Abstract
Sequencing of DNA from 15 expanded-spectrum cephalosporin (e.g., ceftriaxone)-resistant Salmonella isolates obtained in the United States revealed that resistance to ceftriaxone in all isolates was mediated by cmy-2. Hybridization patterns revealed three plasmid structures containing cmy-2 in these 15 isolates. These data suggest that the spread of cmy-2 among Salmonella strains is occurring through mobilization of the cmy-2 gene into different plasmid backbones and consequent horizontal transfer by conjugation.
Salmonellosis is primarily a food-borne disease that affects an estimated 1.4 million people each year in the United States (14). Expanded-spectrum cephalosporins (e.g., ceftriaxone and cefotaxime) are the antimicrobial agents of choice in the treatment of pediatric patients with invasive Salmonella infections (9). Until recently, resistance to expanded-spectrum cephalosporins was rarely reported among Salmonella strains (8). Review of 1996 data from the National Antimicrobial Resistance Monitoring System (NARMS) in the United States identified only 1 (0.1%) ceftriaxone-resistant Salmonella isolate among 1,272 human Salmonella isolates. However, by 1999 almost 2% of Salmonella isolates were ceftriaxone resistant, as determined by review of 1999 NARMS data (6). Comparisons of these ceftriaxone-resistant isolates found divergent strains, indicating multiple probable sources. The isolates either were different serotypes or, among patients infected with Salmonella enterica serotype Typhimurium, were distinguishable by their pulsed-field gel electrophoresis patterns, thus demonstrating that these ceftriaxone-resistant human isolates did not represent the epidemic spread of a clonal strain (6). This study was undertaken to confirm the identity of the β-lactamase conferring resistance to expanded-spectrum cephalosporins and characterize the associated plasmids from the apparently sporadic human Salmonella isolates collected through NARMS from 1996 to 1998.
MATERIALS AND METHODS
The 15 bacterial strains used in the study are listed in Table 1. Thirteen of the isolates were obtained by the Centers for Disease Control and Prevention through NARMS. These 13 isolates represented 87% of the total expanded-spectrum cephalosporin-resistant Salmonella isolates (n = 15) obtained by the Centers for Disease Control and Prevention from 1996 to 1998 (6). Isolate SS034 was isolated in Nebraska, whereas isolate 922 was isolated in Ohio. Susceptibility testing of the Salmonella isolates and the Escherichia coli transconjugants and transformants was performed by the disk diffusion methodology according to NCCLS standards (16). The MIC for the pACYC184 construct containing cmy-2 was tested by the E-test (AB Biodisk, Solna, Sweden) methodology. The MICs of ceftiofur (kindly provided by Pharmacia/Upjohn) were determined by broth microdilution (15, 17). Plasmid DNA was extracted either by the method of Kado and Liu (10) or with the Concert Purification Midi kit (Life Technologies, Milan, Italy) and digested with PstI (Roche, Indianapolis, Ind.). Conjugation and transformation experiments were performed as described previously with E. coli C600N (ampicillin susceptible, nalidixic acid resistant) and E. coli DH5α as hosts (Gibco BRL, Bethesda, Md.) (2, 18, 19). Transformants were selected on Luria-Bertani agar (Difco, Detroit, Mich.) containing 50 μg of ampicillin (Sigma) per ml. All ceftriaxone-resistant C600N and DH5α transconjugants and transformants were subsequently named C6 or DH followed by the appropriate wild-type Salmonella strain designation (e.g., C6/SS034 and DH/4656).
TABLE 1.
Ceftriaxone-resistant Salmonella strains used in the studya
| Strain | Serotype | State in which strain was isolated | Antibiotic resistance phenotype | Tra | Transfer phenotype | Integron | Plasmid type |
|---|---|---|---|---|---|---|---|
| 922 | Typhim. | Ohio | ACSSuTGmToKTpCroXnlFx | Pos | ACSSuTGmToKTpCroXnlFx | ND | A |
| 2039 | Typhim. | California | ACSSuTGmToKTpCroXnlFx | Pos | ACSSuTGmToKTpCroXnlFx | In-t4 | A |
| In-t6 | |||||||
| 2152 | Typhim. | California | ACSSuTGmToKCroXnlFx | NT | In-t4 | A | |
| In-t6 | |||||||
| SS034 | Typhim. | Nebraska | ACSSuTGmToKCroXnlFx | Pos | ACSSuTGmToCroXnlFx | In-t6 | A |
| 4204 | Typhim. | New York | ACSSuTGmToKCroXnlFx | ACSSuTCroXnlFx | In-t6 | A | |
| 2855 | Typhim. | Oregon | ACSSuTKTpCroXnlFx | NT | In-t5 | A | |
| 3977 | Typhim. | Kansas | ACSSuTKCroXnlFx | ACSSuTCroXnlFx | In-t6 | A | |
| 4501 | Typhim. | Colorado | ACSSuTKCroXnlFx | NT | In-t6 | A | |
| 4656 | Typhim. | New York | ACSSuTCroXnlFx | ACSSuTCroXnlFx | Neg | C | |
| 4528 | Newport | Kansas | ACSSuTCroXnlFx | NT | Neg | C | |
| 2668 | Typhim. | Colorado | ACSSuTKCroXnlFx | Pos | ACroXnlFx | In-t6 | B |
| 4255 | Typhim. | Massachusetts | ASCroXnlFx | Pos | ACroXnlFx | Neg | B |
| 3430 | Typhim. | Massachusetts | ACroXnlFx | Pos | ACroXnlFx | Neg | B |
| 4287 | Typhim. | Massachusetts | ACroXnlFx | ACroXnlFx | Neg | B | |
| 1358 | Thompson | Connecticut | ACroXnlFx | Pos | ACroXnlFx | Neg | B |
Abbreviations: A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; T, tetracycline; Gm, gentamicin; To, tobramycin; K, kanamycin; Tp, trimethoprim; Cro, ceftriaxone; Xnl, ceftiofur; Fx, Cefoxitin; Typhim, Typhimurium; Tra, self-transferred plasmids by conjugation; NT, no ceftriaxone-resistant transformants isolated; ND, not done; Pos, positive; Neg, negative
Southern blot hybridizations were performed by standard methods (19) with a cmy-2-specific DNA probe labeled with [α-32P]dCTP with an RTS RadPrimer DNA Labeling kit (Life Technologies). DNA sequencing was performed with primers derived from known sequences and an ABI Prism model 377 sequencer (Perkin-Elmer Biosystems, Foster City, Calif.). The primers and DNA probes used to detect potential class 1 integrons have been described previously (4). Primers 92 (CCGTTTGTCAACACAGTAC [forward]) and 52 (TTGCAGCTTTTCAAGAATGCGCC [reverse]) were used to amplify full-length blacmy. Primer 92 was designed by using the sequence from the intercistronic region between ampC and ampR in Citrobacter freundii (GenBank accession no. X76636). Primer 52 was designed from the known cmy-2 sequence. Plasmid vectors pCRII (Invitrogen, Carlsbad, Calif.) and pACYC184 (5) were used in cloning experiments. Isoelectric focusing was performed at room temperature on a mini isoelectric focusing gel system (model 111; Bio-Rad, Richmond, Calif.) (13). The isoelectric points of unknown β-lactamases were estimated by comparison with those of TEM-1, SHV-3, SHV-5, and CMY-2.
RESULTS AND DISCUSSION
The antibiotic resistance phenotypes of the 15 strains under study are shown in Table 1. All isolates were resistant to ampicillin, ceftriaxone, ceftiofur, and cefoxitin. After mating experiments with C600N, 7 of 15 isolates were able to transfer decreased susceptibilities to ceftriaxone to C600N (Table 1). For those strains for which a transconjugant with decreased susceptibility to ceftriaxone was not isolated, plasmid DNA was isolated and used to transform E. coli DH5α. From the transformation experiments, an additional four transformants with reduced susceptibilities to ceftriaxone were isolated. The MIC of ceftriaxone was 8 to 32 μg/ml for all E. coli C600N transconjugants and DH5α transformants (hereafter these E. coli transconjugants and transformants will be referred to as ceftriaxone resistant). By isoelectric focusing, all ceftriaxone-resistant E. coli transconjugants and transformants expressed a β-lactamase (pI >9.0) that comigrated alongside CMY-2 (data not shown). In addition, primers specific for blacmy amplified an appropriate 631-bp DNA product from all ceftriaxone-resistant E. coli transconjugants and transformants and from the four Salmonella strains for which a transformant or transconjugant was not isolated (strains 2152, 2855, 4501, and 4528) (data not shown). Other resistance factors cotransferred with ceftriaxone resistance in 6 of 11 transconjugants or transformants (Table 1). Two strains (strains 922 and 2039) transferred all resistance factors to their corresponding E. coli transconjugant. The remaining five transconjugants or transformants were resistant only to β-lactam antibiotics.
The sequence of the blacmy gene obtained by PCR amplification was determined. DNA sequencing revealed that all strains encoded cmy-2, and no sequence divergence was detected in any strain. The DNA sequence found in the U.S. isolates was identical to the original cmy-2 sequence described in Klebsiella pneumoniae (3), yet it was different from the cmy-2-like sequence described in a ceftriaxone-resistant Salmonella serotype Senftenberg strain isolated in Algeria (11). Compared with the U.S. isolates, the Algerian isolate had three base pair changes within the first 50 bp, and two of these changes resulted in amino acid changes, suggesting that the cmy-2 gene disseminating throughout the United States is distinct from that in Algeria.
To further demonstrate that CMY-2 alone is responsible for mediating expanded-spectrum cephalosporin resistance in these isolates, CMY-2 was cloned by first amplifying cmy-2 from C6/SS034 with primers 92 and 52 and cloning it into pACYC184. As shown in Table 2, all strains were resistant or intermediate to ceftriaxone, ceftazidime, cefotaxime, ceftiofur, and cefoxitin. Both SS034 and DH/pNF10 were resistant to aztreonam; however, C6/SS034 was susceptible to aztreonam, perhaps due to the lower plasmid copy number or genomic background differences between C600N and DH5α. All strains were susceptible to cefepime and imipenem. Both C6/SS034 and DH/pNF10 were susceptible to piperacillin-tazobactam, as tazobactam is a known inhibitor of cmy-2 (3). The fact that strain SS034 also produces TEM-1 may have contributed to its resistance to piperacillin-tazobactam (7).
TABLE 2.
β-Lactam MICs for CMY-2-containing strains
| Isolate | MIC (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CRO | CAZ | CTX | XNL | FEP | TZP | IPM | ATM | FOX | |
| SS34 | >256 | >256 | >256 | 32 | 2 | >256 | 0.25 | 32 | >256 |
| C6/SS34 | 16 | 32 | 8 | 16 | 0.50 | 4 | 0.25 | 4 | 64 |
| DH/pACYC184 | 0.062 | 0.125 | 0.062 | 0.5 | 0.25 | 0.50 | 0.125 | 0.5 | 2 |
| DH/pNF10 | 64 | >256 | 64 | 8 | 0.5 | 4 | 0.5 | 32 | 64 |
Abbreviations: CRO, ceftriaxone; CAZ, ceftazidime; CTX, cefotaxime; XNL, ceftiofur; FEP, cefepime; TZP, piperacillin-tazobactam; IPM, imipenem; ATM, aztreonam; FOX, cefoxitin.
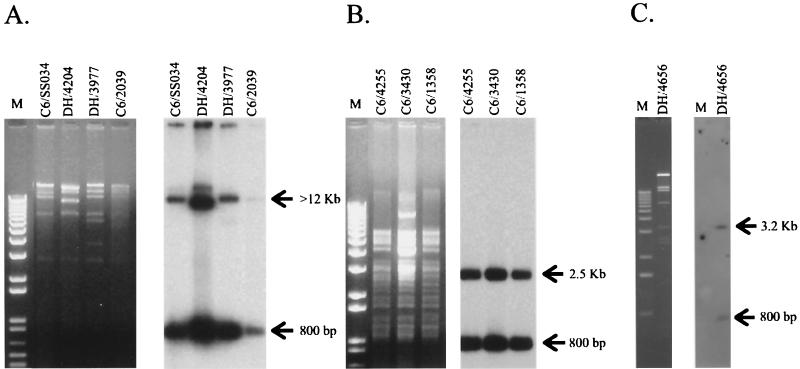
Plasmid DNA was isolated from the 11 E. coli transconjugants and transformants and the 4 wild-type Salmonella strains that did not yield a ceftriaxone-resistant transconjugant or transformant, and the plasmid DNA was probed with a cmy-2-specific probe (amplified with primers 92 and 52). This analysis demonstrated that cmy-2 was encoded on large plasmids (ca. 60 to 75 kb) in each strain (data not shown). The cmy-2-containing plasmids isolated from the E. coli transconjugants and transformants were additionally subjected to restriction endonuclease digestion with PstI (Roche) since a single PstI restriction site is present within cmy-2 (3), and the digests were analyzed by Southern hybridization with cmy-2 as a probe. Three PstI restriction fragment length polymorphism hybridization groups, referred to as types A, B, and C (Fig. 1), were observed. The cmy-2 probe hybridized to bands of approximately 12 kb and 800 bp (type A) and 2.5 kb and 800 bp (type B) for 8 and 5 of 15 cmy-2-containing plasmids, respectively (Fig. 1A, type A, and Fig. 1B, type B). For strains 4528 (wild type) and DH/4656, the cmy-2-specific probe hybridized to a 3.2-kb fragment and an 800-bp fragment (Fig. 1C, type C). PstI digestion of type B plasmids, which encode resistance only to β-lactam antibiotics, suggested that these plasmids were highly related. Plasmids with the type A or C hybridization pattern transferred resistance to at least four antibiotics (streptomycin, chloramphenicol, tetracycline, and sulfonamides), in addition to ceftriaxone (Table 1), but had different PstI restriction fragment length polymorphism patterns (Fig. 1). The significance of the conserved cmy-2 hybridization pattern in these plasmids is not known.
FIG. 1.
Restriction analysis (left panels in the pairs of panels) and cmy-2 Southern hybridization (right panels in the pairs of panels). PstI-digested plasmids were extracted from E. coli C6/SS034, DH/4204, DH/3977, and C6/2039 (type A hybridization pattern) (A); E. coli C6/4255, C6/3430, and C6/1358 (type B hybridization pattern) (B); and E. coli DH/4656 (type C hybridization pattern C) (C). A 1-kb marker (KiloBase DNA marker; Pharmacia Biotech, Milan, Italy) (A and B) and a 12-kb ladder (Gibco BRL) (C) were used as standards.
These data suggest that cmy-2 is being transferred among Salmonella strains by plasmid transfer to different genomic backbones as well as by independent acquisition of cmy-2 by different plasmid backbones, most of which carry multiple antibiotic resistance determinants. The mechanism of transfer and acquisition of cmy-2 is unknown; however, it appears that cmy-2 is not encoded within a cassette that inserts into a class 1 integron. Experiments for the detection of class 1 integrons were performed by both PCR amplification (12) and Southern hybridization by using the integrase gene as a probe (data not shown) (4). Class 1 integrons were detected in eight strains (Table 1); however, the cmy-2 gene was not included as an integron-borne gene cassette. Isolates 2039, SS034, 2152, 4204, 3977, 4501, and 2668 all contained an integron (In-t6) that carries the aadA2 gene cassette, which confers resistance to streptomycin and spectinomycin. Isolates 2039 and 2152 also carried an additional integron (In-t4) that encodes the cmlA and aadB gene cassettes, which confer resistance to chloramphenicol and kanamycin, respectively. One isolate, isolate 2855, contained a larger integron (In-t5) that carries the dfrA1 and aadA2 gene cassettes, which encode trimethoprim and streptomycin-spectinomycin resistance, respectively. Integrons were located on cmy-2-carrying plasmids only in isolates SS034 and 2039.
The results of this study demonstrate the emergence and spread of a CMY-2 β-lactamase in Salmonella strains isolated from humans in the United States. The ceftriaxone resistance reported in porcine, bovine, and human Salmonella isolates in Iowa and Nebraska was also mediated by cmy-2 (20, 7). The emergence of ceftriaxone resistance among Salmonella strains isolated from food animals supports the transfer of ceftriaxone-resistant Salmonella strains from food animals to humans (21, 1). In this study, we demonstrated that CMY-2 alone can mediate resistance to expanded-spectrum cephalosporins, including ceftiofur, by cloning the cmy-2 gene into pACYC184. Although the reasons for the emergence of resistance to expanded-spectrum cephalosporins in humans remain uncertain, the emergence of resistance in food animals may play a role. The increased prevalence of ceftriaxone-resistant Salmonella strains in food animals may in turn be related to the veterinary use of ceftiofur, an expanded-spectrum cephalosporin used only in veterinary medicine. Further studies are warranted to determine the risk factors for dissemination of cmy-2-mediated resistance and to determine whether limiting the use of ceftiofur in food animals, along with improvements in food processing methods, might reduce the potential for dissemination of ceftriaxone resistance.
Acknowledgments
We thank Emma Filetici, Susanna Mariotti, and Susan Greenwood for technical assistance. We also thank the Ohio Public Health Laboratory for Salmonella serotype Typhimurium isolate 922.
This research was partially supported by grants from the Italian Ministry of Health's “Progetto Antibiotico Resistenza” to A.C. and a University of Nebraska Medical Center grant to P.D.F.
REFERENCES
- 1.Angulo, F. J., K. R. Johnson, R. V. Tauxe, and M. L. Cohen. 2000. Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb. Drug Resist. 6:77-83. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, B. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 1190-1219. In F. C. Niedhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, and H. Giamarellou. 1996. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carattoli, A., L. Villa, C. Pezzella, E. Bordi, and P. Visca. 2001. Expanding drug resistance through integron acquisition by Inc plasmids of Salmonella enterica Typhimurium. Emerg. Infect. Dis. 7:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barret, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 7.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infections acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 8.Herikstad, H., P. S. Hayes, J. Hogan, P. Floyd, L. Snyder, and F. J. Angulo. 1997. Ceftriaxone-resistant Salmonella in the United States. Pediatr. Infect. Dis. J. 9:904-905. [DOI] [PubMed] [Google Scholar]
- 9.Hohmann, E. L. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263-269. [DOI] [PubMed] [Google Scholar]
- 10.Kado, C. I., and S. Liu. 1981. Rapid procedure for detection and isolation or large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koeck, J.-L., G. Arlet, A. Philippon, S. Basmaciogullari, H. V. Thien, Y. Buisson, and J. D. Cavallo. 1997. A plasmid-mediated CMY-2 β-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 12.Levesque, C., L. Piche, C. Larose, and P. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew, A., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 14.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffen, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial disk susceptibility tests; approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial disk dilution susceptibility tests for bacteria isolated from animals; approved standard M31-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Pitout, J., K. Thomson, N. Hanson, A. Ehrhardt, E. Moland, and C. C. Sanders. 1998. Beta-lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antmicrob. Agents Chemother. 42:1350-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]