Abstract
Posterior polymorphous corneal dystrophy (PPCD, also known as PPMD) is a rare disease involving metaplasia and overgrowth of corneal endothelial cells. In patients with PPCD, these cells manifest in an epithelial morphology and gene expression pattern, produce an aberrant basement membrane, and, sometimes, spread over the iris and nearby structures in a way that increases the risk for glaucoma. We previously mapped PPCD to a region (PPCD3) on chromosome 10 containing the gene that encodes the two-handed zinc-finger homeodomain transcription factor TCF8. Here, we report a heterozygous frameshift mutation in TCF8 that segregates with PPCD in the family used to map PPCD3 and four different heterozygous nonsense and frameshift mutations in TCF8 in four other PPCD probands. Family reports of inguinal hernia, hydrocele, and possible bone anomalies in affected individuals suggest that individuals with TCF8 mutations should be examined for nonocular anomalies. We detect transcripts of all three identified PPCD genes (VSX1, COL8A2, and TCF8) in the cornea. We show presence of a complex (core plus secondary) binding site for TCF8 in the promoter of Alport syndrome gene COL4A3, which encodes collagen type IV α3, and we present immunohistochemical evidence of ectopic expression of COL4A3 in corneal endothelium of the proband of the original PPCD3 family. Identification of TCF8 as the PPCD3 gene provides a valuable tool for the study of critical gene regulation events in PPCD pathology and suggests a possible role for TCF8 mutations in altered structure and function of cells lining body cavities other than the anterior chamber of the eye. Thus, this study has identified TCF8 as the gene responsible for approximately half of the cases of PPCD, has implicated TCF8 mutations in developmental abnormalities outside the eye, and has presented the TCF8 regulatory target, COL4A3, as a key, shared molecular component of two different diseases, PPCD and Alport syndrome.
Introduction
The cornea, positioned at the interface between the outside world and the internal structures of the eye, protects the eye from environmental insult while maintaining near-perfect transparency, so that light is able to pass undistorted to the lens (Spencer 1996). From the outer to the inner surface, the cornea consists of three main layers: an outer stratified epithelium, a thick middle layer of highly ordered collagenous structures and sparse cellularity called the “corneal stroma,” and an inner single layer of endothelial cells that produce an unconventional basement membrane called “Descemet membrane” and that maintain the hydration and homeostasis of the corneal stroma (Spencer 1996).
Posterior polymorphous corneal dystrophy (PPCD, also known as PPMD [MIM #122000]) is a rare corneal dystrophy that is often inherited in an autosomal dominant manner (Pearce et al. 1969; Heon et al. 1995; Moroi et al. 2003). Symptoms can range from very aggressive to asymptomatic and nonprogressive, even within the same family (Weisenthal and Streeten 1997). The age at diagnosis is, most often, in the 2nd or 3rd decade of life. PPCD usually affects both eyes.
Cellular abnormalities in PPCD include an altered corneal endothelial cell structure, an unusual proliferation of endothelial cells, an aberrant structure of the Descemet membrane, and a disturbed regulation of the protein expression pattern (Weisenthal and Streeten 1997). Metaplastic changes in the cells of the endothelium to cells with characteristics of epithelial cells include multilaminar growth (Boruchoff 1971; Rodrigues et al. 1980) and the expression of a set of cytokeratins that are not normally seen in the corneal endothelium but are typical of epithelial cells (Boruchoff 1971; Rodrigues et al. 1980). In addition to the cellular abnormalities, the posterior two-thirds of the corneal endothelial cell basement membrane, the Descemet membrane, is abnormal (Weisenthal and Streeten 1997).
Several genes have been implicated in PPCD through mapping studies and mutational analysis. PPCD1 was mapped to a 30-cM locus on 20q11 with the use of data from a five-generation family (Heon et al. 1995). Subsequent work on this locus revealed mutations thought to cause PPCD in the gene encoding the homeodomain transcription factor VSX1 (GenBank accession number NM_199425), although no VSX1 mutation was found in the family used for the initial mapping of the PPCD1 locus (Heon et al. 2002). Screening for mutations in the gene encoding collagen type VIII α2 (COL8A2 [GenBank accession number NM_005202]), a gene implicated in the much more prevalent Fuchs endothelial corneal dystrophy (FECD [MIM #136800]), revealed mutations in two related individuals with PPCD (Biswas et al. 2001).
In addition to these PPCD genes, two loci have been mapped for another related disease, congenital hereditary endothelial dystrophy (CHED [MIM #121700]), one on each arm of chromosome 20 (Toma et al. 1995; Hand et al. 1999). Additional insights into PPCD may be gained from observing that mutations in genes encoding several type IV collagens, COL4A3 (Lemmink et al. 1994) [MIM #120070], COL4A4 (Mochizuki et al. 1994) [MIM #120131], and COL4A5 (Barker et al. 1990) [MIM #303630], occur in individuals with Alport syndrome, which is characterized by hereditary nephritis and sensorineural hearing loss and can include PPCD (Alport 1927; Colville and Savige 1997).
We have previously evaluated the role of each of these genes and loci in PPCD in a large multigeneration family (UM:139). Using linkage analysis, we excluded all of the above genes from involvement in PPCD in this family (Moroi et al. 2003). Subsequent linkage analysis led to our recent report (Shimizu et al. 2004) that PPCD in family UM:139 is linked to a new PPCD locus, PPCD3, on 10p11.
One of >30 genes within the PPCD3 genetic inclusion interval is the gene that encodes the two-handed zinc-finger homeodomain transcription factor 8, TCF8 (GenBank accession number NM_030751, also known as AREB6, BZP, Nil-2a, ZEB1, ZFHEP, Zfxh1A, and δEF-1), which was originally reported as a repressor of Il-2 gene expression (Williams et al. 1991). TCF8 plays a critical role in embryonic development, since homozygous null mice exhibit many skeletal defects and do not survive beyond birth, whereas heterozygous null mutant mice exhibit less severe skeletal and T-cell abnormalities (Higashi et al. 1997; Takagi et al. 1998). There are no reports of ocular manifestations in either homozygous or heterozygous Tcf8-null mice, but these studies did not indicate that eyes were examined.
The unusual structure of TCF8, with its homeodomain region flanked on either side by zinc fingers, leads to a diverse set of activities. It has been shown that each zinc finger has slightly different DNA binding specificities, each possibly having a different effect on gene expression (Ikeda and Kawakami 1995). Of particular interest, with regard to evaluation of TCF8 as a candidate PPCD gene, are the previous reports that TCF8 plays a role in the regulation of type I collagen expression (Terraz et al. 2001) and in the repression of the epithelial phenotype (Frisch 1994; Grooteclaes and Frisch 2000).
In the present study, we show TCF8 mutations in both familial and isolate cases of PPCD. We present evidence of ectopic COL4A3 expression in the corneal endothelium of the proband of the original PPCD3 family, and we discuss the possibility that PPCD may include nonocular phenotypic features.
Material and Methods
Subjects
Study participants provided informed consent and blood samples in accordance with a protocol approved by the Institutional Review Board for Human Subject Research of the University of Michigan Medical School. Complete ophthalmologic examinations were performed as described elsewhere (Moroi et al. 2003), and participants were classified as affected with PPCD if they showed any of the following clinical features in at least one eye: posterior vesicles or vesicular, geographic, or band-like lesions at the level of the Descemet membrane (Cibis et al. 1977; Miller 1997). Physicians performing ophthalmologic exams and assigning affection status did not know the subjects’ genotypes. None of the 11 probands in this study reported having hearing problems or kidney disease that could indicate Alport syndrome. Controls are race matched to cases (all white). The ages of these controls ranged from 24 to 96, with an average age of 63.
Molecular Genetic Analysis
Genomic DNA preparation, genotyping, clinical data management, quality control, and data formatting for markers D10S1243, D10S1781, D10S1654, D10S675, and D10S1208 (Invitrogen) were performed as described elsewhere (Shimizu et al. 2004) to confirm haplotyping in the vicinity of TCF8 in family UM:139. Using the same methods, we also genotyped one marker per autosome to confirm the relationships in UM:A01 and UM:797 and to confirm that their mutations were de novo (data not shown).
Sequencing Analysis of TCF8, VSX1 and COL8A2
PCR amplification and sequencing of TCF8 and VSX1 were performed with the primers listed in table 1. COL8A2 was amplified and sequenced with primers described elsewhere (Biswas et al. 2001). PCR amplification was performed with AmpliTaq Gold (Applied Biosystems), with 2.5 mM MgCl2, 2.0 mM dinucleotide triphosphate, and 0.5 μM primers in a 25 μL reaction volume. PCRs were cleaned with the Qiagen PCR purification kit (Qiagen). Sequencing was performed at the University of Michigan Medical School Biomedical Sequencing Core facility with ABI Big Dye Terminator (Applied Biosystems) and read on either an ABI 3700 or 3730. Sequence traces were screened for mutations, both manually and through the use of PolyPhred (Nickerson et al. 1997), in conjunction with Phred/Phrap/Consed (Ewing and Green 1998; Gordon et al. 1998). Reference sequence used was NM_030751 (UCSC Genome Browser).
Table 1.
Sequences of Primers Used to Amplify and Sequence TCF8 and VSX1
| Gene | Exon | Sense | Antisense |
| TCF8 | 1 | AAAGCCGGGAGTGTCGTAA | AGTGCGGAAAGAAGCAACAG |
| 2 | GTTACTCTCTCTCTGCCTTG | TCCTTCCACTCAGCCATAC | |
| 3 | GAGCAAGAGTGTGATGGAAAG | CGGACTAAATTCAGGACTCAC | |
| 4 | CTTAGTGGTGAGATTGCTGTC | CCTATAGTAGAGCAGGTTCC | |
| 5 | GTGGGTAGCACAATATCTGG | AGGCTGCAGATATAGCACTG | |
| 6 | GACTGCTGCAATTTGAGGTCT | GCTAATTGTGCAAAGGAGGC | |
| 7 | CAGTTCTGTCACAAGCATGC | TGCACTGAAATCTGTCCAGC | |
| 7 | GACCTAAAGCAGCCTACTCA | TTGGCTCTACGGGACTGATA | |
| 8 | GATCAGTGTGCTTGCTTTGG | ACTAGAGCCAGACCTTGTCT | |
| 9 | GAGTTTGGGACCTGGAAATG | TACTAGGTCTGAGGTTCAGG | |
| VSX1 | 1 | AAAGCTTCCTCTAAGCTGGG | CAGGGATTTAGGATGCAGCA |
| 2 and 3 | GGCACTAAAAATGCTGGCTC | AAGGGAGCGTGTTGGCTATA | |
| 4 | TCGGGAGCTATTTCCTTCAGAAAT | TGCTTTGGAAATGGCTGCCT | |
| 5 | CTAAGGTGGGAGTTACCTAC | TCCCTAGGTCACTCATCTGT |
Strand-Specific Sequencing
To confirm deletion and insertion mutations observed as mixed sequence, the two alleles were cloned so they could be sequenced separately. Fragments containing insertion or deletion mutations were amplified as described above and subcloned with the pGEM T-Easy Vector System (Promega), according to the manufacturer’s instructions. Cloned DNA was isolated with the Mini-Prep Kit (Qiagen), followed by bidirectional sequencing according to the method described above, with T7 and SP6 sequencing primers.
Mouse RNA Isolation and cDNA Synthesis
Tissues from 180-d-old C57BL/6 mice were stored in TRIzol (Invitrogen) immediately after harvesting. RNA was isolated by the TRIzol method in accordance with the manufacturer’s protocol. Total RNA was purified with an RNeasy Mini Kit (Qiagen). First-strand cDNA synthesis was performed with a SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen).
Quantitation of RNA Levels
Real-time semiquantitative PCR (sqPCR) was performed according to the manufacturer’s protocol, with the iQ SYBR Green Supermix (BioRad) with 250 nM primers for mouse: Tcf8, forward–5′-TGTCTTAGCTCCTCTCTTG-3′ and reverse–5′-ACTGCCCAGTTACCCACAAT-3′; Vsx1, forward–5′-ACTGCCCAGTTACCCACAAT-3′ and reverse–5′-AAGTGGCGTAAGCGAGAGAA-3′; COL8A2, forward–5′-GGTAAAGTATGTGCAGCCCA-3′ and reverse–5′-AGTAATACCTGAGGGACCAG-3′; and RpL19, forward–5′-GGGAAGAGGAAGGGTACTGC-3′ and reverse–5′-GGACGCTTCATTTCTTGGTC-3′ on an iCycler (BioRad), under standard PCR conditions, with an annealing temperature of 59°C. All sqPCR primer pairs were designed so that the primers were located in different exons, resulting in an intron-spanning amplification event that could distinguish amplification of reverse-transcribed RNA from amplification of genomic DNA. Data were analyzed as described elsewhere, with RpL19 as a housekeeping control (Mandal et al. 2004).
Immunohistochemistry
Formalin-fixed, 5-μm paraffin sections were rehydrated to buffer and were treated with Proteinase K for 5 min. After peroxidase blocking, the sections were incubated with serum, to block nonspecific binding, and then with monoclonal antibodies MAB-1, MAB-3, and MAB-5 (Wieslab), each at a concentration of 1:100, at room temperature for 30 min. With use of the DAKO AutoStainer and LSAB+ Kit (DAKO Cytomation), the sections were then exposed to biotinylated secondary antibody, developed with chromagen for 5 min, and counterstained with hematoxylin.
Bioinformatics
Human sequences were annotated with the protein translation and primer sequences with use of the Sequence Manipulation Suite. The sequence upstream of the basement membrane collagen genes and the short-chain collagen gene COL8A2 were searched for the TCF8 core binding site ([T/G][T/G]CACCT[T/G]T) and for proximal downstream secondary TCF8 binding sites (GTTT[C/G]) 100 bp after the core site (Ikeda and Kawakami 1995). This sequence pattern search was performed with use of the fuzznuc module in the EMBOSS package (Rice et al. 2000). Binding site simulations used the Shuffle Utility, developed by Edward H. Trager at the University of Michigan Department of Ophthalmology and Visual Sciences. Shuffle was used to generate 210,000 randomizations of the 20 kb upstream of the start codon of COL4A1, COL4A2, COL4A3, COL4A4, COL4A5, COL4A6, and COL8A2. These randomizations were performed by picking bases at random from the native promoter sequence to fill out the randomized sequence one base at a time, thus retaining the precise sequence composition of the native promoter sequences while scrambling the order. The simulations were completed with the use of fuzznuc to search for the specific binding-site sequences within the randomized sequences.
Statistical Analysis
The significance of the binding-site simulations was evaluated by empirical P values, which were calculated by taking the number of simulations showing binding sites across all seven promoters equal to or greater than the number found in the native promoters and dividing that number by 210,000.
The independence of a particular SNP allele or genotype and the PPCD phenotype was assessed with the use of a χ2 test for independence, with P values corresponding to χ2 with 1 df comparing allele frequency by disease status (table 2). A model-based χ2 test for independence of SNP alleles and the PPCD phenotype was performed by comparing the number of affected individuals with and without the major allele with those in the control sample. One of the two intronic SNPs, c.685A→G, is in dbSNP (accession number rs220060).
Table 2.
SNPs Found in Intronic Regions of TCF8
|
Probands |
Controls |
||||
| Intronic SNP | No. ofReference Alleles | No. of Variant Alleles | No. of Reference Alleles | No. of Variant Alleles | P value |
| c.484+67A→G | 19 | 3 | 219 | 55 | 0.39 |
| c.685−15G→A | 3 | 19 | 13 | 261 | 0.06 |
Results
TCF8 Frameshift Mutation in the Original PPCD3 Family
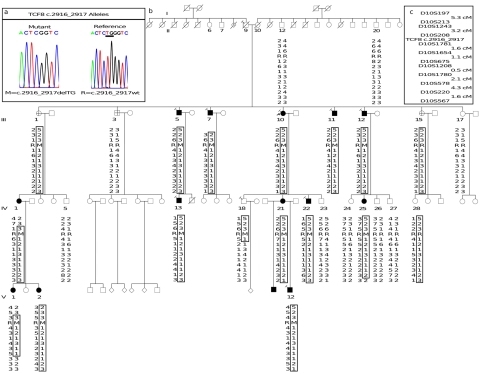
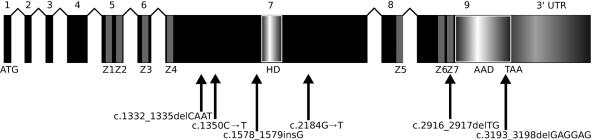
Screening for mutations in TCF8 in the proband of family UM:139 revealed the presence of a 2-bp deletion, 2916_2917delTG (fig. 1a), in the last exon of the gene. This frameshift mutation alters the amino acids downstream of the deletion and maintains an open reading frame for 13 codons before the premature termination signal, which, if a protein is produced, would cause the elimination of most of the last zinc-finger cluster and the acidic activation domain (fig. 2). Follow-up screening of other members of UM:139 showed that the mutation is present in every one of the 13 affected family members who were screened (fig. 1b).
Figure 1.
Haplotype and mutational analysis of the TCF8 2916_2917delTG mutation in family UM:139. a, Based on sequencing of cloned versions of the two alleles present in UM:139, the frameshift mutation (M) sequence trace is shown on the left, and the corresponding reference (R) sequence, NM_030751, is shown on the right. b, Haplotyping over the 8.55-cM PPCD3 region, with the 2916_2917delTG mutation designated M and the reference allele designated R. All relationships of genotyped individuals are correct as depicted, as determined from the previously reported UM:139 genome scans (Moroi et al. 2003; Shimizu et al. 2004). A large number of nonparticipating descendants of members of generation II do not appear in this figure. Open symbols indicate absence of PPCD, filled symbols indicate affected individuals, flags indicate individuals with hernia, and crossed symbols indicate individuals considered to have indeterminate phenotypes, since they do not have PPCD but do have other corneal findings, such as guttae or FECD. c, Traits that did not cosegregate with the mutation include late-onset hearing loss, migraine headaches, lupus, and arthritis. The locus map shows the order of markers and intermarker distances. The germline of II-8 contains three distinct versions of chromosome 10 shown among his progeny: unaffected haplotype without deletion (III-3 and III-17), affected haplotype plus deletion (III-1, III-5, III-7, III-10, III-11, and III-12), and affected haplotype without deletion (III-15 and IV-28). The three possible explanations include back mutation, a double recombination event, and germline mosaicism in individual II-9.
Figure 2.
Structure of the TCF8 transcript. Locations of the mutations and the sequence variant relative to exons encoding functional domains are shown. TCF8 encodes a homeodomain (HD) flanked by two zinc-finger clusters (Z1–Z4 and Z5–Z7). An acidic activation domain (AAD) is encoded after the last zinc-finger domain.
If we consider unaffected family members, we find that penetrance is apparently incomplete. Individual III-1, who has the mutation but does not have PPCD, represents apparent nonpenetrance, since both PPCD and the mutation are present in her child (IV-1) and two grandchildren (V-1 and V-2); however, we cannot rule out the possibility that she will still develop the trait at a later age. Individual IV-18, who was 27 years old at the time of examination and carries the affected haplotype and the deletion but is apparently not affected with PPCD, may represent a case of age-related penetrance, because some affected family members showed onset at a later age.
Data for individual III-15 and her siblings suggest possible germline mosaicism in their father, individual II-9. On the basis of haplotyping performed in coordination with the mapping of the locus, III-15 and her daughter IV-28 originally appeared to be cases of nonpenetrance, because they both carried the entire affected haplotype but were not affected with disease (Shimizu et al. 2004). However, III-15 and IV-28 both possess the affected haplotype without possessing the TCF8 mutation present in their affected relatives. Fine mapping of this region in this family confirms the transmission of the entire affected haplotype from II-9 to III-15 and then to IV-28 (fig. 1), so a double recombination event would have to be confined to a region of ∼1.8 million bases to explain the presence of the normal allele on the affected haplotype. Back mutation in III-15 seems highly unlikely, since it would require reinserting two bases at exactly the correct position in the gene. Germline mosaicism seems a more probable explanation for the fact that II-9 apparently transmitted three distinct versions of this section of chromosome 10 to his children: the affected haplotype with the deletion, the affected haplotype without the deletion, and the normal haplotype without the deletion. However, additional information on genotypes from generation II, especially siblings of II-9, would be needed to take this beyond the realm of speculation.
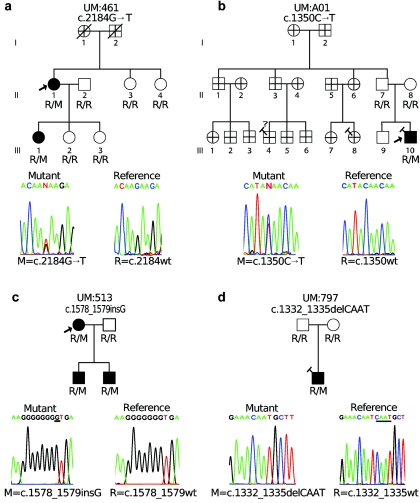
TCF8 Mutations in Additional Families
Screening for TCF8 mutations in samples from the probands of 10 other unrelated PPCD families revealed that 4 of these 10 probands carry mutations that terminate or shift the reading frame (fig. 3). Three of the mutations (c.1332_1335delCAAT, c.1350C→T, and c1578_1579insG) would, if truncated protein were produced, eliminate the homeodomain, the second cluster of zinc fingers, and the acidic activation domain (fig. 2). The c.2184G→T mutation would result in the elimination of the second cluster of zinc fingers and the acidic activation domain (fig. 2). The c.2184G→T and c.1578_1579insG mutations segregate with disease in these families, but the data are not statistically significant because of small family size (fig. 3a and 3c). The other two mutations (c.1350C→T and c.1332_1335delCAAT) are found in the only affected member of their families and are likely examples of de novo mutations (fig. 3b and 3d). To verify parentage in these two trios in the presence of the apparent de novo mutations, one microsatellite marker on each autosome was genotyped, and each trio was found to show inheritance consistent with the expected Mendelian pattern (data not shown).
Figure 3.
Additional PPCD families with TCF8 mutations. The genotype of each individual at the relevant locus is displayed. M indicates the mutant allele and R indicates the reference allele. In each case, the mutant sequence trace is on the left and the sequence trace corresponding to the reference allele is on the right. a and b, Families with nonsense mutations. c and d, Families with frameshift mutations. Symbols are as described in figure 1. Relatives in UM:A01 are shown as having indeterminate ocular phenotype because we have not examined them and have no records on them, but since the proband’s father has been confirmed and has neither PPCD nor the TCF8 mutation, we do not expect to find PPCD or the TCF8 mutation in that branch of the family. Relationships are confirmed as depicted for all genotyped individuals.
There were no nonsense or frameshift mutations among 128 race-matched (white) normal controls. One 69-year-old normal control had a novel in-frame deletion (c.3193_3198delGAGGAG) that was not seen in the PPCD cases. This deletion removes two residues in a repeat of seven glutamic acid residues (E1065_E1066delEE). Whereas the presence of a glutamic acid repeat at this location is well conserved across species, the specific length of the repeat is not conserved (data not shown). Although it is possible that this normal control individual could develop PPCD after the age of 69, these data suggest that this particular deletion is benign.
In addition to the protein-altering changes described above, SNPs were found in PPCD cases and normal controls. One of these SNPs was found 67 bases downstream of the end of exon 4 (c.484+67A→G), and the other was found 15 bases upstream of the start of exon 6 (c.685−15G→A). Neither of these SNPs provides strong evidence of differential distribution among cases compared to controls when comparing the frequency of alleles, genotypes, or the common allele among affected individuals (table 2).
The Role of Other PPCD Genes in These Families
We resequenced all known exons and adjacent splice sites of the previously reported PPCD genes VSX1 (Heon et al. 2002) and COL8A2 (Biswas et al. 2001) in DNA samples from our probands. We found one novel silent change (R166R, c.496C→G) in VSX1 that is located at the same position as a previously reported VSX1 SNP but involves a different substitution (Heon et al. 2002). This change creates an AG, but neither NetGene nor NNSPLICE splice recognition software (Brunak et al. 1991; Hebsgaard et al. 1996; Reese et al. 1997) recognizes this as a potentially new acceptor splice site, although both programs do recognize the actual acceptor splice site for that exon. No protein-altering base changes were found in either gene, indicating that mutations in these genes are unlikely to be the cause of PPCD in our subjects.
Nonocular Phenotypic Features
By family report we find that, among the 14 males with TCF8 mutations, inguinal hernia and/or hydrocele is reported to be present in 11 individuals with PPCD, absent from 1, and uninformative in 2 for whom we lack information regarding hernias or hydroceles (UM:513) (fig. 1 and fig. 2). Among the females with TCF8 mutations, we found one with umbilical hernia subsequent to Cesarean section delivery, one with failure to form a vaginal opening, seven with no report of hernia, and one for whom the information is not available. It is unclear whether any of these traits in women are related to the TCF8 mutation. Some additional reports of hernia among uncharacterized branches of several families raise questions about how simple or complex the situation will appear by the time everyone is fully characterized for both ocular and nonocular characteristics.
We find evidence of orthopedic anomalies among affected individuals in the four TCF8 mutation families for whom information was available. The proband of UM:139, the main person for whom any details are available, reported extra vertebrae in her lower back, bone spurs on vertebrae, and lumps on her kneecaps. Bony lumps on the palms and soles of the feet, associated with Dupuytren’s contractures, are reported for four affected members of UM:139, and the orthopedic trait, Osgood-Schlatter disease, was reported among one affected individual in each of UM:139 and UM:797. Otosclerosis was observed in one member of UM:139, but other cases of late-onset hearing loss that do not cosegregate with PPCD may be unrelated. Other indications of possible orthopedic anomalies include reports of kneecap dislocations and bone spurs which, in some cases, were severe enough to warrant surgery, but radiographs were not available to confirm any of the minor bone anomalies seen in the mice heterozygous for the Tcf8 knockout mutation (Takagi et al. 1998). Evaluation of medical records or examination will be required to allow us to draw firm conclusions about bone anomalies in PPCD.
A review of the records for family UM:139 indicates the presence of ocular phenotypes besides PPCD. We have previously reported the presence of guttae in several individuals in the family (Shimizu et al. 2004), but guttae do not cosegregate with PPCD or TCF8 mutations. A review of records also revealed that one member (III-3) of the family has been diagnosed with the more common corneal dystrophy, Fuchs endothelial corneal dystrophy (FECD), and the proband reports that two others have been diagnosed with FECD (III-11 and III-112), but we do not have recent-enough records to confirm this. Neither guttae nor FECD appear to cosegregate with PPCD or TCF8 mutations.
Evaluation of Expression of PPCD Genes In Ocular and Nonocular Tissues
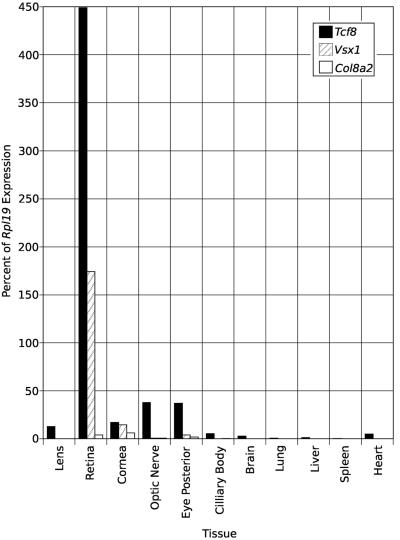
Real-time semiquantitative PCR (sqPCR) on mouse tissue confirms the expression of all three known PPCD genes, Tcf8 (GenBank accession number NM_011546), Vsx1 (GenBank accession number NM_054068), and Col8a2 (GenBank accession number NM_199473), in the eye and, more specifically, in the cornea (fig. 4). Tcf8 and Vsx1 are expressed at rather high levels in the retina, showing mRNA levels at 450% and 178%, respectively, that of the housekeeping control, ribosomal subunit gene Rpl19 (GenBank accession number NM_009078). Col8a2 shows more modest expression in all tissues, including the retina, with expression levels at ∼4% that of Rpl19. All three genes show low-level expression in the cornea, with Tcf8 and Vsx1 showing similar levels of expression at 17% and 14%, respectively, of Rpl19 expression, whereas Col8a2 shows lower levels of expression at ∼6% of Rpl19 expression. Elsewhere in the body, Vsx1 shows very low expression levels in the brain (0.03%) and absent expression in the lung, liver, and spleen. Col8a2 shows very low levels of expression in all of the nonocular tissues screened, with the highest level seen in the heart at 0.2% of Rpl19 expression. Tcf8 shows low-level expression in these organs, with 3% and 5% of Rpl19 expression in the brain and heart, respectively, and >1.5% in the other organs.
Figure 4.
Quantitation of transcript levels. Real-time sqPCR on cDNA from various mouse tissues used primers for mouse genes Tcf8 (NM_011546), Vsx1 (NM_054068), and Col8a2 (NM_199473). Eye posterior includes retinal pigment epithelium, choroid, and sclera.
In Silico Binding Site Search
On the basis of evidence from previous studies that (1) collagen abnormalities occur in the Descemet membrane in PPCD (Terraz et al. 2001), (2) COL8A2 is implicated in both FECD and PPCD (Colville and Savige 1997), and (3) defects in α-type 4 collagen genes are involved in Alport syndrome (Colville and Savige 1997), we did a preliminary evaluation of the potential for TCF8 regulation of expression of these genes by searching the promoter regions of basement membrane collagens COL4A1, COL4A2, COL4A3, COL4A5, and COL4A6, and the short-chain collagen, COL8A2, for possible TCF8 binding sites (Ikeda and Kawakami 1995) (table 3). A search of the 20 kb upstream of the transcription start shows 12 possible TCF8 core binding sites ([T/G][T/G]CACCT[T/G]T) (Ikeda and Kawakami 1995) across all 7 of these genes, with 3 sites in COL4A2, 2 each in COL4A3, COL4A5, COL4A1, and COL8A2, 1 in COL4A6, and none in COL4A4. Statistical analysis of 210,000 in silico simulations, involving searches for this core sequence after scrambling the promoter sequences, indicates that the probability of observing these data across all seven genes by chance for the core sequence alone was unlikely (P=.0001). However, functional studies would be needed to evaluate the biological significance of these observations.
Table 3.
Putative TCF8 Binding Sites in Promoters of Basement Membrane Collagens and COL8A2
| Gene Symbol and Core Site Sequence | Bases from ATG to Core Site | Proximal Downstream Secondary Site Sequence | Bases from ATG to Secondary Site |
| COL4A1: | |||
| GTTACCTGT | 18,539 | … | … |
| TGCACCTGT | 13,723 | … | … |
| COL4A2: | |||
| TGCACCTGT | 11,753 | … | … |
| GTTACCTTT | 10,184 | … | … |
| TGTACCTGT | 7,394 | … | … |
| TTCACCTGT | 910 | … | … |
| COL4A3: | |||
| GTCACCTGT | 3,902 | GTTC | 3,835 |
| TGCACCTGT | 1,161 | … | … |
| COL4A5: | |||
| TGCACCTGT | 11,721 | … | … |
| TGTACCTGT | 6,743 | … | … |
| COL4A6: | |||
| GGCACCTGT | 5,936 | … | … |
| COL8A2: | |||
| TTCACCTGT | 9,119 | … | … |
| TGCACCTGT | 3,481 | … | … |
A secondary sequence (GTTT[C/G]) increases the strength of TCF8 binding when it is found close to the core sequence (Ikeda and Kawakami 1995). A corresponding search of these promoter regions shows that only the COL4A3 promoter contains a complex site consisting of the core and proximal (within 100 bases) downstream secondary sequence.
Immunohistochemical Examination of Type IV Collagen Expression
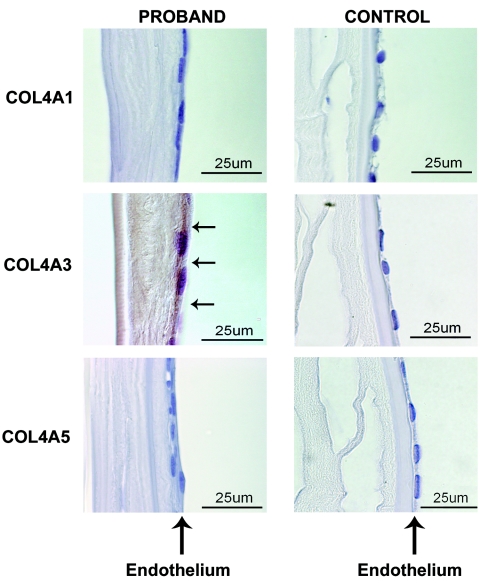
On the basis of the observation that the promoter of COL4A3, one of the Alport syndrome collagen genes, has both a core and a secondary binding site, whereas the other genes tested do not, we evaluated COL4A3 and two related collagens (COL4A1 and COL4A5) for altered expression in corneal endothelium in the presence of a TCF8 mutation. We evaluated the expression of these proteins in the cornea of the proband of family UM:139 and in a control cornea from an individual without PPCD. In figure 5, we show that COL4A1 and COL4A5, the promoters of which lack a secondary binding site, are not expressed in either the PPCD or control cornea. In contrast, COL4A3 is expressed in the corneal endothelium of the UM:139 proband, but not in that of the control eye.
Figure 5.
Immunohistochemical detection of type IV collagens in corneal endothelium. The corneal endothelium, as marked by vertical arrows, is on the right in each panel. Positive intercytoplasmic staining for COL4A3 in the UM:139 proband’s corneal endothelial cells is indicated by horizontal arrows in the sample from the proband COL4A3 panel. In the proband panels, there is a retrocorneal fibrous membrane interposed between the endothelium on the right and the well-defined Descemet membrane on the left. This retrocorneal fibrous membrane is not present in the control panels. COL4A1 and COL4A5 signals are not seen in samples from either the proband or the control.
Discussion
Mutations in TCF8 Cause PPCD
Our data suggest that a large proportion of PPCD is caused by mutations in the gene encoding the two-handed zinc-finger homeodomain transcription factor TCF8. Although the sample size is too small to allow a precise determination of TCF8 mutation frequency, 5 of 11 probands (45%) in this study were found to have PPCD attributable to TCF8 mutations. None of our probands had mutations in PPCD genes VSX1 or COL8A2, but, in previous studies, mutations in VSX1 and COL8A2 account for smaller proportions of the PPCD population (9% and 7%, respectively) (Biswas et al. 2001; Heon et al. 2002). Thus, the causative mutation remains to be accounted for in a sizable fraction of the PPCD population, which suggests that there might be at least one additional PPCD gene remaining to be found.
Both skipped generations and unaffected status among younger mutation carriers must be kept in mind when dealing with PPCD. Penetrance is high but incomplete in families in which PPCD results from TCF8 mutation, and age-related penetrance may be an issue. In three families with TCF8 mutations in which additional affected relatives were available to be tested (UM:139, UM:461, and UM:513), all affected family members each have one copy of the TCF8 mutation found in the probands of those families. However, in the one large family studied (UM:139), we found a case of nonpenetrance (III-1) and another individual whose unaffected status may be the result of age-related penetrance (IV-18).
Two cases, previously reported elsewhere, of apparent nonpenetrance (III-15 and IV-28), on the basis of initial haplotyping in family UM:139 (Shimizu et al. 2004), turn out to lack the deletion mutation present in their affected relatives. Although alternative scenarios, such as local recombination and back mutation, were considered, the most likely explanation seems to be germline mosaicism in individual II-9. This individual appears to have passed three different copies of chromosome 10 to his children: one with the affected haplotype and the deletion, one with the affected haplotype and no deletion, and one with the unaffected haplotype and no deletion. Similar situations have been reported in the literature. A recent Alzheimer disease study presents a situation in which siblings received the identical haplotype from an affected parent, but only one inherited the causative mutation (Beck et al. 2004). Similar situations have also been reported in VATER association (Reardon 2001), von Hippel-Lindau disease (Sgambati et al. 2000), X-linked α-thalassaemia mental retardation syndrome (Bachoo and Gibbons 1999), and hemophilia B (Cutler et al. 2004).
De Novo Mutations
Some of the TCF8 mutations observed in this study are de novo mutations. If we are correct in our hypothesis that II-9 in UM:139 was a germline mosaic, then 60% (3 of 5) of the mutations found in this study are likely de novo mutations, including 25% (1 of 4 probands) of familial cases of PPCD, as well as 29% (2 of 7) of nonfamilial cases. The significance of this unusual level of de novo mutations remains unclear. We did not find a single TCF8 mutation shared by multiple families that might indicate a hotspot for mutation, and amplification of a simple-sequence repeat is not involved. One alternative basis for a high de novo mutation frequency would be reduced reproductive fitness of the mutant allele(s), but PPCD does not cause the kind of death or disability before reproductive age that is seen in some other disorders with many reports of germline mosaicism and de novo mutation such as DMD (Bullock et al. 1996; Mukherjee et al. 2003; Ferreiro et al. 2004). It is interesting to note that, in the one family in which the mutation has been transmitted to progeny across four generations, the deletion is the most C-terminal of the mutations, which would leave most of the functional domains intact if the gene product is, indeed, produced (fig. 2). It may be that the higher-than-expected de novo mutation rate observed here is simply due to chance and that screening a larger PPCD population for mutations in TCF8 will yield a lower de novo mutation rate. This issue is, however, worth further examination, given the apparent co-occurrence of nonocular traits with TCF8 mutations.
Expression of the PPCD Genes
Our finding that all three PPCD genes, Tcf8, Vsx1, and Col8a2, are expressed in the cornea suggests that the endothelial to epithelial changes that characterize PPCD may result from the direct consequences of mutations in these genes in corneal endothelial cells, rather than from indirect processes that originate elsewhere. In the BodyMap database, TCF8 transcripts are reportedly localized to the actual endothelial layer. Our data (fig. 4), plus information from the UniGene Database, indicate that TCF8 is expressed in a variety of organs, and its role in repressing genes such as E-cadherin has been reported in cancers originating from a number of different cell types (Navarro et al. 1993; Ohira et al. 2003). Given the expression pattern of this gene, other nonocular tissues or organs may be susceptible to as-yet unidentified effects of TCF8 protein truncation or haploinsufficiency.
The PPCD3 Phenotype
The ocular findings of the PPCD3 phenotype, as exemplified by the subjects in UM:139, include corneal vesicles and bands in the corneal endothelium, retrocorneal membrane, and syneciae in the drainage angle (Moroi et al. 2003; Shimizu et al. 2004). Although guttae and FECD were also seen in family UM:139, neither cosegregates with the TCF8 mutation or the PPCD phenotype. No unusual ocular features other than PPCD were noted in UM:139 or the other families. The evidence for dysfunction of the cornea affected by TCF8 mutations is also supported by histopathology presented here and elsewhere (Moroi et al. 2003). Thus, the PPCD3 ocular phenotype resulting from TCF8 mutation appears to be a simple PPCD phenotype, although it shows great variation in range of severity (Moroi et al. 2003).
Although most studies of PPCD have focused on the eye, there is also reason to ask whether TCF8 mutations might involve abnormalities in tissues such as the lining of the abdominal cavity, since, when responding to injury, cells lining body cavities can show a pattern of metaplasia similar to that seen in the cornea in PPCD (Waring 1974). Inguinal hernia is a relatively common pediatric condition occurring in 10–20 per 1,000 live births and ∼6 times more often in boys than in girls (Tam 1990; Kapur et al. 1998), but our data suggest a dramatically higher risk of the related conditions hernia or hydrocele among males with TCF8 mutations. It is estimated that as many as 20% of inguinal hernia patients have a positive family history (Tam 1990), pointing to a potential genetic etiology for this common condition. Inguinal hernias are also associated with several inherited connective tissue diseases like Ehlers-Danlos syndrome (Carr et al. 1994) and primary joint hypermobility syndrome, which also includes features of Marfan syndrome, Elhers-Danlos syndrome, and osteogenesis imperfecta (Skoumal et al. 2004). It has previously been proposed elswhere that defects in the level of collagen synthesis, like those seen in PPCD, are involved in the development of hernias (Bendavid 2004). The location and congenital nature of most of the hernias/hydroceles in these families points to a developmental anomaly in which the inguinal ring remains open after the final migration of the sex organs, leading to an inguinal hernia (Tam 1990; Kapur et al. 1998).
Additional evidence for possible developmental effects of TCF8 mutations comes from studies of a mouse Tcf8 knockout model (Takagi et al. 1998). The homozygous animals with both copies of Tcf8 knocked out showed profound abnormalities in bone development. The heterozygous animals that represent a more appropriate model for PPCD showed minor bone abnormalities, including fusion of the smallest ribs. The presence of a variety of orthopedic anomalies among affected members of three of the families include extra vertebrae, bone spurs, otosclerosis, Osgood-Schlatter disease, and Dupetreyn’s contractures, but clinical details and radiographs will be needed to fully evaluate which orthopedic anomalies are part of the phenotype.
Thus, our evidence suggests that PPCD should no longer be considered a strictly ocular trait. The co-occurrence of TCF8 mutation and hernia/hydrocele provides strong evidence of greatly increased risk of at least one nonocular feature of PPCD. The evidence for orthopedic anomalies is less compelling because more information is needed. Given that TCF8 is expressed at many locations in the body, there remains the possibility that there are additional nonocular features to PPCD that were not identified from the limited information available by family report. Clearly, we need not only a more detailed evaluation of nonocular developmental and clinical features that can result from TCF8 mutations but also to evaluate those same features in PPCD in families lacking TCF8 mutations, so that we can determine which are specific to TCF8 mutation cases and which apply to PPCD in general.
Haploinsufficiency
Haploinsufficiency may play a role in the PPCD phenotype because all of the mutations we observed are predicted to truncate the protein, a situation that has sometimes been found to lead to nonsense-mediated decay of the mutant message (McIntosh et al. 1993) (fig. 3). Further studies will be needed to evaluate this hypothesis. Alternatively, haploinsufficiency could result if the protein is made but lacks activity. Indeed, haploinsufficiency is a well-established disease mechanism for other homeodomain transcription factors, such as LMX1B (Vollrath et al. 1998) and PAX6 (Martha et al. 1994). If haploinsufficiency is the mechanism by which TCF8 mutations affect regulation of gene expression, then the retina might be spared the effects of partial loss of TCF8 function because the retinal level of the Tcf8 transcript is >10 times that observed in the cornea. Thus, the levels of TCF8 may be sufficient to maintain retinal function, whereas this may not be the case in the cornea, where expression levels are much lower.
Alternatively, if the protein is indeed made from the mutant allele (fig. 2), a truncated TCF8 protein might lose some functions but not others, rather than simply reducing overall TCF8 functional levels. Elimination of some functional domains while leaving others has the potential to result in novel regulatory activities, with the possibility that dominant negative effects could be involved. Because cell-specific variation in TCF8 functions has been reported (Ikeda and Kawakami 1995; Costantino et al. 2002), novel dominant negative effects could affect the cornea, but not the retina, if the critical functions are relevant to the cornea, but not to the retina.
Models
Before our study, models for PPCD needed only to explain corneal endothelial metaplasia as well as the abnormalities in Descemet membrane. Our study suggests that such models now also need to account for altered gene regulation and developmental events elsewhere in the body. On the basis of genetic evidence from the five families with TCF8 mutations reported here and the molecular evidence from expression studies, we propose that TCF8 mutations produce the full PPCD phenotype through failure to correctly regulate multiple, different genes in more than one cell type in the body.
The findings reported here support our model, in which aberrant regulation of basement membrane collagen synthesis contributes to aberrant formation of Descemet membrane. We showed aberrant expression of COL4A3 in the presence of a heterozygous TCF8 mutation in the corneal endothelium of the proband of UM:139, but not in the normal cornea, which is in keeping with previous immunoelectron microscopy studies (Sawada et al. 1984; Sawada et al. 1987). Mutations in genes encoding the basement membrane collagens COL4A3 and COL4A5 have previously been shown elswhere to disrupt the structure of some basement membranes, leading to PPCD associated with Alport syndrome (Colville and Savige 1997). Thus, we find COL4A3 as a shared molecular component of the pathology of PPCD and Alport syndrome, which can include PPCD as one aspect of the phenotype.
Abnormal basement membrane formation may go beyond ectopic expression of COL4A3. We have previously shown that some cytokeratins that are usually expressed in the corneal epithelium are abnormally present in the diseased corneal endothelium of the UM:139 proband (Moroi et al. 2003), and TCF8 has been implicated in repression of the epithelial cell adhesion genes E-cadherin, desmoglein, and plakoglobin (Grooteclaes and Frisch 2000). In addition, although we do not see evidence of expression of COL4A1 or COL4A5 in either normal or PPCD cornea, we do not yet know whether TCF8 mutations are affecting expression of the other four basement membrane collagens or COL8A2, the PPCD gene that we have shown is expressed in normal cornea. However, it may not be necessary to invoke additional regulatory changes, since simply altering the basement membrane composition apparently can modify epithelial protein expression (Kurpakus et al. 1992).
Yet to be tested is our hypothesis that aberrant regulation of collagens elsewhere in the body will also turn out to be contributing to the phenotype. Support for the idea that collagen regulation might be contributing to bone abnormalities in the mouse or human subjects comes from previous reports of TCF8 repression of expression of type I collagen in mouse osteoblasts through binding to sequences almost 20 kb away from the start codon (Terraz et al. 2001). In addition, hernias have previously been reported to be associated with aberrant regulation of type I and type III collagens (Friedman et al. 1993; Carr et al. 1994; Skoumal et al. 2004), an observation of great interest relative to the finding of hernias in males with PPCD.
In summary, we have shown that mutations in TCF8 apparently cause ∼50% of PPCD. We have identified a shared molecular component of disease etiology for PPCD and Alport syndrome through identification of COL4A3 as a possible target of TCF8 regulation. And, finally, we have identified nonocular features in individuals with PPCD that suggest a need for a more detailed evaluation of the full phenotype.
Acknowledgments
This work was supported by National Institutes of Health grants EY07003 (core grant), EY11671 (J.E.R.), T32 HG00040 (C.M.K., M.P.E.), EY11405 (D.V.), and EY09441 (V.M.E.); and the Lew R. Wasserman Award (J.E.R.), the Career Development Award (S.E.M.), and an unrestricted grant, all from Research to Prevent Blindness, Inc. The authors have no financial or proprietary conflicts relevant to the content of this paper.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BodyMap Database, http://bodymap.ims.u-tokyo.ac.jp/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for c.685A→G [accession number rs220060])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for VSX1 [accession number NM_199425], COL8A2 [accession number NM_005202], TCF8 [accession number NM_030751], Tcf8 [accession number NM_011546], Vsx1 [accession number NM_054068], Col8a2 [accession number NM_199473], Rpl19 [accession number NM_009078])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- The Sequence Manipulation Suite, http://bio.ifom-firc.it/TOOLS/sms/main.html/
- Shuffle Utility (v. 1.01), University of Michigan Medical Center, http://eyegene.ophthy.med.umich.edu/
- UCSC Genome Browser, http://www.genome.ucsc.edu/
- UniProtKB/Swiss-Prot, http://us.expasy.org/cgi-bin/niceprot.pl?P37275/ (for TCF8 [accession number P37275])
References
- Alport AC (1927) Hereditary familial congenital haemorrhagic nephritis. Br Med J 1:504–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo S, Gibbons RJ (1999) Germline and gonosomal mosaicism in the ATR-X syndrome. Eur J Hum Genet 7:933–936 [DOI] [PubMed] [Google Scholar]
- Barker DF, Hostikka SL, Zhou J, Chow LT, Oliphant AR, Gerken SC, Gregory MC, Skolnick MH, Atkin CL, Tryggvason K (1990) Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248:1224–1227 [DOI] [PubMed] [Google Scholar]
- Beck JA, Poulter M, Campbell TA, Uphill JB, Adamson G, Geddes JF, Revesz T, Davis MB, Wood NW, Collinge J, Tabrizi SJ (2004) Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Hum Mol Genet 13:1219–1224 [DOI] [PubMed] [Google Scholar]
- Bendavid R (2004) The unified theory of hernia formation. Hernia 8:171–176 [DOI] [PubMed] [Google Scholar]
- Biswas S, Munier FL, Yardley J, Hart-Holden N, Perveen R, Cousin P, Sutphin JE, Noble B, Batterbury M, Kielty C, Hackett A, Bonshek R, Ridgway A, McLeod D, Sheffield VC, Stone EM, Schorderet DF, Black GC (2001) Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet 10:2415–2423 [DOI] [PubMed] [Google Scholar]
- Boruchoff SA, Kuwabara T (1971) Electron microscopy of posterior polymorphous degeneration. Am J Ophthalmol 72:879–887 [DOI] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S (1991) Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 220:49–65 [DOI] [PubMed] [Google Scholar]
- Bullock S, Felix C, Iskander-Gabra S, Davison V (1996) Detection of germinal mosaicism in a DMD family. Biochem Soc Trans 24:273S [DOI] [PubMed] [Google Scholar]
- Carr AJ, Chiodo AA, Hilton JM, Chow CW, Hockey A, Cole WG (1994) The clinical features of Ehlers-Danlos syndrome type VIIB resulting from a base substitution at the splice acceptor site of intron 5 of the COL1A2 gene. J Med Genet 31:306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibis GW, Krachmer JA, Phelps CD, Weingeist TA (1977) The clinical spectrum of posterior polymorphous dystrophy. Arch Ophthalmol 95:1529–1537 [DOI] [PubMed] [Google Scholar]
- Colville DJ, Savige J (1997) Alport syndrome: a review of the ocular manifestations. Ophthalmic Genet 18:161–173 [DOI] [PubMed] [Google Scholar]
- Costantino ME, Stearman RP, Smith GE, Darling DS (2002) Cell-specific phosphorylation of Zfhep transcription factor. Biochem Biophys Res Commun 296:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler JA, Mitchell MJ, Smith MP, Savidge GF (2004) Germline mosaicism resulting in the transmission of severe hemophilia B from a grandfather with a mild deficiency. Am J Med Genet A 129:13–15 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- Ferreiro V, Szijan I, Giliberto F (2004) Detection of germline mosaicism in two Duchenne muscular dystrophy families using polymorphic dinucleotide (CA)n repeat loci within the dystrophin gene. Mol Diagn 8:115–121 [DOI] [PubMed] [Google Scholar]
- Friedman DW, Boyd CD, Norton P, Greco RS, Boyarsky AH, Mackenzie JW, Deak SB (1993) Increases in type III collagen gene expression and protein synthesis in patients with inguinal hernias. Ann Surg 218:754–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM (1994) E1a induces the expression of epithelial characteristics. J Cell Biol 127:1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- Grooteclaes ML, Frisch SM (2000) Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823–3828 [DOI] [PubMed] [Google Scholar]
- Hand CK, Harmon DL, Kennedy SM, FitzSimon JS, Collum LM, Parfrey NA (1999) Localization of the gene for autosomal recessive congenital hereditary endothelial dystrophy (CHED2) to chromosome 20 by homozygosity mapping. Genomics 61:1–4 [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24:3439–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heon E, Greenberg A, Kopp KK, Rootman D, Vincent AL, Billingsley G, Priston M, Dorval KM, Chow RL, McInnes RR, Heathcote G, Westall C, Sutphin JE, Semina E, Bremner R, Stone EM (2002) VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet 11:1029–1036 [DOI] [PubMed] [Google Scholar]
- Heon E, Mathers WD, Alward WL, Weisenthal RW, Sunden SL, Fishbaugh JA, Taylor CM, Krachmer JH, Sheffield VC, Stone EM (1995) Linkage of posterior polymorphous corneal dystrophy to 20q11. Hum Mol Genet 4:485–488 [DOI] [PubMed] [Google Scholar]
- Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H (1997) Impairment of T cell development in deltaEF1 mutant mice. J Exp Med 185:1467–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kawakami K (1995) DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem 233:73–82 [DOI] [PubMed] [Google Scholar]
- Kapur P, Caty MG, Glick PL (1998) Pediatric hernias and hydroceles. Pediatr Clin North Am 45:773–789 [DOI] [PubMed] [Google Scholar]
- Kurpakus MA, Stock EL, Jones JC (1992) The role of the basement membrane in differential expression of keratin proteins in epithelial cells. Dev Biol 150:243–255 [DOI] [PubMed] [Google Scholar]
- Lemmink HH, Mochizuki T, van den Heuvel LP, Schroder CH, Barrientos A, Monnens LA, van Oost BA, Brunner HG, Reeders ST, Smeets HJ (1994) Mutations in the type IV collagen alpha 3 (COL4A3) gene in autosomal recessive Alport syndrome. Hum Mol Genet 3:1269–1273 [DOI] [PubMed] [Google Scholar]
- Mandal MN, Ambasudhan R, Wong PW, Gage PJ, Sieving PA, Ayyagari R (2004) Characterization of mouse orthologue of ELOVL4: genomic organization and spatial and temporal expression. Genomics 83:626–635 [DOI] [PubMed] [Google Scholar]
- Martha A, Ferrell RE, Mintz-Hittner H, Lyons LA, Saunders GF (1994) Paired box mutations in familial and sporadic aniridia predicts truncated aniridia proteins. Am J Hum Genet 54:801–811 [PMC free article] [PubMed] [Google Scholar]
- McIntosh I, Hamosh A, Dietz HC (1993) Nonsense mutations and diminished mRNA levels. Nat Genet 4:219 [DOI] [PubMed] [Google Scholar]
- Miller CA, Krachmer JH (1997) Endothelial dystrophies. In: Kaufman HE, Barron BA, McDonald MB (eds) The cornea. Butterworth-Heinemann, Boston, pp 453–475 [Google Scholar]
- Mochizuki T, Lemmink HH, Mariyama M, Antignac C, Gubler MC, Pirson Y, Verellen-Dumoulin C, et al (1994) Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat Genet 8:77–81 [DOI] [PubMed] [Google Scholar]
- Moroi SE, Gokhale PA, Schteingart MT, Sugar A, Downs CA, Shimizu S, Krafchak C, Fuse N, Elner SG, Elner VM, Flint A, Epstein MP, Boehnke M, Richards JE (2003) Clinicopathologic correlation and genetic analysis in a case of posterior polymorphous corneal dystrophy. Am J Ophthalmol 135:461–470 [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Chaturvedi LS, Srivastava S, Mittal RD, Mittal B (2003) De novo mutations in sporadic deletional Duchenne muscular dystrophy (DMD) cases. Exp Mol Med 35:113–117 [DOI] [PubMed] [Google Scholar]
- Navarro P, Lozano E, Cano A (1993) Expression of E- or P-cadherin is not sufficient to modify the morphology and the tumorigenic behavior of murine spindle carcinoma cells: possible involvement of plakoglobin. J Cell Sci 105 (Pt 4):923–934 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, Zeng C, Baron A, Bemis L, Erickson P, Wilder E, Rustgi A, Kitajewski J, Gabrielson E, Bremnes R, Franklin W, Drabkin HA (2003) WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci USA 100:10429–10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce WG, Tripathi RC, Morgan G (1969) Congenital endothelial corneal dystrophy: clinical, pathological, and genetic study. Br J Ophthalmol 53:577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Zhou XP, Eng C (2001) A novel germline mutation of the PTEN gene in a patient with macrocephaly, ventricular dilatation, and features of VATER association. J Med Genet 38:820–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in Genie. J Comput Biol 4:311–323 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277 [DOI] [PubMed] [Google Scholar]
- Rodrigues MM, Sun TT, Krachmer J, Newsome D (1980) Epithelialization of the corneal endothelium in posterior polymorphous dystrophy. Invest Ophthalmol Vis Sci 19:832–835 [PubMed] [Google Scholar]
- Sawada H, Furthmayr H, Konomi H, Nagai Y (1987) Immunoelectronmicroscopic localization of extracellular matrix components produced by bovine corneal endothelial cells in vitro. Exp Cell Res 171:94–109 [DOI] [PubMed] [Google Scholar]
- Sawada H, Konomi H, Nagai Y (1984) The basement membrane of bovine corneal endothelial cells in culture with beta-aminopropionitrile: biosynthesis of hexagonal lattices composed of a 160 nm dumbbell-shaped structure. Eur J Cell Biol 35:226–234 [PubMed] [Google Scholar]
- Sgambati MT, Stolle C, Choyke PL, Walther MM, Zbar B, Linehan WM, Glenn GM (2000) Mosaicism in von Hippel-Lindau disease: lessons from kindreds with germline mutations identified in offspring with mosaic parents. Am J Hum Genet 66:84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Krafchak C, Fuse N, Epstein M, Schteingart M, Sugar A, Downs C, Rozsa F, Trager E, Reed D, Boehnke M, Moroi S, Richards J (2004) A locus for posterior polymorphous dystrophy (PPCD3) maps to chromosome 10. Am J Med Genet A 130:372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoumal M, Haberhauer G, Mayr H (2004) Concomitant diseases in primary joint hypermobility syndrome. Med Klin 99:585–590 [DOI] [PubMed] [Google Scholar]
- Spencer WH (1996) Ophthalmic pathology: an atlas and textbook. Vol 1. American Academy of Ophthalmology, Toronto [DOI] [PubMed] [Google Scholar]
- Takagi T, Moribe H, Kondoh H, Higashi Y (1998) DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 125:21–31 [DOI] [PubMed] [Google Scholar]
- Tam P (1990) Inguinal hernia. In: Lister J, Irving I (eds) Neaonatal surgery. Butterworth, London, pp 367–375 [Google Scholar]
- Terraz C, Toman D, Delauche M, Ronco P, Rossert J (2001) delta Ef1 binds to a far upstream sequence of the mouse pro-alpha 1(I) collagen gene and represses its expression in osteoblasts. J Biol Chem 276:37011–37019 [DOI] [PubMed] [Google Scholar]
- Toma NM, Ebenezer ND, Inglehearn CF, Plant C, Ficker LA, Bhattacharya SS (1995) Linkage of congenital hereditary endothelial dystrophy to chromosome 20. Hum Mol Genet 4:2395–2398 [DOI] [PubMed] [Google Scholar]
- Vollrath D, Jaramillo-Babb VL, Clough MV, McIntosh I, Scott KM, Lichter PR, Richards JE (1998) Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum Mol Genet 7:1091–1098 [DOI] [PubMed] [Google Scholar]
- Waring GO, Laibson PR, Rodrigues M (1974) Clinical and pathologic alterations of Descemet’s membrane: with emphasis on endothelial metaplasia. Surv Ophthalmol 18:325–363 [Google Scholar]
- Weisenthal RW, Streeten B (1997) Posterior membrane dystrophies. In: Krachmer J, Mannis M, Holland E (eds) Cornea-corneal and external disease: clinical diagnosis and management. Vol 2. Mosby, Chicago, pp 1063–1090 [Google Scholar]
- Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher FJ 3rd, Kant JA (1991) Identification of a zinc finger protein that inhibits IL-2 gene expression. Science 254:1791–1794 [DOI] [PubMed] [Google Scholar]