Abstract
The growth of an individual is deeply influenced by the regulation of cell growth and division, both of which also contribute to a wide variety of pathological conditions, including cancer, diabetes, and inflammation. To identify a major regulator of human growth, we performed positional cloning in an autosomal recessive type of profound short stature, anauxetic dysplasia. Homozygosity mapping led to the identification of novel mutations in the RMRP gene, which was previously known to cause two milder types of short stature with susceptibility to cancer, cartilage hair hypoplasia, and metaphyseal dysplasia without hypotrichosis. We show that different RMRP gene mutations lead to decreased cell growth by impairing ribosomal assembly and by altering cyclin-dependent cell cycle regulation. Clinical heterogeneity is explained by a correlation between the level and type of functional impairment in vitro and the severity of short stature or predisposition to cancer. Whereas the cartilage hair hypoplasia founder mutation affects both pathways intermediately, anauxetic dysplasia mutations do not affect B-cyclin messenger RNA (mRNA) levels but do severely incapacitate ribosomal assembly via defective endonucleolytic cleavage. Anauxetic dysplasia mutations thus lead to poor processing of ribosomal RNA while allowing normal mRNA processing and, therefore, genetically separate the different functions of RNase MRP.
Introduction
The growth of an individual is maintained by many factors and depends, for the most part, on the size and number of cells the individual contains. Both parameters are deeply influenced by the regulation of cell growth and division. Dysfunction of the signaling pathways that control cell growth results in cells of altered size or number and can lead to developmental errors and contribute to a wide variety of pathological conditions, including cancer, diabetes, and inflammation (Hall et al. 2004).
Research on common types of human growth deficiencies in recent years has led to the elucidation of genetic etiologies for isolated growth hormone deficiencies, combined pituitary hormone deficiencies (Rosenfeld and Hwa 2004), and a variety of skeletal dysplasias revealing molecular pathways, including defects in extracellular structural proteins, macromolecules, metabolic pathways, signal transduction mechanisms, nuclear proteins, oncogenes, and RNA and DNA processing (Superti-Furga et al. 2001). To identify a major contributor to cell growth regulation, we performed positional cloning in a rare autosomal recessive type of extreme short stature, named “anauxetic” dysplasia after the Greek term for “to not let grow” (Horn et al. 2001). Anauxetic dysplasia (MIM 607095) is an autosomal recessive spondylometaepiphyseal dysplasia characterized by the prenatal onset of extreme short stature, an adult height of <85 cm, hypodontia, and mild mental retardation (Horn et al. 2001) (fig. 1). Major radiographic characteristics are late-maturating, ovoid vertebral bodies with concave dorsal surfaces in the lumbar region; small capital femoral epiphyses; hypoplastic femoral necks; hypoplastic iliac bodies and shallow acetabulae; irregular metaphyseal mineralization and demarcation of the long tubular bones; short metacarpals with widened shafts (I and V); very short and broad phalanges with small, late-ossifying epiphyses and bullet-shaped middle phalanges; and midfacial hypoplasia. The number of chondrocytes is severely reduced in the resting and proliferating cartilage, with diminished columnization of the hypertrophic zone. We report here the identification of RMRP gene mutations as the underlying genetic defect of anauxetic dysplasia, which, therefore, is allelic to cartilage hair hypoplasia (CHH [MIM 250250]) (Ridanpaa et al. 2001), as well as metaphyseal dysplasia without hypotrichosis (MDWH [MIM 250460]) (Bonafe et al. 2002; Ridanpaa et al. 2003b). The RMRP gene encodes the untranslated RNA subunit of the ribonucleoprotein endoribonuclease, RNase MRP. The disease-causing functional impairment of the RMRP gene product in CHH mutations in humans is unknown (Ridanpaa et al. 2003a). The RMRP ortholog in yeast, nme1, is involved in ribosome synthesis, the generation of RNA primers for mitochondrial DNA replication (Welting et al. 2004), and the degradation of cell cycle–regulated mRNA (Gill et al. 2004). We performed functional studies in yeast and humans to provide the first physiological demonstration of a function for RNase MRP in mammalian cells. Furthermore, we explain clinical heterogeneity by a correlation between the level and type of functional impairment in vitro and the severity of short stature or the presence of cancer predisposition.
Figure 1.
Clinical and x-ray characteristics of anauxetic dysplasia. A, far left and near left, Patient, aged 16.5, from family 2 (Horn et al. 2001), showing severe short stature with hyperlordosis (height of 74 cm). Upper right, X-ray of the left arm at age 7, showing extremely retarded carpal ossification corresponding to ∼3 mo bone age, as well as short and broad tubular bones. Lower right, X-ray of spine and pelvis at age 7, showing decreased vertical dimension of the ilia, unossified femoral necks, and small, irregular capital femoral epiphyses. B, X-rays of lower extremities at age 7 mo (top) and pelvis at age 2 (bottom) of patient 3, showing δ-shaped metaphyses with irregular borders and pelvic changes similar to those in panel A.
Material and Methods
Patients
This study was approved by the ethical review board of the Friedrich-Alexander University in Erlangen-Nürnberg, Germany. After receiving informed consent, we obtained peripheral blood samples from members of two previously published families (Menger et al. 1996; Horn et al. 2001) and from one new sporadic patient with spondylometaepiphyseal dysplasia Menger type, or anauxetic dysplasia. From the Jordanian family 1 (Menger et al. 1996), three affected children, six unaffected children, and their consanguineous parents were available. From the German family 2 (Horn et al. 2001), two affected children (one living and one deceased) and their nonconsanguineous parents were recruited. From the living child of family 2, a fibroblast culture from a skin biopsy and RNA samples from peripheral blood collected in PAXgene tubes (PreAnalytiX) were also obtained. The PAXgene tube contains a proprietary reagent that immediately stabilizes intracellular RNA for days at room temperature and for weeks at 4°C. Specific consent was obtained from the respective parents to publish photographs of their children. The living patient from family 2 (Horn et al. 2001), who showed severe short stature with hyperlordosis (height of 74 cm, weight of 11.8 kg, and head circumference of 50.2 cm) was re-evaluated at age 16.5 (fig. 1A). Because of atlantoaxial subluxation, he underwent cervical fusion. Puberty was somewhat delayed but was otherwise normal. The patient is mildly mentally retarded (capable of some reading, writing, and calculating up to the number 20).
The sporadic case, patient 3, of German origin, (fig. 1B) was the deceased offspring of nonconsanguineous, healthy parents, who have two additional healthy children. Extreme short stature was evident at term birth, when body length was 37 cm, head circumference was 31 cm, and weight was 2.02 kg. As in the deceased patient from family 2, there was spasticity caused by cervical cord compression. This child also showed mental retardation and severe growth failure, with a body length of only 53 cm at age 7. Radiographic signs included extremely slow epiphyseal maturation, metaphyseal irregularities, hypoplastic femoral necks, thick and short tubular bones of the hands, and ovoid vertebral bodies. Recurrent pneumonias due to a progressive thoracic deformity finally led to lethal respiratory insufficiency.
Linkage Analysis
Genomic DNA was extracted from peripheral blood lymphocytes by standard salt precipitation methods and, from the deceased patient of family 2, with the use of a dried blood spot. Homozygosity mapping was performed with a genomewide set of 385 evenly distributed microsatellite markers, with an average distance of 11 cM, on the basis of the Généthon final linkage map (Dib et al. 1996) and as described elsewhere (Horn et al. 2000). Markers were labeled with fluorescent dyes and amplified by PCR. PCRs were pooled for genotyping on a Prism 377 Genetic Analyzer (Applied Biosystems) running Genescan (v. 2.1) and Genotyper (v. 2.0) software (Applied Biosystems).
We used the MLINK program of the LINKAGE software package and the LINKRUN software to calculate two-point LOD scores between the disease phenotype and each of the markers, assuming a recessive mode of inheritance with full penetrance (Lathrop and Lalouel 1984), and we used Genehunter (v. 1) for multipoint linkage analysis. A set of additional markers (D9S1679, D9S1833, D9S126, D9S1114, D9S161, D9S263, D9S746, D9S1678, D9S270, D9S1875, D9S2025, D9S1868, D9S52, D9S251, D9S43, D9S1845, D9S78, D9S1817, D9S1804, D9S1791, D9S1879, and D9S175) for fine-mapping analysis was genotyped in families 1 and 2 on an ABI 3100 capillary sequencer. We manually reconstructed haplotypes for the region flanked by the markers D9S1679 and D9S1874, with marker order based on the draft human genomic sequence (National Center for Biotechnology Information).
Mutation Analysis
Exons of positional candidate genes were amplified by PCR with the use of intronic and additional exonic primers for larger exons. Concerning the RMRP gene, we amplified the coding region, the promoter region, and the TATA box in all members of family 1 and 2 and in patient 3. The PCR products were purified with Filter plates LSKMPCR50 (Millipore) on a Tecan liquid handling system, and sequencing reactions were performed on both strands with the use of the BigDye Terminator Cycle Sequencing Kit (v. 3.1 [Applied Biosystems]), in accordance with the manufacturer’s instructions. After purification with filter plates LSKS09624 (Millipore), the products were analyzed on an ABI Genetic Analyzer 3730 (Applied Biosystems) with SeqMan II software (DNASTAR).
The nucleotide sequence of the human RMRP gene was aligned with the sequences of orthologs from other species with use of the ClustalW Multiple Sequence Alignment algorithm (Baylor College of Medicine) and AlignX of the Vector NTI Suite 6.0 (InforMax). RNA was isolated from the fibroblasts and lymphocytes of one patient and the parents of family 2, as well as from 10 controls with normal growth development, with use of the RNeasy Mini Kit (Qiagen). The synthesis of cDNA was performed with the use of a Superscript II Reverse Transcriptase Kit with random hexamer primers (Invitrogen), in a total volume of 20 μl.
Analysis of Mutant RMRP Constructs and Real-Time PCR Assays
By PCR, we amplified wild-type and mutant RMRP, using lymphocyte DNA from a healthy control and from affected patients as template, and we subcloned the RMRP into pcDNA3.1 with the Directional TOPO Expression Kit vector (Invitrogen). To generate the cartilage hair hypoplasia founder mutation +70A→G, we modified the wild-type RMRP construct, using the QuikChange site-directed mutagenesis kit (Stratagene), in accordance with the manufacturer's instructions.
We transiently transfected fibroblast cells from a healthy donor, with 500 ng of each RMRP construct (wild-type, +14G→A, +90_91AG→GC, ins111_112ACGTAGACATTCCT, +254C→G, and +70A→G), using Lipofectamine 2000 and Plus Reagent (Invitrogen). We also transfected dermal fibroblasts from the affected, living child of family 2, with 500 ng of the RMRP wild-type construct. As a negative control, the empty vector was also transfected. We collected cells at 12 h, 36 h, 48 h, and 60 h after transfection and extracted RNA and DNA, using TRIzol Reagent (Invitrogen). RMRP, CCNA2, CCNB2, 5.8S ribosomal RNA (rRNA), and ITS-1–bound 5.8S rRNA expression was quantified by real-time PCR (fig. 2). TaqMan primers and probes were newly designed with the Primer Express Software v2.0 (Applied Biosystems) for RMRP (GenBank accession number M29916), human ribosomal DNA complete repeating unit (GenBank accession number U13369), CCNB2 (GenBank accession number AY864066), and CCNA2 (GenBank accession number NM_001237). We used minor-grove-binder probes containing a fluorophore 5′ FAM modification as reporter and a 3′ nonfluorescent quencher. The multiplex assay for each sample was performed in 384-well optical plates (Applied Biosystems) with a final volume of 20 μl each. All measurements, each from three different cultures, were performed in quadruplicate. Quantitative RT-PCR was performed with the QuantiTect Probe RT-PCR Kit (Qiagen) with an ABI Taqman 7900HT. After the evaluation of normalization to three different endogenous controls (18s-RNA, β-2-microglobulin, and RNA-II-polymerase), we performed experiments with RNA-II-polymerase (RPII) as the internal standard. We verified the relative cell count by targeting albumin genomic DNA, normalizing it to the level at 12 h after transfection (Thiel et al. 2003).
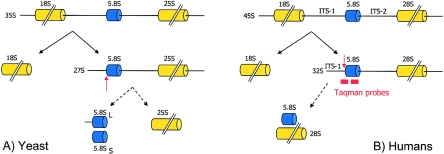
Figure 2.
Scheme of rRNA processing in yeast and humans. A, Deletions of nme1, the RMRP RNA gene homolog in yeast, impacts late-60S ribosomal assembly via defective endonuclease cleavage of the precursor subunit 5.8S rRNA at the ITS-1 A3 site (red arrow). The eukaryotic pre-60S particles undergo maturation via several endonucleolytic cleavages, resulting in 25S, 5.8S, and 5S rRNA components in yeast and 28S, 5.8S, and 5S rRNA components in humans. In yeast, the 5.8S rRNA consists of two main species, 5.8SL and 5.8SS, with a ratio of 1:10 long to short, generated by two different pathways. The nme1 gene product is required for the production of the 5.8SS species; hence, depletion leads to increased levels of 5.8SL and reduced levels of 5.8SS. B, Assumed cleavage site of human RNase MRP (red arrow) and location of Taqman probes used to measure endonucleatic cleavage in mutants.
Analysis of Mutant nme1 Constructs in Saccharomyces cerevisiae
Plasmid DNA pMES140 carrying the full-length nme1 gene was used as a template (Schmitt 1999). We modified the wild-type nme1 construct, using the QuikChange site-directed mutagenesis kit (Stratagene) to generate the +28T→C (M1, corresponding to human +14G→A by sequence alignment), +279C→G (M2, corresponding to human +254C→G by sequence alignment), +18T→C (M3, corresponding to human +14G→A by structural homology), and +319T→G (M4, corresponding to human +254C→G by structural homology) variants. Mutant variants were exchanged for the wild type with use of a standard plasmid shuttle (Shadel et al. 2000). Yeast was grown on YPD medium (1% yeast extract, 2% peptone, and 2% dextrose), RNA was extracted, and 5.8S rRNA was examined directly as described elsewhere (Shadel et al. 2000).
Mutant yeast strains were analyzed for temperature-conditional growth on YPD or YPG (1% yeast, 2% peptone, and 3% glycerol) plates and were compared with congenic wild-type strain (Shadel et al. 2000). Metabolic activity was indicated by adenine consumption (Shadel et al. 2000) and the resulting development of a red color.
Mitomycin C Resistance Analysis
Skin fibroblasts from the living patient of family 2 were cultured on microscopic slides in Quadriperm dishes in DMEM/Ham’s F12 medium containing 20% fetal calf serum and supplemented with Ultraser (4%). The cultures were kept at 37°C in a 5% CO2 atmosphere. Mitomycin C was added in various final concentrations (0.025 μg/ml, 0.05 μg/ml, 0.1μg/ml, and 0.2 μg/ml) 24 h before harvesting. One slide was cultured without mitomycin C. As a control, an identical experiment was performed in parallel on fibroblast cultures derived from the skin of a healthy individual. Chromosomal breakage was analyzed on Giemsa-stained metaphase preparations.
Results
Positional Cloning
Genomewide homozygosity mapping in the large, consanguineous family 1 with both parents, three affected children, and five unaffected children revealed a 18.7-cM region on chromosome bands 9p13–9p21, with a maximum multipoint LOD score of 3.012 between markers D9S171 and D9S1874 (fig. 3). Fine mapping with additional markers and family members in families 1 and 2 refined the linkage region to 17.1 cM between markers D9S1114 and D9S1874, resulting in a 11.15-Mb candidate interval, with a maximum LOD score of 3.158.
Figure 3.
Summary of the genome scan for anauxetic dysplasia or spondylometaphyseal dysplasia (SMED) with use of GENEHUNTER-PLUS (Kong and Cox 1997). Upper panel, Multipoint parametric LOD-score analysis across all autosomes, from pter to qter, with use of a fully penetrant autosomal recessive model. For clarity, only LOD scores >−2 are shown. Maximum LOD score was 3.012 at markers D9S171 and D9S1874 on chromosome 9p. Lower panel, Nonparametric multipoint Z score. Again, the maximum Z-score was obtained for markers in the same region on chromosome 9p. Dotted lines indicate chromosome boundaries.
According to the National Center for Biotechnology Information Map Viewer and the UCSC Genome Browser, the critical interval contained 77 known genes. Only when using GeneSeeker (van Driel et al. 2003) did we identify the RMRP gene to also be located in the interval. Sequencing of RMRP in the anauxetic dysplasia families revealed a homozygous insertion mutation, ins111_112ACGTAGACATTCCT, in the three affected patients from family 1 and revealed compound heterozygous mutations in both affected members of family 2 (+14G→A and +90_91AG→GC), as well as in patient 3 (+90_91AG→GC and +254C→G) (fig. 4A–4E). All four novel mutations affect evolutionarily highly conserved regions and were absent in 378 control chromosomes.
Figure 4.
RMRP gene mutations identified in anauxetic dysplasia patients of families 1–3 and their evolutionary conservation in various orthologs. Affected members of family 1 were homozygous for a 14-bp insertion at position 111_112 (A), affected members of family 2 were compound heterozygous for the mutations +90_91AG→GC (B) and +14G→A (C), and patient 3 was compound heterozygous for the +90_91AG→GC mutation (B) and for a +254C→G mutation (D). E, Significant decrease in RMRP expression of the +14G→A mutant allele (*), as revealed by RT-PCR in the patient of family 2.
The +90_91AG→GC mutation occurred in family 2 and in patient 3, both of whom originated from the same German region. Accordingly, the mutation was found to reside on a common ancestral haplotype in both families, confirming a founder effect (SNP haplotype: −56G, −48A, −6A, +156C, +177T, and +274C).
Functional Studies in Yeast
In yeast, the anauxetic dysplasia point mutations resulted only in mild growth retardation and reduced metabolic activity, whereas the 5.8SL/5.8SS ratio was unaltered and colony size was near normal on different carbon sources and at different temperatures (fig. 5C).
Figure 5.
Scheme of the two-dimensional structure of RMRP and nme1 RNA and the results of nme1 mutant analysis in S. cerevisiae. A, Predicted two-dimensional structure of human RMRP RNA, showing the position of the anauxetic dysplasia mutations +14G→A and +254C→G. B, Predicted two-dimensional structure (Shadel et al. 2000) of yeast nme1 RNA, showing positions considered equivalent by sequence alignment (M1=+14G→A and M2=+254C→G) or structural alignment (M3=+14G→A and M4=+254C→G). C, Reduced colony growth and metabolic activity in M1–M4 colonies grown on YPD and YPG plates at 30°C and 34°C, as compared with wild-type colonies that show red color due to an ade2 marker. Because of a mutation in the ade2 gene, adenine biosynthesis is impaired, which leads to the accumulation of an oxidized (red) precursor in yeast with normal metabolic activity.
Human Functional Analyses
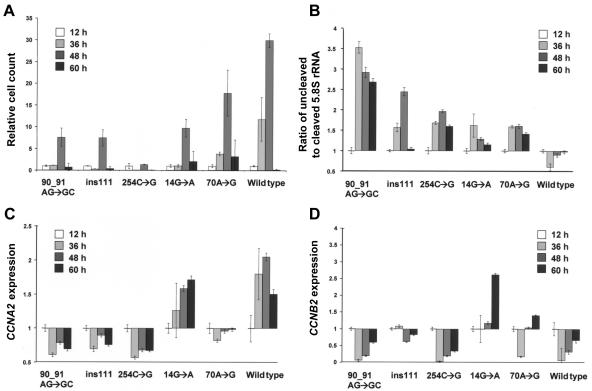
Concerning the +14G→A mutation, sequencing of the patients' RT-PCR products, as well as allele-specific RT-PCR, showed a lack of expression of this mutant RMRP allele (fig. 4E). Expression levels in the cells of a patient from family 2 (carrying the +90_91AG→GC and +14G→A mutations) showed a 1.8-fold increase in uncleaved pre-5.8S rRNA bound to ITS-1 in fibroblasts and a 1.44-fold increase in lymphocytes, indicating defective rRNA processing (fig. 6A–6D). Transfection of the patient's very poorly growing fibroblasts with the wild-type RMRP construct significantly increased the growth rate and production of the 5.8S rRNA (fig. 6E and 6F).
Figure 6.
A–D, Assessment of RMRP expression and endonucleolytic cleavage for ribosomal assembly in native cells of one patient from family 2 (P) with anauxetic dysplasia and compound heterozygosity for the RMRP mutations +90_91AG→GC and +14G→A. E and F, Rescue of phenotype by wild-type hyperexpression. A, Expression of RMRP was decreased 13-fold in the patient's lymphocytes and threefold in his fibroblasts (B), compared with average levels in 11 healthy control individuals with (C) and without outliers (C′). Decreased endonucleolytic cleavage in the patient, demonstrated by the increased ratio of 5.8S rRNA bound to ITS-1 versus cleaved 5.8S rRNA of 1.44-fold in lymphocytes (C) and 1.8-fold in fibroblasts (D). E, Rescue of phenotype by wild-type hyperexpression, as shown by the dramatically increasing cell count up to 48 h after transfection (dark bars) in comparison to transfection with the empty vector (pale bars). F, Increase of cell count correlates with increasing endonucleolytic cleavage in ribosomal assembly as measured by the ratio of 5.8S rRNA bound to ITS-1 versus cleaved 5.8S rRNA.
Human fibroblast cells transfected with the RMRP wild type showed a 30-fold increase in cell count in 48 h, cells transfected with the anauxetic dysplasia mutations showed only a minor increase in cell count, and the CHH founder mutation showed an intermediate reduction (fig. 7A). Accordingly, the wild-type transfected cells showed a shift to a slightly higher proportion of cleaved 5.8S rRNA, whereas all cells transfected with mutant constructs demonstrated diminished endonucleolytic cleavage at the ITS-1/5.8S rRNA junction site (fig. 7B). This effect was the most pronounced in the anauxetic dysplasia mutations.
Figure 7.
Comparison of different RMRP mutants overexpressed in transiently transfected human wild-type fibroblasts, with reference to relative cell counts and endonucleolytic cleavage activity in ribosomal assembly and cyclin levels. A, Relative cell count normalized to the level at 12 h after transfection in anauxetic dysplasia mutants and the CHH founder mutation +70A→G, as compared with the wild-type transfected cells. B, Impaired growth correlating with decreased endonucleolytic cleavage activity investigated via the ratio of 5.8s rRNA bound to ITS-1 versus cleaved 5.8s rRNA normalized to 12 h after transfection. C, Decreased CCNA2 expression in naturally expressed anauxetic dysplasia mutations and less severe in the CHH founder mutation. D, Significantly increased CCNB2 expression in the +70A→G mutation. Note that the +14G→A mutation is not expressed in vivo.
With respect to cyclin-dependent cell cycle function in human RNase MRP, except for cells with the naturally nonexpressed +14G→A mutation, all cells overexpressing RMRP mutants showed significantly decreased cyclin A2 levels correlating with the reduction in the growth rate and the clinical phenotype (fig. 7C and 7D). In contrast to the anauxetic dysplasia mutations, the CHH founder mutation +70A→G had significantly increased cyclin B2 mRNA levels. Mitomycin C sensitivity testing in fibroblasts of the patient of family 2 revealed no evidence for chromosomal instability (data not shown).
Discussion
Linkage analysis in two families mapped anauxetic dysplasia to a 17.1-cM interval in 9p13–9p21. These 11.15 Mb spanning the region between D9S1114 and D9S1874 contained 77 known genes without an outstanding candidate. Since untranslated RNA genes are not annotated in the commonly used databases, only when using GeneSeeker (van Driel et al. 2003) did we identify the RMRP gene to also be located in the interval. Mutations of the RMRP gene are known to cause cartilage hair hypoplasia (CHH [MIM 250250]) (Ridanpaa et al. 2001), as well as metaphyseal dysplasia without hypotrichosis (MDWH [MIM 250460]) (Bonafe et al. 2002; Ridanpaa et al. 2003b). Both are milder types of autosomal recessive skeletal dysplasias (adult heights ranging from 103.7 cm to 149.0 cm), with proliferative bone marrow dysfunction and predisposition to malignant tumors in patients with CHH (Makitie et al. 1995). In contrast to those in patients with anauxetic dysplasia, vertebral bodies in patients with CHH and MDWH are only mildly affected, the pelvic appearance is normal, and the epiphyses are not dysplastic. Mutational analysis in the RMRP gene in our patients with anauxetic dysplasia subsequently revealed homozygous or compound heterozygous mutations in every patient from the three unrelated families (fig. 4A–4E). All of these novel mutations affect evolutionarily highly conserved regions and were absent in healthy controls. One mutation (+90_91AG→GC), which occurred in two families originating from the same region, was shown to reside on a common ancestral haplotype, confirming a founder effect.
The RMRP gene encodes the untranslated RNA subunit of the ribonucleoprotein endoribonuclease, RNase MRP. In yeast, it is involved in ribosome synthesis, the generation of RNA primers for mitochondrial DNA replication (Welting et al. 2004), and the degradation of cell cycle–regulated mRNA (Gill et al. 2004). The disease-causing functional impairment of the RMRP gene product in CHH mutations in humans is unknown (Ridanpaa et al. 2003a). The founder mutation +70A→G contributes to 92% of Finnish and 48% of non-Finnish patients with CHH (Ridanpaa et al. 2002). When we compared mutations found in the different RMRP gene phenotypes with sites of known RNA protein or RNA-RNA interaction (Welting et al. 2004), we were unable to identify any obvious genotype-phenotype correlation. Therefore, we assume that the clinical heterogeneity of RMRP mutations might be explained by different effects on the overall structure or activity of the enzyme.
In accordance with previous predictions of the RMRP structure published elsewhere (Topper and Clayton 1990; Schmitt et al. 1993; Pluk et al. 1999), the present model (fig. 5A) suggests that the site of the +90_91AG→GC mutation is located in a loop. The size of the loop and the pairing of the adjacent bases, however, show some degree of variation among the different predictions. To obtain a more detailed picture of the geometry of the respective region, we compared the sequence properties of the loop to RNA for which a three-dimensional structure is available.
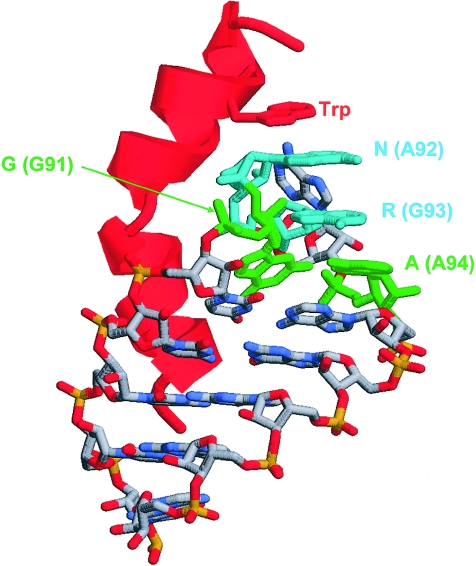
Nucleotides 91–94 match the consensus sequence of a GNRA tetraloop, a motif that adopts a well-defined three-dimensional structure (Heus and Pardi 1991) and that has been shown previously to be involved in RNA-protein interaction, for example, in the complex of the nutRboxB RNA with the N-protein (Schärpf et al. 2000). In the latter structure, the first and last nucleotide of the GNRA tetraloop form a noncanonical (“sheared”) base pair (fig. 8). Mutation of +91G→C destroys this base pair and, thus, may severely alter the structure of this loop and its ability to interact with proteins.
Figure 8.
Structural properties of a GNRA tetraloop and implications for RNA-protein interaction, as evident from the nutboxB-RNA structure in complex with the N-protein (Schärpf et al. 2000). The RNA is shown in stick presentation and the bound protein is schematically shown in red. Key nucleotides of the GNRA tetraloop are colored in green and cyan, and the respective sequence positions in RMRP are given in parentheses. The G and the A of the tetraloop form a sheared base pair which will be disrupted by a G→C mutation in RMRP. The role of the tetraloop structure for RNA-protein interactions is emphasized by the stacking of a tryptophan in the nutboxB-RNA complex with the N-protein.
The +254C→G mutation is located at the end of a large loop containing the LRI-I sequence. The exchange of +254C→G might promote base pairing in the respective region between U252-U253-G254 and the complementary stretch A237-A236-C235, which is located at the other end of this loop (fig. 5A). This enhanced base pairing might result in a distortion of the LRI-I sequence and, thus, hamper its interaction with the LRI-I sequence located around nucleotide 74. The LRI-1 forms the central core of domain 1, which also includes helixes I–V and is responsible for cleavage activity (Schmitt 1999). Therefore, disruption of LRI-I may severely hamper the cleavage activity of RNase MRP.
The insertion after position +111 is located within helix VII, belonging to the second domain of RNase MRP RNA (fig. 5A), which is thought to provide enzyme-specific functions such as substrate specificity (Schmitt 1999). Its location within a loop might explain why such a large insertion of 14 nucleotides does not result in a complete disruption of the RMRP structure, which should be lethal.
Lack of expression of the +14G→A mutation may indicate a yet-unknown regulatory or intragenic promoter region in the evolutionarily conserved region +3 to +31. However, this finding is unexpected, since RNA polymerase III promoters containing a TATA box were believed to not contain gene-internal elements (Schramm and Hernandez 2002). Alternatively, this mutation may lead to extremely unstable RNA.
Since the rate of growth depends on the synthesis of proteins, and the synthesis of proteins depends on ribosomes, the regulation of growth must ultimately depend on the regulation of the synthesis of new ribosomes (Hall et al. 2004). Therefore, we assumed that the novel RMRP mutations observed in anauxetic dysplasia would have a more pronounced impact on ribosomal assembly than the known CHH and MDWH mutations.
Mutation of nme1, the RMRP RNA gene homolog in yeast, impacts late-60S ribosomal assembly via defective endonuclease cleavage of the precursor subunit 5.8S rRNA at the ITS-1 A3 site (Schmitt and Clayton 1993; Henry et al. 1994). The severity of the 5.8S rRNA–processing defect is directly proportional to the severity of the growth defect in nme1 mutant yeast (Shadel et al. 2000). Deficiency of a single 60S subunit not only can lead to the breakdown of the entire particle but also can disrupt regulatory signals that feed back on ribosome biogenesis from the secretory machinery (Zhao et al. 2003). To investigate the functional impairment resulting from the anauxetic dysplasia mutations, we introduced them in the analogous positions of the yeast nme1 gene (fig. 5A and 5B). In yeast, the 5.8S rRNA consists of two main species, 5.8SL and 5.8SS, with a ratio of 1:10 long to short, generated by two different pathways (Lindahl et al. 1992; Schmitt and Clayton 1993) (fig. 2). The nme1 gene is required for production of the 5.8SS species; hence, depletion leads to increased levels of 5.8SL and reduced 5.8SS (Lindahl et al. 1992; Schmitt and Clayton 1993; Henry et al. 1994).
In yeast, the anauxetic dysplasia point mutations resulted only in mild growth retardation and reduced metabolic activity, whereas the 5.8SL/5.8SS ratio was unaltered and the colony size was near normal (fig. 5C). However, in contrast to gross deletions, single point mutations in the nme1 gene have resulted only once in a strong phenotype (Shadel et al. 2000). This may be explained by the fact that unicellular yeast has an excess RNase MRP capacity, whereas humans are much more sensitive to altered activity levels.
Accordingly, expression levels in the cells of a patient in family 2 (carrying the +90_91AG→GC and +14G→A mutations) showed an increase in uncleaved pre-5.8S rRNA bound to ITS-1 in fibroblasts and lymphocytes, indicating defective rRNA processing. To ensure that the effect was RNase MRP dependent, we transfected the patient's very poorly growing fibroblasts with a wild-type RMRP construct, thereby significantly increasing the growth rate and production of the 5.8S rRNA (fig. 6E and 6F). This is the first physiological demonstration of a function for RNase MRP in mammalian cells.
To compare the effects of different RMRP mutations, we investigated 5.8S rRNA processing in cell constructs overexpressing the novel anauxetic dysplasia mutations or the CHH founder mutation (+70A→G), respectively (fig. 7A). Whereas cells transfected with the RMRP wild type showed a 30-fold increase in cell count in 48 h, cells transfected with the anauxetic dysplasia mutations showed only a minor increase in cell count, suggesting that each of these mutations leads to a nearly complete disruption of the wild-type function. As the cell count in the CHH founder mutation showed an intermediate reduction, the milder clinical phenotype of this mutation correlates with a minor effect on cell growth in vitro.
As expected, the wild-type transfected cells showed a shift to higher proportions of cleaved 5.8S rRNA, whereas all cells transfected with mutant constructs demonstrated diminished endonucleolytic cleavage at the ITS-1/5.8S rRNA junction site (fig. 7B). This effect was most pronounced in the anauxetic dysplasia mutations.
In addition to its role in rRNA processing, yeast nme1 is involved in cell cycle regulation through degradation of cyclin B mRNA (Gill et al. 2004). Mutant yeast cells show increased cyclin B2 mRNA levels and accumulate in late mitosis (Cai et al. 2002). Cyclin synthesis is essential for entry into mitosis (G2/M transition), and its degradation marks M-phase exit. Whereas cyclin A2 encoded by CCNA2 promotes both G1/S and G2/M transitions in the mammalian cell cycle (Pagano et al. 1992), cyclin B2 encoded by CCNB2 is involved in G2/M transition (Liu et al. 1999). To investigate a similar function in human RNase MRP, we analyzed CCNA2 and CCNB2 mRNA levels in fibroblasts transfected with the RMRP mutants (fig. 7C and 7D). Except for cells with the naturally nonexpressed +14G→A mutation, all cells overexpressing RMRP mutants showed significantly decreased cyclin A2 levels, indicating a prolonged cell cycle and correlating with the reduction in the growth rate and the clinical phenotype. In contrast to the anauxetic dysplasia mutations, the CHH founder mutation +70A→G had significantly increased cyclin B2 mRNA levels, indicating a mitotic delay due to decreased mRNA degradation, as seen in yeast mutants. Since cyclin B2 overexpression from different mechanisms contributes through alterations of the spindle checkpoint to the chromosomal instability observed in some cancers (Cai et al. 1999; Sarafan-Vasseur et al. 2002), our findings could also explain why only CHH, and not anauxetic dysplasia, is associated with proliferative bone marrow dysfunction and susceptibility to cancer. Accordingly, mitomycin C sensitivity testing in fibroblasts of the patient in family 2 revealed no evidence for chromosomal instability in anauxetic dysplasia. Anauxetic dysplasia mutations may, therefore, genetically separate the different functions of RNase MRP, processing mRNA normally while poorly processing rRNA (fig. 9).
Figure 9.
Simplified overview of the cell cycle–related pathways impaired in anauxetic dysplasia and CHH. Right, The hereby affected RMRP gene encodes the untranslated RNA subunit of the ribonucleoprotein endoribonuclease, RNase MRP, which is essential for cell growth and division in yeast and, as our data suggest, also in humans. One function of the RNase MRP complex is the processing of the precursor of 5.8S rRNA, which is a subunit of the 60S ribosomal particle. Therefore, severe and moderate disruption (red flashes) of RNase MRP function in anauxetic dysplasia and CHH, respectively, impacts late-60S ribosomal assembly, resulting in a reduced capacity to synthesize proteins. As a secondary effect, cyclin A2, which promotes G1/S and G2/M phase transitions, is diminished, correlating with the magnitude of delay in the cell cycle. Left, The second function of RNase MRP complex in yeast and, apparently, in humans is the degradation of cyclin B2 mRNA, which is important for the exit of mitosis. In contrast to anauxetic dyplasia, the latter pathway is also impacted in CHH, as shown by increased cyclin B2 mRNA levels. Since cyclin B2 overexpression from different mechanisms contributes through alterations of the spindle checkpoint to the chromosomal instability observed in some cancers, our findings could also explain why only CHH, and not anauxetic dysplasia, is associated with proliferative bone marrow dysfunction and susceptibility to cancer.
We further assume that other phenotypes, including idiopathic short stature, unclassified skeletal dysplasias with spondylo-, meta-, and epiphyseal involvement, and cancer predisposition, might be caused by distinct mutations in the RMRP gene. With reference to other, so far etiologically unexplained types of profound short stature or dwarfism, we believe that the underlying defects are also likely to affect the basic pathways of cell growth or division.
In conclusion, we show that anauxetic dysplasia is caused by RMRP mutations and is allelic to CHH and MDWH. We explain clinical differences by the magnitude and type of alteration in ribosomal assembly and cyclin-dependent cell cycle regulation. Whereas the CHH founder mutation affects both pathways intermediately, anauxetic dysplasia mutations do not affect B-cyclin mRNA levels but severely incapacitate ribosomal assembly via defective endonucleolytic cleavage.
Acknowledgments
We thank the families for their kind cooperation. This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) network grant “SKELNET” GFGM01141901 (to A.Ra., A.Re., and B.Z.) and by the National Institutes of Health grant GM063798 (to M.E.S.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- ClustalW, http://www.ebi.ac.uk/clustalw/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for RMRP [accession number M29916], human ribosomal DNA complete repeating unit [accession number U13369], CCNB2 [accession number AY864066], and CCNA2 [accession number NM_001237]
- GeneSeeker, http://www.cmbi.kun.nl/GeneSeeker/
- UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway/
- National Center for Biotechnology Information Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Bonafe L, Schmitt K, Eich G, Giedion A, Superti-Furga A (2002) RMRP gene sequence analysis confirms a cartilage-hair hypoplasia variant with only skeletal manifestations and reveals a high density of single-nucleotide polymorphisms. Clin Genet 61:146–151 10.1034/j.1399-0004.2002.610210.x [DOI] [PubMed] [Google Scholar]
- Cai T, Aulds J, Gill T, Cerio M, Schmitt ME (2002) The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics 161:1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Reilly TR, Cerio M, Schmitt ME (1999) Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol Cell Biol 19:7857–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 10.1038/380152a0 [DOI] [PubMed] [Google Scholar]
- Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME (2004) RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol 24:945–953 10.1128/MCB.24.3.945-953.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MN, Raff M, Thomas G (2004) Cell growth: control of cell size. Cold Spring Harbor Laboratory Press, Woodbury, New York [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. Embo J 13:2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus HA, Pardi A (1991) Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253:191–194 [DOI] [PubMed] [Google Scholar]
- Horn D, Krebsova A, Kunze J, Reis A (2000) Homozygosity mapping in a family with microcephaly, mental retardation, and short stature to a Cohen syndrome region on 8q21.3-8q22.1: redefining a clinical entity. Am J Med Genet 92:285–292 [DOI] [PubMed] [Google Scholar]
- Horn D, Rupprecht E, Kunze J, Spranger J (2001) Anauxetic dysplasia, a spondylometaepiphyseal dysplasia with extreme dwarfism. J Med Genet 38:262–265 10.1136/jmg.38.4.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Archer RH, Zengel JM (1992) A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res 20:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Wei S, Burnette PK, Gamero AM, Hutton M, Djeu JY (1999) Functional association of TGF-beta receptor II with cyclin B. Oncogene 18:269–275 10.1038/sj.onc.1202263 [DOI] [PubMed] [Google Scholar]
- Makitie O, Sulisalo T, de la Chapelle A, Kaitila I (1995) Cartilage-hair hypoplasia. J Med Genet 32:39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger H, Mundlos S, Becker K, Spranger J, Zabel B (1996) An unknown spondylo-meta-epiphyseal dysplasia in sibs with extreme short stature. Am J Med Genet 63:80–83 [DOI] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G (1992) Cyclin A is required at two points in the human cell cycle. EMBO J 11:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJ, van Venrooij WJ (1999) RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA 5:512–524 10.1017/S1355838299982079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpaa M, Jain P, McKusick VA, Francomano CA, Kaitila I (2003a) The major mutation in the RMRP gene causing CHH among the Amish is the same as that found in most Finnish cases. Am J Med Genet C Semin Med Genet 121:81–83 10.1002/ajmg.c.20006 [DOI] [PubMed] [Google Scholar]
- Ridanpaa M, Sistonen P, Rockas S, Rimoin DL, Makitie O, Kaitila I (2002) Worldwide mutation spectrum in cartilage-hair hypoplasia: ancient founder origin of the major70A→G mutation of the untranslated RMRP. Eur J Hum Genet 10:439–447 10.1038/sj.ejhg.5200824 [DOI] [PubMed] [Google Scholar]
- Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, Makitie O, Kaitila I, de la Chapelle A (2001) Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104:195–203 10.1016/S0092-8674(01)00205-7 [DOI] [PubMed] [Google Scholar]
- Ridanpaa M, Ward LM, Rockas S, Sarkioja M, Makela H, Susic M, Glorieux FH, Cole WG, Makitie O (2003b) Genetic changes in the RNA components of RNase MRP and RNase P in Schmid metaphyseal chondrodysplasia. J Med Genet 40:741–746 10.1136/jmg.40.10.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld RG, Hwa V (2004) Toward a molecular basis for idiopathic short stature. J Clin Endocrinol Metab 89:1066–1067 10.1210/jc.2004-0092 [DOI] [PubMed] [Google Scholar]
- Sarafan-Vasseur N, Lamy A, Bourguignon J, Pessot FL, Hieter P, Sesboue R, Bastard C, Frebourg T, Flaman JM (2002) Overexpression of B-type cyclins alters chromosomal segregation. Oncogene 21:2051–2057 10.1038/sj.onc.1205257 [DOI] [PubMed] [Google Scholar]
- Schärpf M, Sticht H, Schweimer K, Boehm M, Hoffmann S, Rosch P (2000) Antitermination in bacteriophage lambda: the structure of the N36 peptide-boxB RNA complex. Eur J Biochem 267:2397–2408 10.1046/j.1432-1327.2000.01251.x [DOI] [PubMed] [Google Scholar]
- Schmitt ME (1999) Molecular modeling of the three-dimensional architecture of the RNA component of yeast RNase MRP. J Mol Biol 292:827–836 10.1006/jmbi.1999.3116 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Bennett JL, Dairaghi DJ, Clayton DA (1993) Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J 7:208–213 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA (1993) Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13:7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620 10.1101/gad.1018902 [DOI] [PubMed] [Google Scholar]
- Shadel GS, Buckenmeyer GA, Clayton DA, Schmitt ME (2000) Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene 245:175–184 10.1016/S0378-1119(00)00013-5 [DOI] [PubMed] [Google Scholar]
- Superti-Furga A, Bonafe L, Rimoin DL (2001) Molecular-pathogenetic classification of genetic disorders of the skeleton. Am J Med Genet 106:282–293 10.1002/ajmg.10233 [DOI] [PubMed] [Google Scholar]
- Thiel CT, Kraus C, Rauch A, Ekici AB, Rautenstrauss B, Reis A (2003) A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2–12 duplications and deletions leading to HMSN/HNPP. Eur J Hum Genet 11:170–178 10.1038/sj.ejhg.5200920 [DOI] [PubMed] [Google Scholar]
- Topper JN, Clayton DA (1990) Secondary structure of the RNA component of a nuclear/mitochondrial ribonucleoprotein. J Biol Chem 265:13254–13262 [PubMed] [Google Scholar]
- van Driel MA, Cuelenaere K, Kemmeren PP, Leunissen JA, Brunner HG (2003) A new web-based data mining tool for the identification of candidate genes for human genetic disorders. Eur J Hum Genet 11:57–63 10.1038/sj.ejhg.5200918 [DOI] [PubMed] [Google Scholar]
- Welting TJ, van Venrooij WJ, Pruijn GJ (2004) Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res 32:2138–2146 10.1093/nar/gkh539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sohn JH, Warner JR (2003) Autoregulation in the biosynthesis of ribosomes. Mol Cell Biol 23:699–707 10.1128/MCB.23.2.699-707.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]