Abstract
Revertant mosaicism by somatic reversion of inherited mutations has been described for a number of genetic diseases. Several mechanisms can underlie this reversion process, such as gene conversion, crossing-over, true back mutation, and second-site mutation. Here, we report the occurrence of multiple corrections in two unrelated probands with revertant mosaicism of non-Herlitz junctional epidermolysis bullosa, an autosomal recessive genodermatosis due to mutations in the COL17A1 gene. Immunofluorescence microscopy and laser dissection microscopy, followed by DNA and RNA analysis, were performed on skin biopsy specimens. In patient 1, a true back mutation, 3781T→C, was identified in the specimen from the arm, and a second-site mutation, 4463-1G→A, which compensated for the frameshift caused by the inherited 4424-5insC mutation, was identified in the 3′ splice site of exon 55 in a specimen from the middle finger. Patient 2 showed—besides two distinct gene conversion events in specimens from the arm and hand sites, both of which corrected the 1706delA mutation—a second-site mutation (3782G→C) in an ankle specimen, which prevented the premature ending of the protein by the 3781C→T nonsense mutation (R1226X). Thus, both inherited mutations, paternal as well as maternal, reverted at least once by different reversion events in distinct cell clusters in the described patients. The occurrence of multiple correcting mutations within the same patient indicates that in vivo reversion is less unusual than was generally thought. Furthermore, in the male patient, mosaic patterns of type XVII collagen–positive keratinocytes were present in clinically unaffected and affected skin. This latter observation makes it likely that reversion may be overlooked and may happen more often than expected.
Introduction
Genetic mosaicism is defined as the presence of two or more genetically distinct populations of cells that differ from each other in their DNA but are derived from a single zygote within one individual (Hall 1988). If an inherited disease-causing mutation is corrected in a somatic cell by means of a second genetic event, gene function can be completely or partially restored in these so-called revertant cells. Correction of a stem cell will lead, in this case, to life-long repair. This phenomenon of “natural gene therapy” has led to revertant mosaicism in several human inherited diseases, including epidermolysis bullosa (EB), tyrosinemia type I (MIM 276700), Bloom syndrome (MIM 210900), Fanconi anemia (FA [MIM 227650]), and primary immunodeficiency syndromes, such as Wiskott-Aldrich syndrome (WAS [MIM 301000]), X-linked severe combined immunodeficiency (X-linked SCID [MIM 300400]), and adenosine deaminase deficiency (MIM 102700) (for reviews, see Hirschhorn [2003] and Jonkman et al. [2003]). Restoration of gene function is obtained by different genetic mechanisms, including back mutation, intragenic crossover, mitotic gene conversion, and second-site mutation (Kvittingen et al. 1994; Ellis et al. 1995; Jonkman et al. 1997). Among the reported second-site mutations are a reading-frame–restoring insert of 2 bp and a frameshift mutation that nullifies a dominant-negative allele (Darling et al. 1999; Smith et al. 2004). Despite this apparent range of reversion mechanisms available to the human body, only a single type has been found in each individual patient, apart from one very recently described individual with tyrosinemia type I (Bliksrud et al. 2005). This suggests either that such in vivo reversion is a one-time single event or that a particular mechanism is preferred for correction of a specific type of mutation.
Generalized atrophic benign epidermolysis bullosa (GABEB [MIM 226650])—which is the generalized variant of non-Herlitz junctional epidermolysis bullosa (nH-JEB), an autosomal recessive bullous genodermatosis—is characterized by generalized skin blistering from birth onward, dental anomalies, universal alopecia, and nail dystrophy (Hintner and Wolff 1982; Jonkman et al. 1996). The underlying defect is mutation of the COL17A1 gene, which encodes the 180-kDa type XVII collagen (Jonkman et al. 1995; McGrath et al. 1995) (human COL17A1 mRNA sequence: GenBank accession number NM_000494; human COL17A1 transcript sequence: GenBank accession number NP_000485). The COL17A1 gene spans 52 kb of the genome on chromosome 10 and encompasses 56 exons that range in size from 27 to 390 bp (Li et al. 1991; Gatalica et al. 1997). Type XVII collagen is a glycosylated type II transmembrane protein of 1,497 aa. Its COOH-terminal extracellular part, comprising approximately two-thirds of the protein, contains 15 interrupted collagenous domains, designated “COL1”–“COL15” (Li et al. 1993). The protein is expressed in basal epithelial cells and is part of the hemidesmosome, a critical structure for attachment of basal epithelial cells to the underlying matrix. Although type XVII collagen almost certainly has signal-transduction properties, its main function is to act as a transmembrane link in the molecular chains that anchor the cytoskeleton of the basal keratinocytes to the basement membrane.
Previously, we demonstrated that the in vivo reversion in keratinocytes cultured from a revertant skin patch of the hand of a patient with GABEB was the result of mitotic gene conversion, the nonreciprocal transfer from one chromosome to another (Jonkman et al. 1997). Here, we expand these studies by analyzing separate revertant patches within individual patients. Using laser dissection microscopy (LDM), we now find that different correcting COL17A1 mutations are responsible for the restoration of type XVII collagen expression in distinct cell clusters within the same patient.
Material and Methods
Biopsy Sites
For immunofluorescence (IF) microscopy, six 4-mm punch-biopsy specimens were obtained and were snap frozen: two from clinically unaffected skin of the right middle finger of a 75-year-old man (patient 1), three from nonlesional affected skin of his right upper arm, and one from nonlesional affected skin of his shoulder. Biopsy specimens from unaffected skin of the left forearm and the right upper arm of a 45-year-old woman (patient 2) were also analyzed. In addition to specimens from these biopsy locations described by Jonkman et al. (1997), another biopsy specimen was obtained from unaffected skin of the left ankle.
For electron microscopy, three 2-mm punch-biopsy specimens were obtained from patient 1: two from lesional affected skin of the right upper arm and one from unaffected skin of the right middle finger.
Immunomorphological Analysis of Skin and Cultured Keratinocytes
Details of IF microscopy, electron microscopy, and keratinocyte culturing have been described extensively elsewhere (Jonkman et al. 1995). For detection of type XVII collagen, the following monoclonal antibodies were used: 1A8C, specific to the endodomain (epitope located between amino acids 155 and 160), and 1D1 and 233, both specific to the ectodomain (1D1 epitope located between amino acids 1357 and 1387; 233 epitope located between amino acids 1118 and 1143) (all were gifts from Dr. K. Owaribe [Nagoya, Japan]) (Di Zenzo et al. 2004). The Alexa488-conjugated goat anti-mouse IgG antibody (Brunschwig Molecular Probes) was used for a secondary step.
Mutation Detection Strategy
The detection of mutations in the genomic DNA of patient 2 has been published previously (Jonkman et al. 1997). For identification of the mutations in the DNA of patient 1, all 56 exons of COL17A1 were amplified using the primers described by Gatalica et al. (1997) and subsequently sequenced (GenBank accession numbers U76564–U76604), except the primer for exon 46. For exon 46, we used instead the sense primer 5′-GTGCTTCAGGTCACCTCCGT-3′ and the antisense primer 5′-ACGAGGAGATGAGGCTCTGG-3′. Unfortunately, genomic DNA from the parents or from other first-degree relatives of patient 1 was no longer available.
Verification of Mutations
Verification of the 1706delA mutation in exon 18 and the 3781C→T mutation in exon 51 by restriction analysis was done as described elsewhere (Jonkman et al. 1997). The 4424-5insC mutation in exon 54 does not create or abolish a restriction site and was therefore verified by repeated sequencing.
LDM
For DNA recovery by LDM, skin cryosections of 4 μm were mounted on 1.35-μm-thin polyethylene-naphthalene membranes attached to normal 1-mm slides and were stained with monoclonal antibody 1D1 by use of the same staining procedure described elsewhere (Jonkman et al. 1997), except for the absence of the bisbenzimid step in the protocol. For selection of the sites of epidermal basement membrane that stained either positive or negative for type XVII collagen, IF images were acquired using a digital camera and were processed with P.A.L.M. RoboSoftware 1.2 (P.A.L.M. Microlaser Technology AG). Cells were dissected using the Laser Robot Microbeam System (P.A.L.M. Microlaser Technology AG). Dissected areas were directly collected in the cap of a 0.2-ml thin-wall reaction tube with a flat cap containing 30 μl of one-time PCR buffer (Amersham Pharmacia Biotech) and 1 μl of proteinase K (Invitrogen). For RNA recovery, staining with monoclonal antibody 1D1 was omitted because we noted that, after staining, almost all the RNA was lost. Therefore, serial cryosections were prepared, and every fourth section was mapped by IF microscopy. On the basis of the staining pattern of these sections, the epidermis of five to seven skin sections was collected in the cap of a 0.5-ml P.A.L.M. tube (P.A.L.M. Microlaser Technology) that contained 30 μl of lysis buffer (Stratagene Europe) and was used for RNA isolation.
DNA Isolation of LDM Samples
Microdissected epidermis from ∼200 cells was collected in a 0.2-ml reaction tube. During digestion by proteinase K, the tubes remained inverted for 60 min at 55°C; subsequent heating to 98°C for 15 min inactivated the proteinase K. The final aliquots were used for PCR.
Cytospin
To analyze DNA of cultured revertant keratinocytes from the hand of patient 2, cytospin was conducted; 1.35-μm-thin polyethylene-naphthalene membranes attached to normal 1-mm slides (P.A.L.M. Microlaser Technology AG) were coated with 10 μl of polylysine and were dried for 30 min at 37°C. Cytospin was performed with a number of cells sufficient to allow LDM isolation of reverted cells. The glass slides were then dried. Subsequently, the cells were fixed for 10 min at room temperature by use of 1% formaldehyde in phosphate-buffered saline, were washed, and were incubated for 5 min in 0.5% Triton X-100 in phosphate-buffered saline. The cells were stained with monoclonal 1A8C, as described elsewhere (Jonkman et al. 1995), and were then used for LDM.
RNA Isolation
Tubes were centrifuged for 1 min at 10,000 g, and 70 μl of lysis buffer containing 0.7 μl of β-mercaptoethanol was added. Total RNA was then prepared using the Stratagene RNA nanoprep kit (Stratagene), for a final volume of 10 μl.
cDNA Synthesis
For cDNA synthesis, 1 μl of 300-ng/μl random primers (Invitrogen) and 1 μl of 10 mM dNTP mix (Fermentas) were added to 10 μl of total RNA and were incubated for 5 min at 65°C. The tubes were then immediately put on ice, and 5 μl of 5 × First Strand buffer, 2 μl of 0.1 M dithiothreitol, and 1 μl of 10-units/μl RNase Inhibitor Cloned (all Invitrogen) were added. Incubation for 10 min at 25°C was followed by 2 min at 42°C. Finally, 1 μl of 1-unit/μl Superscript II RNase H− Reverse Transcriptase (Invitrogen) was added. Tubes were incubated for 50 min at 42°C, followed by 15 min at 70°C to inactivate the enzyme. For PCR, 10 μl of cDNA was used in a total volume of 50 μl.
Identification of Mutations in LDM Samples
For detection of mutations in LDM-isolated DNA, we used nested PCR. For the second PCR, 1 μl of the first PCR product was used. PCR cycling conditions were 5 min at 94°C, followed by 30 cycles at 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min and 30 s and a final extension at 72°C for 7 min. Water, instead of DNA, was used as a negative control. Primer sequences and sizes of PCR products are listed in table 1. After PCR, aliquots of 14 μl were examined using 4% agarose gel electrophoresis.
Table 1.
PCR Amplification of COL17A1 Genomic DNA
|
Primer(5′→3′) |
|||
| Position and PCRa | Forward | Reverse | Product Size(bp) |
| Exon 18, 1706delA: | |||
| 1 | taggagcgagtgtatggaag | ggatgatggtactcacagca | 385 |
| 2 | agaggatcaggaggagcata | cttcctcacgaacatccaga | 155 |
| Intron 19, 1822+14T/C: | |||
| 1 | agatgtgaagctgactcagg | agtcattcagctcttcatcc | 187 |
| 2 | agatgtgaagctgactcagg | aactcacttccaactgcagg | 125 |
| Intron 28, 2268+120T/C: | |||
| 1 | acaggagttcacggatacag | ctccttatcctccactccag | 327 |
| 2 | cggatacagtgagaactacc | ctccttatcctccactccag | 316 |
| Exon 37, 2700T/C: | |||
| 1 | agaggtgagaagtgtggcag | gcagaagagagcaaggaaga | 263 |
| 2 | tgagaagtgtggcagagagc | gtgcattgaatctcctgtcc | 180 |
| Exon 43, 2988A/C: | |||
| 1 | tacagggctgttagtgggtg | acttgctgcatctcttcagg | 345 |
| 2 | gctgactgcactgtgtgtgt | tcaccaccttctcactgctc | 138 |
| Exon 51, 3781C→T: | |||
| 1 | tttctctcctcccatcaccc | tgtccctttaagtgcctccc | 374 |
| 2 | ccagcctcatcccctccttt | ctccggaagctgtcgctgtt | 236 |
| Exon 52, 4214A/G: | |||
| 1 | caagtctttctctccaccga | ccacaaacaagaaagccagt | 527 |
| 2 | ctcctacagctcttccatga | gctgcatgctctctgacacc | 242 |
| Exon 54, 4424-5insC: | |||
| 1 | ttgatgtcccaagcttccag | ctaatgagcgtcagccttgc | 419 |
| 2 | tcaagactctaagggccagt | tggagcagtcaacacttacc | 352 |
The numbers 1 and 2 refer to the first PCR and the second/nested PCR, respectively.
Also, the cDNA samples from dissected cells were subjected to nested PCR. All primers were designed in such a way that the PCR product contained sequences from multiple exons. Primer sequences for amplification of COL17A1 cDNA and sizes of PCR products are listed in table 2. All PCRs were repeated with templates from at least three separate nucleic-acid isolations obtained by LDM, and all products were sequenced.
Table 2.
PCR Amplification of COL17A1 cDNA
|
Primer(5′→3′) |
|||
| Position and PCRa | Forward | Reverse | Product Size(bp) |
| Exon 18, 1706delA: | |||
| 1 | ctcttcggcctcattgctct | tcttcctcacgaacatccag | 220 |
| 2 | agaggatcaggaggagcata | cttcctcacgaacatccaga | 155 |
| Exon 51, 3781C→T: | |||
| 1 | ttcagaggcatcgttggacc | agctgcgcacatcaggactt | 268 |
| 2 | tgtggtccagcatcagcgtg | ctccggaagctgtcgctgtt | 169 |
| Exon 54, 4424-5insCb: | |||
| 1 | gcatcagcaaggtcttctct | cacggcttgacagcaatact | 262 |
| 2 | cggacctcatggacttcttc | cggcttgacagcaatacttc | 221 |
| 1b | tggcaatggcggactattgg | aggttggctgtgctgtctca | 547 |
| 2b | tacaatgagctggctgtgag | tctggagaccttggacctaa | 450 |
The numbers 1 and 2 refer to the first PCR and the second/nested PCR, respectively.
To investigate the possible presence of alternative spliced mRNA transcripts due to the splice-site mutation in intron 54 (4463-1G→A), additional PCRs were performed for exon 54, 4424-5insC, by use of developed primers, with the sense primer located in exon 52 and the antisense primer after exon 56.
Identification of SNPs in LDM Samples
For the identification of SNPs, nested PCRs and, in a few cases, seminested PCRs were performed. Primer sequences and sizes of PCR products for 1822+14T/C in intron 19 (dbSNP identifier rs17821926), 2268+120T/C in intron 28 (Gatalica et al. 1997), 2700T/C in exon 37 (rs4918079), 2988A/C in exon 43 (rs2296219), and 4214A/G in exon 52 (rs17116350) are listed in table 1. Like the identification of the mutations in LDM samples, all PCRs for SNPs were repeated with templates from at least three separate nucleic-acid isolations obtained by LDM, and products were sequenced afterward.
Microsatellite Analysis
To detect possible crossovers in the DNA of patient 2, the alleles flanking the COL17A1 gene on chromosome 10q24.3 were analyzed using microsatellite polymorphisms (Gyapay et al. 1994). The flanking markers D10S603 and D10S597 (8 cM apart) were used. D10S566 and D10S554 were not analyzed, since these were homozygous in the patient’s DNA and were thus not informative (Jonkman et al. 1997). For analysis of the microsatellites D10S603 and D10S597, we also used nested PCRs, because of the small amount of LDM-isolated DNA. Primers for the first PCR for D10S603 were sense primer 5′-GCTGGATTATCTCGGTAACC-3′ and antisense primer 5′-GTTCATCTCCCAAGGCAATG-3′. Primers for the first PCR for D10S597 were sense primer 5′-AGAAGAGGAAGGCTGTCAGA-3′ and antisense primer 5′-GCCACAAGACTTGTGTTTGC-3′. Primers for the second PCRs were as described elsewhere (Gyapay et al. 1994).
Cloning
The nested PCR product of exon 51 from the ankle specimen of patient 2 was cloned into the pCR4-TOPO (Invitrogen) vector to investigate whether the mutations 3781C→T and 3782G→C were located on the same chromosome. Five clones were selected and sequenced.
Results
Patient 1
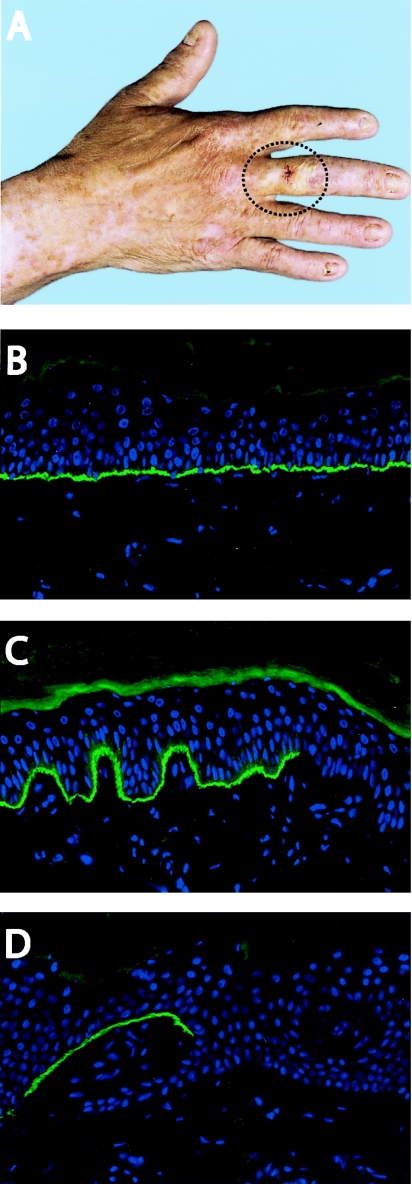
The first proband was a 75-year-old Dutch male who had the GABEB variant of nH-JEB. Since birth, he had generalized blistering after minor trauma, without evidence of scarring or milia formation. He also had universal alopecia, small rudimentary nails, and enamel hypoplasia. His nonconsanguineous parents and his sister were healthy. On clinical examination, he presented with a circular patch of clinically unaffected skin, ∼2 cm2 on his right middle finger, which tolerated the wearing of his wedding ring (fig. 1A). This skin looked perfectly healthy and sturdy, and no blisters had ever occurred there. The size of this unaffected area had remained stable throughout his life. This clinical presentation was suggestive of revertant mosaicism, since the nH-JEB symptoms were generalized, with the rare exception of the patch on the middle finger. Two biopsy specimens of the unaffected skin of the right middle finger, three of the nonlesional affected skin of the right upper arm, and one of the lesional affected skin of the shoulder were studied using IF microscopy. Staining with monoclonal antibodies against type XVII collagen showed a mosaic pattern in both biopsy specimens of the middle finger. The epidermal basement membrane stained positive for type XVII collagen, showing the same intensity as normal human control skin, with one interruption of ∼150 μm that was not stained, which resembled 10% of the basal cells of this biopsy specimen (fig. 1B and 1C). In contrast, the specimen from the nonlesional affected skin of the shoulder and two specimens from the upper arm were completely negative for type XVII collagen. However, the third upper-arm biopsy specimen of affected skin demonstrated an interruption in the dermal-epidermal junction: ∼500 μm (25% of the basal cells) of type XVII collagen–expressing cells (fig. 1D).
Figure 1.
Clinical and cellular mosaicism in the skin of patient 1. A, Patch of skin on the right hand where blisters had never occurred (circled). A biopsy specimen was obtained from the middle of this unaffected area. B, IF staining of type XVII collagen with monoclonal antibody 1D1 in human healthy control skin. C, Mosaic pattern along the dermal-epidermal junction of type XVII collagen stained with monoclonal antibody 1D1, observed in the right middle finger. D, Basal cells that stained positive for type XVII collagen, observed in the right upper arm despite the affected skin of the upper arm.
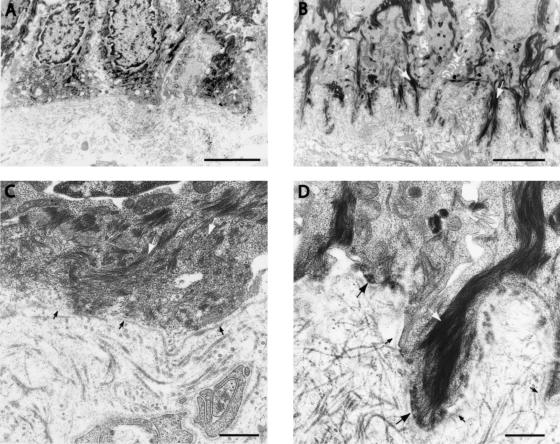
Electron microscopy of gently rubbed affected skin showed a cleavage plane through the lamina lucida, thereby confirming the JEB diagnosis (data not shown). In nonlesional affected skin, hemidesmosomes were low in number (<5 per 40 μm of basement membrane vs. 49–98 per 40 μm in normal control skin) and appeared to be hypoplastic (fig. 2A and 2C). The unaffected skin of the middle finger showed a larger number of hemidesmosomes (41 per 40 μm) than the affected skin (fig. 2B and 2D), and these hemidesmosomes were normal in shape.
Figure 2.
Electron microscopy of affected and revertant skin of patient 1. A and C, Affected skin of the arm, showing basal keratinocytes with intermediate filaments that do not project to the flattened basilar cell periphery, which almost completely lacks hemidesmosomes. B and D, Revertant skin of the middle finger, revealing pronounced rootlets of basal keratinocytes containing intermediate filaments that connect to normal hemidesmosomes in the basilar cell periphery. In keratin, white arrows indicate intermediate filaments, large black arrows indicate hemidesmosomes, and small black arrows indicate the lamina densa. Calibration bars represent 2 μm in panels A and B and 500 nm in panels C and D.
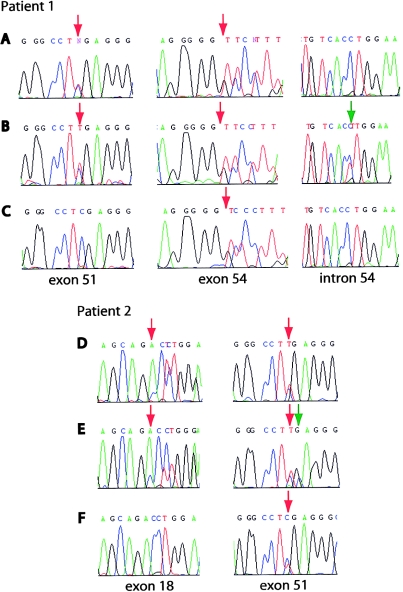
To determine mutations in COL17A1, DNA was isolated from peripheral blood lymphocytes and was sequenced with intron primers (Gatalica et al. 1997). Two heterozygous mutations were identified: a nonsense mutation in exon 51, 3781C→T (R1226X), and an insertion of a C in exon 54 between nucleotides 4424 and 4425. This insC mutation caused a frameshift, resulting in a novel sequence beginning at amino acid 1441 and leading to a premature termination codon (PTC) at position 1453.
To study the cells re-expressing type XVII collagen in the mosaic biopsy specimens, we needed a technique that could isolate separately the different cell populations: type XVII collagen–negative keratinocytes and type XVII collagen–positive keratinocytes. LDM allows for recovery of DNA and RNA directly from biopsy material and eliminates the necessity of culturing the keratinocytes with the risk of selection in vitro. Furthermore, in vivo spliced mRNA is analyzed, which excludes possible deviant splicing due to culturing conditions, as was demonstrated for the hemidesmosomal protein integrin α6β4 (Chavanas et al. 1999).
Interestingly, the type XVII collagen–positive cells of the middle finger, isolated by LDM, carried an additional mutation, 4463-1G→A, in the intron 54–exon 55 border, on the same allele as the 4424-5insC mutation (fig. 3B). This second-site mutation changes the consensus sequence of the 3′ splice site, which ends with the invariant AG. In this case, the invariant G is replaced with an A, and, since exon 55 starts with a G, a possible new splice site is created 1 nt further downstream. Subsequently, RNA was isolated from epidermis that stained positive, and this RNA indeed showed a deletion of G at position 4463 (fig. 4A and 4B). This deletion restores the frameshift caused by the 4424-5insC mutation, beginning at amino acid 1453, and results in a protein of correct size but with a stretch of 13 different amino acids (GQKGEMGTPGPKG→WAKRRDGHSRTQS) in the first collagenous domain (COL1). Since the patient's middle finger displays healthy skin, this protein is functional enough to restore normal skin strength, which also implies that, despite the disruption of the Gly-X-Y collagen motif, the changed amino acids are not essential for interactions with other hemidesmosomal proteins.
Figure 3.
Identification of the different correcting COL17A1 mutations. A, Nonsense mutation 3781C→T in exon 51 and insertion 4424-5insC in exon 54, present in keratinocytes of patient 1 that stained negative for type XVII collagen. Note that exon 54 was sequenced with a reverse primer to circumvent the problem of the double sequence due to the insertion. B, Additional mutation in the 3′ splice site of exon 55, 4463-1G→A, in keratinocytes of the middle finger that stained positive. C, Absence of the exon 51 nonsense mutation (3781C→T) in keratinocytes of the upper arm that stained positive. D, Inherited mutations 1706delA in exon 18 and 3781C→T in exon 51, present in keratinocytes of patient 2 that stained negative. E, Additional mutation 3782G→C in keratinocytes of the ankle that stained positive. F, Absence of the exon 18 deletion (1706delA) in keratinocytes of the forearm and upper arm that stained positive. Red arrows indicate the position of the mutations, and green arrows indicate the second-site mutations.
Figure 4.
Effects of the second-site mutations on mRNA. A, Additional mutation 4463-1G→A, which results in an mRNA transcript with a deletion of 1 nt, 4463delG. B, Nucleotides 4459–4467 of mRNA of normal human skin, which served as a control. C, Compensatory mutation 3782G→C, which changes the PTC caused by the 3781C→T mutation into the amino acid serine. D, Nucleotides 3778–3786 of mRNA of normal human skin, which served as a control. Red arrows indicate the position of the mutations, and green arrows indicate the second-site mutations.
Unexpectedly, the analysis of the revertant cells of the upper arm showed something completely different. The frameshift caused by the 4424-5insC mutation was not corrected, because the restoring mutation 4463-1G→A was lacking. However, the 3781C→T mutation on the other allele was missing, thus revealing another reversion mechanism in these keratinocytes (fig. 3C). The absence of this nonsense mutation in exon 51, in combination with the presence of immunoreactive type XVII collagen, could be caused by various genetic mechanisms: gene conversion, single crossing-over, or true back mutation. These mechanisms can result in somatic cells becoming homozygous at loci during mitotic recombination. To differentiate between these possibilities, SNPs around the exon 51 mutation in the COL17A1 gene were investigated for loss of heterozygosity in revertant keratinocytes of the upper arm (table 3) (Gatalica et al. 1997; Jonkman et al. 1997). A true back mutation would only revert the nonsense mutation, leaving the SNPs in intron 28 (2268+120T/C) and exon 52 (4214A/G) unaffected. In contrast, single crossing-over and gene conversion would result in loss of heterozygosity of one or both SNPs surrounding exon 51. Since the SNPs in intron 28 and exon 52 were found to be still heterozygous, a single crossing-over can be excluded as the genetic mechanism. The heterozygosity of the SNPs in intron 28 and exon 52 indicates instead that true back mutation (3781T→C) is the underlying mechanism. Nevertheless, we cannot completely rule out gene conversion, because this can occur even when limited to tens of nucleotides (Smithies and Powers 1986).
Table 3.
Inherited and Somatic Mutations in the COL17A1 Gene[Note]
|
SNPs in |
|||||||||||||
| Patient and Exon or Intron of COL17A1 | Nucleotide Position | Blood | Upper Arm | Middle Finger | Forearm | Ankle | Handa | ||||||
| Patient 1: | |||||||||||||
| Intron 19 | 1822+14 | T | C | T | C | T | C | … | … | … | … | … | … |
| Intron 28 | 2268+120 | T | C | T | C | T | C | … | … | … | … | … | … |
| Exon 51 |
3781 |
C | T |
C | C | C | T |
… | … | … | … | … | … |
| Exon 52 | 4214 | G | A | G | A | G | A | … | … | … | … | … | … |
| Exon 54 |
4424–4425 |
insC |
… | insC |
… | insC |
… | … | … | … | … | … | … |
| Intron 54 | 4463-1 | G | G | G | G | A | G | … | … | … | … | … | … |
| Patient 2: | |||||||||||||
| Exon 18 |
1706 |
A | del |
A | A | … | … | A | A | A | del |
A | A |
| Intron 19 | 1822+14 | C | T | C | C | … | … | C | C | C | T | C | T |
| Intron 28 | 2268+120 | C | T | C | T | … | … | C | T | C | T | C | T |
| Exon 37 | 2700 | T | C | T | C | … | … | T | C | T | C | T | C |
| Exon 43 | 2988 | A | C | A | C | … | … | A | C | A | C | A | C |
| Exon 51 |
3781 |
T |
C | T |
C | … | … | T |
C | T |
C | T |
C |
| Exon 51 | 3782 | G | G | G | G | … | … | G | G | C | G | G | G |
Note.— SNPs within the COL17A1 gene are indicated by nucleotides. The pathogenic mutations are underlined, and the compensatory second-site mutations are in bold italics.
Data for the hand were obtained from Jonkman et al. (1997).
Patient 2
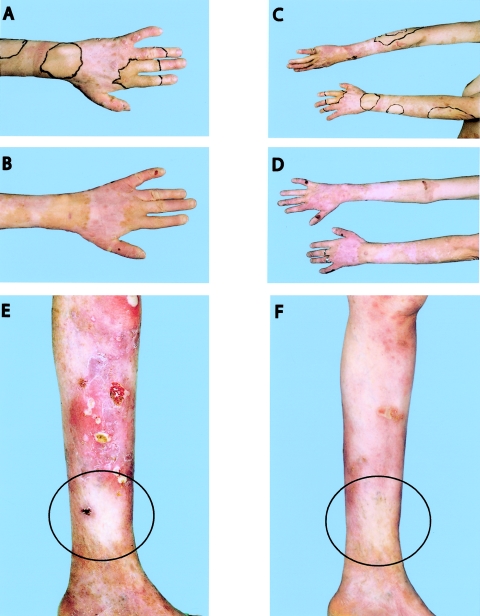
The second proband, a 45-year-old female with revertant mosaicism, who is compound heterozygous for a paternal mutation in exon 51, 3781C→T (the same mutation as in patient 1), and who has a maternal deletion in exon 18, 1706delA, has been described elsewhere (Jonkman et al. 1997). She had unaffected nonconsanguineous parents and the same constellation of symptoms as patient 1, which classified her condition too as the GABEB variant of JEB. About 10% of her total body surface was covered with revertant skin. Extensive photo documentation from 1994 and 2005 showed the stability of the patches and excluded the possibility of expansion of the clinically unaffected skin (fig. 5A–5F). We demonstrated elsewhere that, in cultured revertant keratinocytes derived from the dorsum of the patient's left hand, a gene conversion had occurred, resulting in the loss of the maternal deletion (Jonkman et al. 1997). For the current study, with the intriguing results of patient 1 in mind and with the possibility of directly analyzing stored skin-biopsy specimens by use of the LDM technique, we reinvestigated this patient. Besides the patches of unaffected skin on her arms and hands, the patient indicated another unaffected patch on the ankle, which is also visible in photographs taken in 1969. IF microscopy showed that, in the ankle specimen, in contrast to the interrupted pattern in the biopsy specimens of the forearm and the right upper arm (50% of cells positive for type XVII collagen), the whole epidermal basement membrane stained positive for type XVII collagen, with a similar intensity as that of normal human control skin. Unexpectedly, in these LDM-isolated, type XVII collagen–positive keratinocytes from the ankle, both of the inherited mutations, 1706delA and 3781C→T, were still present, but a second mutation was detected on the 3781C→T allele. At position 3782, a transversion from G to C (3782G→C) had occurred, 1 nt behind the 3781C→T mutation (fig. 3E). This compensatory mutation results in the replacement of the nonsense mutation at position 1226 to a missense mutation (R1226X→R1226S). Figure 4C shows that the RNA isolated from the same cells originates mainly from the corrected allele. The 1706delA allele hardly contributes, most likely because of degradation by nonsense-mediated RNA decay, a conserved posttranscriptional mechanism that prevents the synthesis of truncated proteins that could be deleterious to the cell. Considering the absence of blister formation after hard rubbing of the skin on the ankle and the IF staining with the intensity of normal control skin, we conclude that the produced type XVII collagen with one altered residue is functional.
Figure 5.
Depiction of the area of unaffected patches remaining stable over the years. A, Left hand of patient 2 in 1994. The unaffected skin was outlined with a black marker. B, Left hand in 2005, with the same pigmentation as in 1994, demonstrating the stability of the healthy skin area. C and D, Forearms of patient 2 in 1994 (C) and 2005 (D), showing the stability of the healthy zones. E and F, Leg of patient 2 in 1994 (E) and 2005 (F). Unaffected skin is located in the middle of the black circle. In panel E, the difference in the erosions situated above this unaffected area is clearly visible.
A third reversion event was found in the biopsy specimens of the forearm and upper arm. Like the revertant skin of the hand, the LDM-isolated, type XVII collagen–positive keratinocytes of both specimens showed loss of the maternal 1706delA mutation (fig. 3F). Also, data from the SNP and microsatellite analysis with D10S597, on the telomeric side of the COL17A1 gene, and D10S603, on the centromeric side of COL17A1, were similar to the data for the hand. However, the SNP in intron 19 (1822+14C/T) showed a loss of heterozygosity (table 3) (Gatalica et al. 1997). To confirm our original data, we reinvestigated the keratinocytes originally isolated from the hand. New cultures were cytospinned and immunostained, and type XVII collagen–positive cells were selected by LDM. Results of PCR and subsequent sequencing were identical to our original data and again showed heterozygosity of the intron 19 SNP. Thus, in the left forearm and right upper arm of this female patient, the correction results from another larger gene conversion of at least 511 bp, including 14 nt of intron 19. The RNA isolated from the type XVII collagen–positive cells of the upper arm could be attributed solely to the maternal allele (not shown). The 3781C→T mutation of the paternal allele was not present, most likely because of nonsense-mediated RNA decay.
Discussion
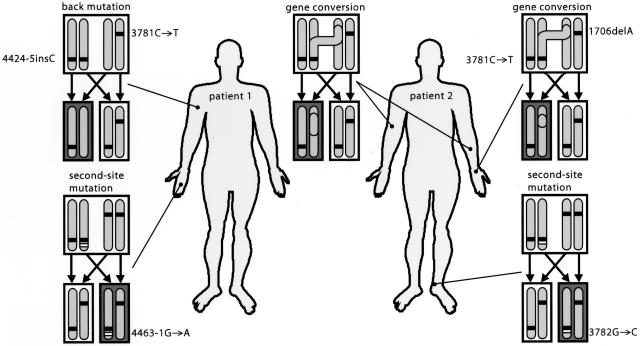
This study reveals that, in two compound heterozygous, unrelated probands, multiple distinct somatic reversions of pathogenic COL17A1 mutations have occurred that clinically result in discrete patches of functionally restored skin. In the patients described, both of the inherited mutations, paternal as well as maternal, reverted at least once by different reversion events (fig. 6).
Figure 6.
Multiple reversion mutations in patients 1 and 2. In mitosis in a diploid cell after DNA replication and normal segregation, each daughter cell obtains one chromosome from the father and one from the mother. Homologous chromosomes carry the paternal or maternal mutation in the COL17A1 gene (black bars). Cells devoid of the protein are depicted in white, and cells with a revertant phenotype are depicted in gray. The dots in the picture refer to the biopsy sites, and the accompanying drawings show the underlying mechanism of the reverse mutation of that skin patch. Although the type XVII collagen produced in the cells with a second-site mutation (white bars) was different from the wild-type protein, the skin displayed a revertant phenotype.
Despite the low number of reversions described in humans, three cases—one case of FA and two cases of WAS—were reported in which patients with revertant mosaicism who were from the same family independently developed the same compensatory second-site mutation (Waisfisz et al. 1999; Wada et al. 2003, 2004). Because of the occurrence of the same correcting mutation in the siblings, it was suggested that these second-site mutations resulted from a specific molecular mechanism, rather than from a randomized process. Waisfisz et al. (1999) showed, in two brothers, the functional correction of the inherited 1749T→G mutation in the FANCC gene by the 1748C→T nucleotide change, creating a cysteine codon (L496C) instead of an arginine codon (L496R). This transversion was assumed to be mediated by methylation. In addition, Wada et al. (2003, 2004) implied that DNA polymerase slippage was the cause of the compensatory deletions in two of their families. In one family, the function of the WAS protein was restored in two brothers by the same second-site mutation, a deletion of 19 bp that included the original 1305insC mutation in the WASP gene (Wada et al. 2003). In the other family, DNA polymerase slippage was also hypothesized to underlie the inherited mutation, a 6-bp sequence insertion, as well as the correction, the deletion of the same 6-bp sequence (Wada et al. 2004). Our data are fascinatingly different. The mutation 3781C→T, present in both patients, reverted in two different ways. In patient 1, the reversion was most likely a true back mutation, whereas, in patient 2, the second-site mutation 3782G→C resulted in a serine codon (R1226S) instead of a stop codon (R1226X). In contrast to the described siblings with FA and WAS, our patients are unrelated. Thus, if correction mutations result from a specific molecular mechanism, then other factors also influence the preference for a certain reversion mechanism.
In patient 2, gene conversion was demonstrated as the correcting mechanism for the 1706delA mutation in three of the four biopsy specimens studied. The specimens of the left forearm and right upper arm, although located far apart and in separate clusters, had the same DNA pattern. Therefore, they could have originated either from a single early correction event that took place before lateralization of the embryo or from two independent but identical correction events. Since no other heterozygous SNPs were noticed before exon 18 or between intron 19 and intron 28, the answer remains elusive. It might be that, in this patient, gene conversion around the exon 18 mutation is favored as the correcting mechanism because 1706delA is located close to the COL17A1-CA1 microsatellite in intron 18, consisting of GT repeats. For chromosome 22, an association between the recombination frequency and the GT microsatellite distribution has been demonstrated (Majewski and Ott 2000).
The fifth type of correction found was the 4463-1G→A splice-site mutation that corrected the 4424-5insC mutation. This 3′ splice-site mutation in the revertant keratinocytes of the middle finger is a new example of a second-site mutation in humans. Although splice-site mutations are well known, they have not yet been described as a mechanism for restoring a frameshift. The 4463-1G→A at the 3′ end of intron 54 results in a new hypothetical splice site 1 base downstream that, by RNA analysis, was demonstrated to be functional. The analysis of this new hypothetical acceptor splice site of exon 55 by use of the NetGene2 prediction server was in line with a confidence value of 0.83.
Multiple Correcting Mutations in Other Hereditary Diseases
Recently, a patient with revertant mosaicism of hereditary tyrosinemia was described who had multiple reversion mutations in the FAH gene in different liver nodules (Bliksrud et al. 2005). This patient was compound heterozygous for a missense mutation in exon 12 (1009G→A; G337S) and a splice-site mutation in intron 12 (IVS12+5G→A). Three immunopositive liver nodules were investigated. In one nodule, a compensatory mutation (1061C→A) 6 bp upstream of the inherited splice-site mutation was detected, whereas, in the other two nodules, this splice-site mutation reverted to the wild-type sequence. True mutation reversion of the FAH gene most often underlies the restoration of the enzymatic activity. However, in this patient, besides true mutation reversion, a second-site mutation that restored the 5′ splice site of exon 12 was also identified. The presence of multiple correcting mutations in patients with EB and tyrosinemia type I suggests that, in the future, various mutations within one patient will be detected for other genetic diseases also.
Random Mutagenesis versus Directed Mutagenesis
One other patient with revertant nH-JEB with correction of COL17A1 mutation has been described, although, in this patient, no regions of clinically unaffected skin were present (Darling et al. 1999). Darling et al. (1999) found, among three biopsy specimens taken from nonlesional affected skin, two with focal expression of type XVII collagen. Keratinocytes independently grown from a third site also demonstrated restored protein synthesis. LDM analysis of one biopsy specimen revealed correction of the homozygous 4003delTC deletion by a frame-restoring insertion of two Gs, 4080insGG, in one chromosome. The resulting type XVII collagen molecule was of normal length but had a stretch of 25 altered amino acids. Thus, at the moment, three patients with mosaic nH-JEB have been described from among ∼3,000 patients with nH-JEB worldwide, established on the basis of the prevalence of 0.5 per million persons observed in the United States (Fine et al. 1999). Most likely, the number of patients with mosaicism is larger than that reported, since revertant keratinocytes can be present in affected skin and therefore will often remain unnoticed. An interesting question, to which the answer is yet unclear, is whether these reversions are occurring with a higher frequency than would be expected by chance. Recently, in the Arabidopsis plant, it was shown that the genetic restoration events that took place occurred with a higher frequency, in the order of a few percent, than could be explained by random mutagenesis (Lolle et al. 2005). Those authors postulated that the precise reversions are the result of a template-directed process that makes use of an ancestral RNA-sequence cache that comes from previous generations. Such a mechanism, if we assume that it would be present in other eukaryotes as well, would not, however, explain the presence of the second-site mutations detected in our patients.
Patient 2 has one patch on her left hand that includes both the middle finger and the pointing finger (fig. 5A and 5B). The reversion event that occurred in these keratinocytes should have taken place before the evolvement of the fingers, in the 8th week of pregnancy. If we assume that the fetus, at the time of the reversion event, was not >2.5 cm in length and had a similar skin constitution as normal human skin, with a keratinocyte count of 47,000/mm2 (Bergstresser et al. 1978), then ∼4.7×107 keratinocytes were present. With this amount of cells, given the average human mutation rate observed by Nachman and Cromwell (2000) of 2.5×10-8 per nucleotide per generation, the chance that such a specific reverse mutation would occur in one of the fetus’s cells is 0.39 per cell division (Nachman and Cromwell 2000). This number indicates that such a specific mutation could have occurred by chance and that the mosaic patches could have been the result of random mutagenesis. Nevertheless, it still leaves open the possibility that the reverse mutations are the result of a directed process.
Between different genetic disorders, a large variety in the frequency of mosaicism exists—for example, it is present in ∼85% of patients with tyrosinemia type I and ∼20% of patients with Bloom syndrome (German et al. 1977; Demers et al. 2003). The higher frequency of reversion in tyrosinemia type I, caused by deficiency of the enzyme fumarylacetoacetate hydrolase (FAH), was suggested to arise from the accumulation of the mutagenic metabolites fumarylacetoacetate and its precursor maleylacetoacetate in the cell. It is interesting to note that the extent of FAH mutation reversion, which ranges from 0.1% to 85% in the studied patients, was inversely correlated with the clinical severity of the disease (Demers et al. 2003). In disorders in which the genetic defect has an effect on the maintenance of genomic stability, such as FA and Bloom syndrome, the high reversion frequencies can be explained by the increased overall mutation rate. For example, in the autosomal recessive Bloom syndrome, loss-of-function mutations in the BLM gene, which encodes the RecQ family DNA helicase BLM, lead to high rates of exchange between sister chromatids. Ellis et al. (1995) showed that somatic intragenic recombination in compound heterozygous patients could generate a functionally wild-type BLM, resulting in cells with a low sister-chromatid exchange rate and, accordingly, a high percentage of individuals with mosaicism. In concert with this reversion type is the fact that patients with a common ancestor very rarely exhibit mosaicism. Because bilateral cell trafficking between the mother and fetus was suggested by a number of studies, Tanguay and colleagues (Bergeron et al. 2004) decided to investigate whether, in tyrosinemia type I, such an event was responsible for the commonly observed liver mosaicism. However, the corrected liver nodules in their patients were not of maternal origin, and therefore the data did not support this hypothesis.
Functional Restoration of the Skin
Our patients demonstrate that reversion of pathogenic COL17A1 mutations, either to the wild type or to a slightly altered sequence, may lead to functionally healthy skin. To obtain this restoration, only one corrected allele is needed, since nH-JEB is a recessive disease and heterozygous carriers display a normal skin phenotype. Furthermore, not all cells need to be reverted, as shown by the presence of interruptions in the staining of the epidermal basement membrane in the biopsy specimens of unaffected skin. However, these type XVII collagen–positive cells have to be present over a sufficiently large area. In patient 1, we found, in one specimen, wild-type expression of type XVII collagen over ∼0.25 mm2, but this area was obviously too small.
The earlier that reversion takes place in the embryonic development in vivo, the more likely that revertant descendant stem cells may finally spread as larger clusters and the more beneficial for clinical severity this may be. Because the correcting mutations were undetectable in leukocytes derived from mesoderm, all reversion events in the described patients took place after the development of the three-layered embryo in the 2nd week. Although the phylloid mosaic pattern of patient 2 was bilaterally distributed, the correcting mutations could have taken place after lateralization of the embryo, because the revertant patches were not symmetrical and the reversion events were caused by different mechanisms. The later the reversion occurred during embryogenesis, the smaller the patch. It may be that the focal correction observed in the upper arm of patient 1 and the focal correction observed by Darling et al. (1999) reflect such late-embryonic events.
The somatic mutations in our patients occurred in areas of the skin exposed to sunlight—in particular, the ankle and the extensor surfaces of arms and hands—and, for this reason, it was suggested that the acquired somatic mutations could have been environmentally induced early in life. Ultraviolet irradiation may cause two adjacent pyrimidine residues (C or T) to form a dimer. If not repaired, mutations in the form of C→T and CC→TT transitions can be introduced in the DNA sequence. In our patients, three mutations were introduced: 3781T→C, 3782G→C, and 4463-1G→A. Since the formation of pyrimidine dimers and (6–4) photoproducts cannot result in a T→C or G→C nucleotide change, the involvement of ultraviolet irradiation can be excluded for the 3781T→C and 3782G→C mutations. In the case of the 4463-1G→A mutation, a possible effect of ultraviolet irradiation is not ruled out, because the DNA sequence on the opposite strand contains adjacent pyrimidine residues.
Stability of the Healthy Skin Patches
Both our patients claimed that their patches of healthy skin had been there as long as they could remember and that they had not changed in size. Extensive photo documentation of patient 2 in 1969, 1994, and 2005 confirmed the stability of the unaffected patches (fig. 5). Thus, epidermal stem cells with corrected COL17A1 mutation do not spread in favor of their uncorrected counterparts. Since blistering and subsequent wound healing frequently occurred on the adjacent type XVII collagen–negative patches of skin, the expression of type XVII collagen apparently does not offer a selective advantage in repopulating stripped epidermis. This may be the result of hair follicles and sebaceous glands that are sheltered from subepidermal blistering, and, in this case, residing mutant stem cells may rapidly cover the wound area before a reverted stem cell from the periphery can invade. Our observations seemingly contrast with the mouse model of epidermolysis bullosa simplex, Dowling-Meara type (EBS-DM), with an inducible keratin 14 mutation in which wild-type stem cells replaced mutant keratinocytes in the wound bed (Cao et al. 2001). It also contradicts the report by Smith et al. (2004) of a revertant mosaic case with a keratin 14 mutation compensated for by a second 1-bp insertion that nullified the dominant-negative allele; the patient had experienced significant improvement in the skin-blistering phenotype from her early teens. It was suggested that revertant mosaicism could be responsible for the commonly observed amelioration with age in EBS-DM, either by multiple reversions occurring over a relatively short time or by replacement of mutant cells with revertant cells under suitable selective pressure. Such pressure is obviously absent in the mouse model for epidermolytic hyperkeratosis with mutations in keratin 10, since no spreading of wild-type cells into the mutated area was observed (Arin et al. 2001). Here, the mutant stem cells in the basal layer escape destruction because the epidermolytic process is suprabasal at the site of keratin 10 expression. All these results together indicate that the healing over time of an inherited disease probably depends on the benefit the stem cells experience from re-expressing the protein in their competition with mutant stem cells. Since these mutant stem cells may be located in the hair follicle, interfollicular epidermis, and sebaceous glands, the spreading of the revertant stem cells probably must also include destruction of the deeper-located stem cells. In addition, selective growth advantage was also demonstrated for FAH-positive hepatocytes in the FAH knockout mouse model of tyrosinemia type I, in which transplanted FAH-expressing cells effectively repopulated the diseased liver, because, in the FAH-deficient cells, the accumulation of the mutagenic fumarylacetoacetate was inducing cell-cycle arrest and apoptosis (Overturf et al. 1996; Jorquera and Tanguay 1999). Furthermore, in WAS, X-linked SCID, and adenosine deaminase deficiency, a selective growth advantage over their protein-deficient counterparts has also been observed for revertant lymphocytes (Hirschhorn et al. 1996; Stephan et al. 1996; Wada et al. 2003).
In summary, we demonstrate that 2 of 11 Dutch patients with GABEB (A.M.G.P., unpublished results) have multiple correcting mutations in COL17A1. Given this observed high frequency, the occurrence of more than one reversion event per individual, and the observation that reversion clusters may be too small to fully restore function, it is conceivable that reversions have often been overlooked in other patients. Studying this “natural gene therapy” will benefit experimental research of gene and cell therapy and will bring successful therapies for genetic diseases closer to actuality.
Acknowledgments
We are grateful to the patients for their participation in this study. We thank Dr. H. H. Lemmink and G. Jansen, for assistance in automated sequencing and microsatellite analysis; H. Sharafbayani for assistance in electron microscopy; and T. Blokzijl for support with the LDM technique. We also thank Dr. K. Owaribe (Nagoya, Japan) for the generous gift of anti–type XVII collagen monoclonal antibodies.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNPs rs17821926, rs4918079, rs2296219, and rs17116350)
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for human COL17A1 mRNA sequence [accession number NM_000494], human COL17A1 transcript sequence [accession number NP_000485], and sequences of exons 1–56 of COL17A1 [accession numbers U76564–U76604])
- NetGene2 Server, http://www.cbs.dtu.dk/services/NetGene2/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for tyrosinemia type I, Bloom syndrome, FA, WAS, X-linked SCID, adenosine deaminase deficiency, and GABEB)
References
- Arin MJ, Longley MA, Wang X-J, Roop DR (2001) Focal activation of a mutant allele defines the role of stem cells in mosaic skin disorders. J Cell Biol 152:645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron A, Lettre F, Russo P, Morissette J, Tanguay RM (2004) No evidence of maternal cell colonization in reverted liver nodules of tyrosinemia type I patients. Gastroenterology 127:1381–1385 [DOI] [PubMed] [Google Scholar]
- Bergstresser PR, Pariser RJ, Taylor JR (1978) Counting and sizing of epidermal cells in normal human skin. J Invest Dermatol 70:280–284 [DOI] [PubMed] [Google Scholar]
- Bliksrud YT, Brodtkorb E, Andresen PA, van den Berg IET, Kvittingen EA (2005) Tyrosinaemia type I—de novo mutation in liver tissue suppressing an inborn splicing defect. J Mol Med 83:406–410 [DOI] [PubMed] [Google Scholar]
- Cao T, Longley MA, Wang X-J, Roop DR (2001) An inducible mouse model for epidermolysis bullosa simplex: implications for gene therapy. J Cell Biol 152:651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S, Gache Y, Vailly J, Kanitakis J, Pulkkinen L, Uitto J, Ortonee J-P, Meneguzzi G (1999) Splicing modulation of integrin β4 pre-mRNA carrying a branch point mutation underlies epidermolysis bullosa with pyloric atresia undergoing spontaneous amelioration with ageing. Hum Mol Genet 8:2097–2105 [DOI] [PubMed] [Google Scholar]
- Darling TN, Yee C, Bauer JW, Hintner H, Yancey KB (1999) Revertant mosaicism: partial correction of a germ-line mutation in COL17A1 by a frame-restoring mutation. J Clin Invest 103:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers SI, Russo P, Lettre F, Tanguay RM (2003) Frequent mutation reversion inversely correlates with clinical severity in a genetic liver disease, hereditary tyrosinemia. Hum Pathol 34:1313–1320 [DOI] [PubMed] [Google Scholar]
- Di Zenzo G, Grosso F, Terracina M, Mariotti F, De Pita O, Owaribe K, Mastrogiacomo A, Sera F, Borradori L, Zambruno G (2004) Characterization of the anti-BP180 autoantibody reactivity profile and epitope mapping in bullous pemphigoid patients. J Invest Dermatol 122:103–110 [DOI] [PubMed] [Google Scholar]
- Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J (1995) Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am J Hum Genet 57:1019–1027 [PMC free article] [PubMed] [Google Scholar]
- Fine J-D, Bauer EA, McGuire J, Moshell A (1999) Epidermolysis bullosa: clinical, epidemiologic, and laboratory advances, and the findings of the National Epidermolysis Bullosa Registry. Johns Hopkins University Press, Baltimore [Google Scholar]
- Gatalica B, Pulkkinen L, Li K, Kuokkanen K, Ryynanen M, McGrath JA, Uitto J (1997) Cloning of the human type XVII collagen gene (COL17A1), and detection of novel mutations in generalized atrophic benign epidermolysis bullosa. Am J Hum Genet 60:352–365 [PMC free article] [PubMed] [Google Scholar]
- German J, Schonberg S, Louie E, Chaganti RSK (1977) Bloom’s syndrome. IV. Sister-chromatid exchanges in lymphocytes. Am J Hum Genet 29:248–255 [PMC free article] [PubMed] [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J (1994) The 1993–94 Généthon human genetic linkage map. Nat Genet 7:246–339 [DOI] [PubMed] [Google Scholar]
- Hall JG (1988) Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am J Hum Genet 43:355–363 [PMC free article] [PubMed] [Google Scholar]
- Hintner H, Wolff K (1982) Generalized atrophic benign epidermolysis bullosa. Arch Dermatol 118:375–384 [PubMed] [Google Scholar]
- Hirschhorn R (2003) In vivo reversion to normal of inherited mutations in humans. J Med Genet 40:721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn R, Yang DR, Puck JM, Huie ML, Jiang C-K, Kurlandsky EL (1996) Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat Genet 13:290–295 [DOI] [PubMed] [Google Scholar]
- Jonkman MF, Castellanos Nuijts M, van Essen AJ (2003) Natural repair mechanisms in correcting pathogenic mutations in inherited skin disorders. Clin Exp Dermatol 28:625–631 [DOI] [PubMed] [Google Scholar]
- Jonkman MF, de Jong MCJM, Heeres K, Pas HH, van der Meer JB, Owaribe K, Martinez de Velasco AM, Niessen CM, Sonnenberg A (1995) 180-kD bullous pemphigoid antigen (BP180) is deficient in generalized atrophic benign epidermolysis bullosa. J Clin Invest 95:1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, de Jong MCJM, Heeres K, Steijlen P, Owaribe K, Küster W, Meurer M, Gedde-Dahl TJ, Sonnenberg A, Bruckner-Tuderman L (1996) Generalized atrophic benign epidermolysis bullosa: either 180-kd bullous pemphigoid antigen or laminin-5 deficiency. Arch Dermatol 132:145–155 [DOI] [PubMed] [Google Scholar]
- Jonkman MF, Scheffer H, Stulp R, Pas HH, Nijenhuis M, Heeres K, Owaribe K, Pulkkinen L, Uitto J (1997) Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell 88:543–551 [DOI] [PubMed] [Google Scholar]
- Jorquera R, Tanguay RM (1999) Cyclin B-dependent kinase and caspase-1 activation precedes mitochondrial dysfunction in fumarylacetoacetate-induced apoptosis. FASEB J 13:2284–2298 [DOI] [PubMed] [Google Scholar]
- Kvittingen EA, Rootwelt H, Berger R, Brandtzaeg P (1994) Self-induced correction of the genetic defect in tyrosinemia type I. J Clin Invest 94:1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Sawamura D, Giudice GJ, Diaz LA, Mattei M-G, Chu M-L, Uitto J (1991) Genomic organization of collagenous domains and chromosomal assignment of human 180-kDa bullous pemphigoid antigen-2, a novel collagen of stratified squamous epithelium. J Biol Chem 266:24064–24069 [PubMed] [Google Scholar]
- Li K, Tamai K, Tan EM, Uitto J (1993) Cloning of type XVII collagen: complementary and genomic DNA sequences of mouse 180-kilodalton bullous pemphigoid antigen (BPAG2) predict an interrupted collagenous domain, a transmembrane segment, and unusual features in the 5′-end of the gene and the 3′-untranslated region of the mRNA. J Biol Chem 268:8825–8834 [PubMed] [Google Scholar]
- Lolle SJ, Victor JL, Young JM, Pruitt PR (2005) Genome-wide non-Mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434:505–509 [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J (2000) GT repeats are associated with recombination on human chromosome 22. Genome Res 10:1108–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA, Gatalica B, Christiano AM, Owaribe K, McMillan JR, Eady RAJ, Uitto J (1995) Mutations in the 180-kD bullous pemphigoid antigen (BPAG2), a hemidesmosomal transmembrane collagen (COL17A1), in generalized atrophic benign epidermolysis bullosa. Nat Genet 11:83–86 [DOI] [PubMed] [Google Scholar]
- Nachman MW, Cromwell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou C-N, Finegold M, Grompe M (1996) Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet 12:266–273 [DOI] [PubMed] [Google Scholar]
- Smith FJD, Morley SM, McLean WHI (2004) Novel mechanism of revertant mosaicism in Dowling-Meara epidermolysis bullosa simplex. J Invest Dermatol 122:73–77 [DOI] [PubMed] [Google Scholar]
- Smithies O, Powers PA (1986) Gene conversions and their relation to homologous chromosome pairing. Philos Trans R Soc Lond B Biol Sci 312:291–302 [DOI] [PubMed] [Google Scholar]
- Stephan V, Wahn V, Le Deist F, Dirksen U, Bröker B, Müller-Fleckenstein I, Horneff G, Schroten H, Fischer A, de Saint Basile G (1996) Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N Engl J Med 335:1563–1567 [DOI] [PubMed] [Google Scholar]
- Wada T, Konno A, Schurman SH, Garabedian EK, Anderson SM, Kirby M, Nelson DL, Candotti F (2003) Second-site mutation in the Wiskott-Aldrich syndrome (WAS) protein gene causes somatic mosaicism in two WAS siblings. J Clin Invest 111:1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Schurman SH, Jagadeesh GJ, Garabedian EK, Nelson DL, Candotti F (2004) Multiple revertant patients in a single Wiskott-Aldrich syndrome family. Blood 104:1270–1272 [DOI] [PubMed] [Google Scholar]
- Waisfisz Q, Morgan NV, Savino M, de Winter JP, van Berkel CGM, Hoatlin ME, Ianzano L, Gibson RA, Arwert F, Savoia A, Mathew CG, Pronk JC, Joenje H (1999) Spontaneous functional correction of homozygous Fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat Genet 22:379–383 [DOI] [PubMed] [Google Scholar]