PI salvage therapy.
In summary, our data show that most primary PI resistance mutations in the protease gene, with the possible exception of D30N, play a role in the future development of further resistance to multiple PIs. Isolates from patients with HIV-1 containing G48V, V82A/F/T, I84V, or L90M who experience virologic failure on a new PI regimen continue to harbor the primary resistance mutation that was originally present. Virologic failure during PI salvage therapy leads to the accumulation of new primary and secondary resistance mutations onto a viral backbone containing the original primary resistance mutation.
REFERENCES
- 1.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condra, J. H., C. J. Petropoulos, R. Ziermann, W. A. Schleiff, M. Shivaprakash, and E. A. Emini. 2000. Drug resistance and predicted virologic responses to human immunodeficiency virus type 1 protease inhibitor therapy. J. Infect. Dis. 182:758-765. [DOI] [PubMed] [Google Scholar]
- 3.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks, S. G., N. S. Hellmann, R. M. Grant, N. T. Parkin, C. J. Petropoulos, M. Becker, W. Symonds, M. Chesney, and P. A. Volberding. 1999. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J. Infect. Dis. 179:1375-1381. [DOI] [PubMed] [Google Scholar]
- 5.Devereux, H. L., M. Youle, M. A. Johnson, and C. Loveday. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123-F127. [PubMed] [Google Scholar]
- 6.Dulioust, A., S. Paulous, L. Guillemot, A. M. Delavalle, F. Boue, and F. Clavel. 1999. Constrained evolution of human immunodeficiency virus type 1 protease during sequential therapy with two distinct protease inhibitors. J. Virol. 73:850-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falloon, J., S. Piscitelli, S. Vogel, B. Sadler, H. Mitsuya, M. F. Kavlick, K. Yoshimura, M. Rogers, S. LaFon, D. J. Manion, H. C. Lane, and H. Masur. 2000. Combination therapy with amprenavir, abacavir, and efavirenz in human immunodeficiency virus (HIV)-infected patients failing a protease-inhibitor regimen: pharmacokinetic drug interactions and antiviral activity. Clin. Infect. Dis. 30:313-318. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales, M. J., R. N. Machekano, and R. W. Shafer. 2001. Human immunodeficiency virus type 1 reverse-transcriptase and protease subtypes: classification, amino acid mutation patterns, and prevalence in a northern California clinic-based population. J. Infect. Dis. 184:998-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. Van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6,000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 11.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korber, B. 2000. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo and G. H. Learn (ed.), Computational and evolutionary analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Lawrence, J., J. Schapiro, M. Winters, J. Montoya, A. Zolopa, R. Pesano, B. Efron, D. Winslow, and T. C. Merigan. 1999. Clinical resistance patterns and responses to two sequential protease inhibitor regimens in saquinavir and reverse transcriptase inhibitor-experienced persons. J. Infect. Dis. 179:1356-1364. [DOI] [PubMed] [Google Scholar]
- 14.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markland, W., B. G. Rao, J. D. Parsons, J. Black, L. Zuchowski, M. Tisdale, and R. Tung. 2000. Structural and kinetic analyses of the protease from an amprenavir-resistant human immunodeficiency virus type 1 mutant rendered resistant to saquinavir and resensitized to amprenavir. J. Virol. 74:7636-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, V. 2001. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S34-S50. [DOI] [PubMed]
- 18.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 19.Parkin, N. T., S. G. Deeks, M. T. Wrin, J. Yap, R. M. Grant, K. H. Lee, D. Heeren, N. S. Hellmanna, and C. J. Petropoulos. 2000. Loss of antiretroviral drug susceptibility at low viral load during early virological failure in treatment-experienced patients. AIDS 14:2877-2887. [DOI] [PubMed] [Google Scholar]
- 20.Schapiro, J. M., M. A. Winters, J. Lawrence, and T. C. Merigan. 1999. Clinical cross-resistance between the HIV-1 protease inhibitors saquinavir and indinavir and correlations with genotypic mutations. AIDS 13:359-365. [DOI] [PubMed] [Google Scholar]
- 21.Shafer, R. W., K. Hertogs, A. R. Zolopa, A. Warford, S. Bloor, B. J. Betts, T. C. Merigan, R. Harrigan, and B. A. Larder. 2001. High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J. Clin. Microbiol. 39:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shafer, R. W., M. A. Winters, S. Palmer, and T. C. Merigan. 1998. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated patients. Ann. Intern. Med. 128:906-911. [DOI] [PubMed] [Google Scholar]
- 23.Tebas, P., A. K. Patick, E. M. Kane, M. K. Klebert, J. H. Simpson, A. Erice, W. G. Powderly, and K. Henry. 1999. Virologic responses to a ritonavir-saquinavir-containing regimen in patients who had previously failed nelfinavir. AIDS 13:F23-F28. [DOI] [PubMed]
- 24.Verhofstede, C., F. V. Wanzeele, B. Van Der Gucht, N. De Cabooter, and J. Plum. 1999. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS 13:2541-2546. [DOI] [PubMed] [Google Scholar]
- 25.Walter, H., B. Schmidt, A. Rascu, M. Helm, B. Moschik, C. Paatz, M. Kurowski, K. Korn, K. Uberla, and T. Harrer. 2000. Phenotypic HIV-1 resistance correlates with treatment outcome of nelfinavir salvage therapy. Antivir. Ther. 5:249-256. [PubMed] [Google Scholar]
- 26.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, F. Ariasi, F. Volot, J. A. Gastaut, H. Gallais, J. Moreau, and J. Fantini. 1999. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 se-quences. J. Clin. Microbiol. 37:4099-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]