Abstract
Polymorphisms of the vitamin D receptor gene (VDR) have been shown to be associated with several complex diseases, including osteoporosis, but the mechanisms are unknown and study results have been inconsistent. We therefore determined sequence variation across the major relevant parts of VDR, including construction of linkage disequilibrium blocks and identification of haplotype alleles. We analyzed 15 haplotype-tagging SNPs in relation to 937 clinical fractures recorded in 6,148 elderly whites over a follow-up period of 7.4 years. Haplotype alleles of the 5′ 1a/1e, 1b promoter region and of the 3′ untranslated region (UTR) were strongly associated with increased fracture risk. For the 16% of subjects who had risk genotypes at both regions, their risk increased 48% for clinical fractures (P = .0002), independent of age, sex, height, weight, and bone mineral density. The population-attributable risk varied between 1% and 12% for each block and was 4% for the combined VDR risk genotypes. Functional analysis of the variants demonstrated 53% lower expression of a reporter construct with the 1e/1a promoter risk haplotype (P = 5 × 10−7) in two cell lines and 15% lower mRNA level of VDR expression constructs carrying 3′-UTR risk haplotype 1 in five cell lines (P = 2 × 10−6). In a further analysis, we showed 30% increased mRNA decay in an osteoblast cell line for the construct carrying the 3′-UTR risk haplotype (P = .02). This comprehensive candidate-gene analysis demonstrates that the risk allele of multiple VDR polymorphisms results in lower VDR mRNA levels. This could impact the vitamin D signaling efficiency and might contribute to the increased fracture risk we observed for these risk haplotype alleles.
Introduction
The vitamin D receptor (VDR [MIM 601769]) (12q13) is a steroid receptor acting as a transcription factor responding to the biologically active form of the secosteroid vitamin D hormone. The vitamin D endocrine system is pleiotropic and plays an important role in skeletal metabolism, including intestinal calcium absorption and regulation of osteoblast differentiation, but it has also been shown to modulate immune response, insulin secretion, the renin/angiotension system, and growth of cancer cells (Haussler et al. 1998).
Rare, deleterious mutations in the VDR gene cause the well-known 1,25-dihydroxyvitamin D–resistant rickets (rickets type II), a rare monogenetic disease characterized by osteomalacia, alopecia, and increased 1,25-(OH)2D3 levels. Some isolated, more common VDR SNPs have previously been associated with several complex diseases and traits, such as osteoporosis (Morrison et al. 1994; Young et al. 1996; Uitterlinden et al. 2001; Fang et al. 2003). However, the relationship between disease and haplotype alleles across VDR has not been systematically analyzed, and their effects on VDR function are poorly understood.
The interpretation of polymorphic variations in VDR is severely hindered by the fact that, until now, only a few polymorphisms in this large gene have been studied and that most of these are anonymous (nonfunctional) polymorphisms. To explain the associations observed with complex diseases, they should be in linkage disequilibrium (LD) with truly functional polymorphisms. Haplotype-based methods offer a powerful approach to studying the association of genetic variation with complex disease (Gabriel et al. 2002). Following a recent study of VDR polymorphisms (Nejentsev et al. 2004), we here present a detailed description of the genomic organization of the VDR gene region, the identification of 62 polymorphisms across relevant areas of the gene, and an analysis of LD and haplotype diversity of VDR variations in different ethnic groups. We use this information in a large-scale association analysis of haplotype-tagging SNPs (htSNPs) in relation to osteoporosis in a group of 6,148 white elderly men and women. We analyzed functionality of relevant polymorphisms in the promoter and 3′ UTR to help understand the underlying mechanism of the association we observed.
Material and Methods
Subjects
We sequenced genomic DNA (see supplementary material and methods in online-only appendix A) from 15 young white individuals, including five homozygotes for each major VDR 3′-UTR haplotype—that is, 11, 22, and 33, as defined in a previous study (Uitterlinden et al. 1996) on the basis of BsmI, ApaI, and TaqI RFLPs. We studied LD in 234 random white blood-bank donor samples and in DNA from 107 Asian and 58 African individuals (from the Coriell Institute: 90 Chinese Han [group HD100], 9 Chinese [HD02], 8 Japanese [HD07], 47 African Americans [HD04 and HD50], and 9 Africans from south of the Sahara [HD12]). The association of VDR genotype with fracture risk was analyzed in the Rotterdam Study with 7,983 subjects (Hofman et al. 1991); DNA was collected from 6,580 of them, and 6,148 DNA samples were available for which genotyping of all SNPs was successful. Genomic DNA was isolated from blood in accordance with standard procedures.
Genotyping
We genotyped 47 SNPs (table 1) in three ethnic groups and 14 htSNPs and FokI, BsmI, ApaI, and TaqI RFLPs in 6,148 white subjects from the Rotterdam Study, with the use of high-throughput TaqMan allelic discrimination assays. The assay mixes (including unlabeled PCR primers, FAM and VIC dye-labeled TaqMan MGB probes) of three Assays-on-Demand and 36 Assays-by-Design were designed and provided by Applied Biosystems (ABI). The reaction system contained 1–5 ng of dried genomic DNA, 2.5 μl of TaqMan Universal PCR Master Mix, 2 × No AmpErase UNG, 0.125 μl 40 × Assay Mix or 0.0625 μl 80× Assay Mix and was adjusted with Milli-Q H2O for a total volume of 5 μl. The genotyping results were analyzed using independent end-point readings by two operators of the ABI Prism 7900HT, and a random 5% of samples were independently repeated to confirm genotyping results. The disagreement rate varied from 0.3% to 1.2% for five htSNPs, whereas the genotype results for all other htSNPs were completely consistent.
Table 1.
Allele Frequencies of 47 SNPs in Different Ethnic Groups
|
MAF (%) in |
|||||
| Region and SNP Codea | SNP ID b | Minor Allelec | Whites (468 Chromosomes) | Asians (214 Chromosomes) | Africans (116 Chromosomes) |
| Intergenic region of COL2A1 and VDR (5 SNPs): | |||||
| A | hCV2615323 (G/A) | A | 10 | 38 | 4 |
| B | hCV8724908 (G/C) | C | 44 | 15 | 27 |
| C | hCV2626648 (G/A) | A | 11 | 33 | 4 |
| D | hCV8724903 (C/G) | G | 7 | 1 | 5 |
| E | hCV8724902 (C/T) | T | 12 | 34 | 28 |
| Promoter (24 SNPs): | |||||
| 1 | 1f-G−1904A | A | 46 | 5 | 10 |
| 3 | 1f-C−1570T | T | 27 | 78 | 66d |
| 5 | 1f-G−1344A | A | 46d | 5d | 10d |
| 6 | 1f-T−1198G | G | 28d | 17d | 12d |
| 7 | 1f-G−777A | A | 13d | 29d | 23d |
| F | 1e-T−3743C | C | 17 | 48 | 76 |
| 10 | 1e-C−2090T | T | 43 | 3 | 6d |
| 12 | 1e-G−1739A (Cdx-2) | A | 17d | 49d | 75d |
| 15 | 1e-C−577A | A | 42 | 3 | 10d |
| 17 | 1a-G−1521C | C | 43 | 3 | 6 |
| 18 | 1a-A−1012G (GATA) | G | 43d | 3 | 6d |
| 22 | 1b-T−2746C | C | 34 | 3 | 13d |
| 23 | 1b-G−2528A | A | 28 | 66 | 75d |
| 24 | 1b-C−2481A | A | 10d | .0 | 3 |
| 25 | 1b-A−2225G | G | 27 | 64 | 74 |
| 26 | 1b-T−1748A | A | 38 | 31 | 10 |
| 28 | 1b-G−886A | A | 25d | 44 | 9d |
| 29 | 1b-C−673T | T | 10d | 0 | 2 |
| 30 | 1b-T−391C | C | 39 | 31d | 10d |
| 31 | 1b-C25A (Thr/Lys) | A | 32d | 3 | 12d |
| 33 | 1c-T−1930C | C | 40 | 34 | 14 |
| 34 | 1c-G−1633C | C | 28d | 20d | 30d |
| 35 | 1c-C−1453T | T | 41d | 33d | 51d |
| 36 | 1c-G−1156A | A | 29 | 17 | 11d |
| Coding (8 SNPs): | |||||
| 38 | E2-C4T (FokI) | T | 34d | 51d | 21d |
| 41 | E4-A−62G | G | 42 | 75 | 35d |
| 44 | E7-D+75G | G | 2 | 1 | 22d |
| # | E8-G+284A (BsmI) | A | 42 | 6 | 36d |
| 46 | E9-G-111C | C | 13 | 17 | 10 |
| 47 | E9-G-94A | A | 2 | 1 | 14d |
| 48 | E9-T−48G (ApaI) | G | 44 | 76 | 26 |
| 49 | E9-T32C (TaqI) | C | 43 | 8 | 31d |
| UTR (7 SNPs): | |||||
| 50 | U-A311C | C | 45d | 76d | 31d |
| 51 | U-C440G | G | 2 | .5 | 1 |
| 52 | U-G464T | T | 13d | 17d | 10d |
| 53 | U-D796T | T | 44 | 24 | 59d |
| 57 | U-A1909C | C | 44 | 75 | 29d |
| 61 | U-G2795A | A | 2 | 1 | 16d |
| 62 | U-A2978T | T | 2d | .5 | 1 |
| Intergenic region of VDR and HDAC7A (3 SNPs): | |||||
| G | hCV3290614 (A/T) | T | 35 | 75 | 23 |
| H | hCV3290610 (T/C) | C | 46 | 64 | 42 |
| I | hCV16253844 (T/C) | T | 46 | 36 | 33 |
The SNP codes from A to I indicate SNPs that are selected from the Celera database. SNP A is a missense mutation in exon 53 of COL2A1; # indicates the BsmI RFLP, E8-G+284A (Morrison et al. 1992). The code numbers correspond to those in table 3.
The SNP IDs are derived from the Celera database and the resequencing results given in table 4.
Minor alleles were determined on the basis of frequency in the white population.
VDR tagSNPs for association study of different ethnic groups.
Sequence Analysis, LD, and Haplotype Analyses
Human and mouse VDR genomic sequences (>105 kb from the Celera database) were analyzed using the Vista program (Vista Tools Web site) to visualize the pairwise percentage identity as calculated for every 100 bp. We used the TFSEARCH program (TFSEARCH: Searching Transcription Factor Binding Sites Web site) from the TRANSFAC databases developed by GBF-Braunschweig and the MetInspector from Genomatix software GmbH to search for potential transcription-factor binding sites (TFBSs) around promoter polymorphisms.
We first analyzed LD among all 62 polymorphisms determined on the basis of 15 sequenced white samples, to select 37 SNPs; we also included an additional 9 SNPs (A to I in fig. 1A and table 1) that flank VDR from the Celera database and E8-G+284A (a BsmI RFLP) (Morrison et al. 1992). (For polymorphism nomenclature, the first element of the polymorphism ID is the designation of exon [coding exons begin with capital “E”] or 3′-UTR [“U”]; in most IDs, this is followed by a hyphen and the major nucleotide allele [for whites]; then, the nucleotide location is designated as plus [+] or minus [−], relative to the first or last base of the nearest exon, or is shown without a sign if it is in the exon; and the last element of the polymorphism ID is the minor nucleotide allele [for whites].) Selection criteria for VDR SNPs in the LD analysis is based on whether the SNP (1) has a minor-allele frequency (MAF) >10% (3/30 alleles), as determined by our sequence analysis; (2) is in a potential promoter TFBS or destabilizing element in the 3′ UTR; (3) is in a highly conserved region; or (4) is a tagging SNP (tagSNP) (on the basis of data from the 15 sequenced subjects).
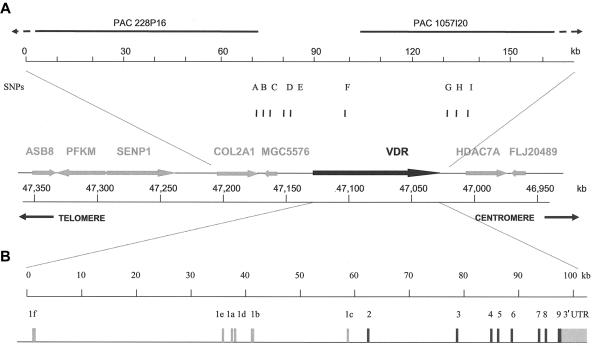
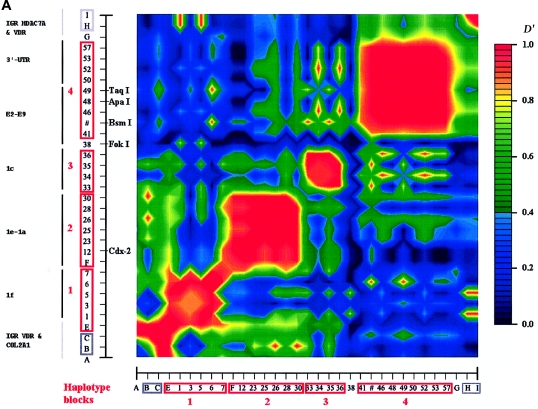
Figure 1.
Genomic structure and LD map of the human VDR gene. a, Physical organization of the 12q12 area containing VDR, mostly based on the Celera database (47032–47145 kb at chromosome 12q12). The arrows for each gene indicate the transcription direction, and distances are in kb. b, Genomic structure of the human VDR gene. Black bars indicate the coding exons of VDR, and the gray bars indicate 5′ exons and the 3′ UTR. c, Sequenced areas and positions of the 62 variations. Gray bars in the 3′-UTR indicate destabilizing elements (DE 1, 2, and 3 [Durrin et al. 1999]). The sequence-variation numbers refer to those given in table 3. d, Haplotype map of VDR in whites, Asians, and African Americans, based on SNPs with an MAF ⩾5% in each of the different ethnic populations. Common haplotype alleles in each block with a frequency >3% are presented below the blocks. SNPs and alleles in red indicate htSNPs. Fracture-risk haplotype alleles are underlined. The correspondence is shown for whites to haplotypes in block 5 of the BsmI-ApaI-TaqI haplotype alleles we defined elsewhere (Uitterlinden et al. 1996).
We then genotyped 47 SNPs in whites, Han Chinese, and African Americans, to calculate allele frequencies (table 1). We determined the race-specific SNPs, whose MAF is >3% in any of the ethnic study populations, and identified 42 SNPs for whites, 33 for Asians, and 41 for African Americans. We constructed haplotype structure by use of the PHASE program and then used the PHASE outcome to calculate the pairwise standardized disequilibrium coefficient (D′) with Haploxt, to estimate the linkage magnitude between two SNPs, and depicted the graphic overviews of LD by use of the GOLD program (GOLD Web site). We identified haplotype blocks and calculated haplotype frequencies in each block by use of the HaploBlockFinder program (HaploBlockFinder Web site). Selection of the htSNPs in each ethnic group was based on the condition that (1) the minimal combination of htSNPs in each haplotype block represents ⩾95% of the haplotypes; (2) there is potential functionality in the 3′ UTR or promoter; or (3) the SNP is unlinked to any block—for example, E2-C4T (the FokI RFLP).
Epidemiological Analysis
Genotype distribution was tested for Hardy-Weinberg equilibrium. Anthropometric measurements, BMI (weight in kg/height in m2), bone mineral density (BMD) (g/cm2; determined by dual energy x-ray absorptiometry [Lunar DPX-L densitometer]) at the femoral neck and lumbar spine (L2–L4), and other variables were measured as described elsewhere (Burger et al. 1994; Schuit et al. 2004). BMD measurements were available for 5,027 subjects (82% of the cohort with genotypes). Sex-specific T scores were calculated from the femoral neck BMD by use of the NHANES reference population (Looker et al. 1998) of white males and females aged 20–29 years. For assessment of the incidence of fracture, follow-up ended before January 1, 2002 (mean follow-up period 7.4±3.3 years). We defined “clinical fracture” to include all fractures confirmed by general practitioners or at hospitals, as described elsewhere (Fang et al. 2003; Schuit et al. 2004), and we excluded head, foot, hand, pathological, postprocedural, skull, and face fractures. The presence of vertebral fracture was analyzed as described elsewhere (McCloskey et al. 1993; Van der Klift et al. 2002).
We applied the Pearson χ2 test to estimate differences in fracture frequency by genotype. We calculated the relative risk (RR) and 95% CI by logistic regression models and calculated hazard ratio (HR) for incidence of clinical, wrist, and hip fractures by Cox regression model. Both the logistic and the Cox regression models were adjusted for potential confounders, such as age, sex, height, weight, BMD, and bone loss. The population-attributable risk (PAR) was calculated for genetic and other markers for clinical fracture risk. All statistical analyses were done using SPSS (version 11.0). The PAR was calculated as Pe×(HR-1)/[Pe×(HR-1)+1], where Pe is the proportion of the study population that is exposed to the risk factor for fracture and where the HR is for the risk factor.
Functional Experiments
Details on the electrophoretic gel mobility shift assay (EMSA) and the transactivation assays for promoter polymorphisms as well as the mRNA levels and stability measurements for the 3′-UTR polymorphisms are provided in online-only appendix A. The oligonucleotides used for EMSA are provided in table 2.
Table 2.
Oligonucleotides for EMSA
| Site and Oligonucleotide Name | Sequence |
| Cdx-2: | |
| 1e-2090-C sense | 5′-AAGTACTGGGATTACAGGCCTGAGCCACT-3′ |
| 1e-2090-C antisense | 5′-AGTGGCTCAGGCCTGTAATCCCAGTACTT-3′ |
| 1e-2090-T sense | 5′-AAGTACTGGGATTATAGGCCTGAGCCACT-3′ |
| 1e-2090-T antisense | 5′-AGTGGCTCAGGCCTATAATCCCAGTACTT-3′ |
| 1e-1739-G sense | 5′-TAAACTAGGTCACAGTAAAAACTTATTTC-3′ |
| 1e-1739-G antisense | 5′-GAAATAAGTTTTTACTGTGACCTAGTTTA-3′ |
| 1e-1739-A sense | 5′-TAAACTAGGTCACAATAAAAACTTATTTC-3′ |
| 1e-1739-A antisense | 5′-GAAATAAGTTTTTATTGTGACCTAGTTTA-3′ |
| SIF sense | 5′-GAGGGTGCAATAAAACTTTATGAGTAGGT-3′ |
| SIF antisense | 5′-ACCTACTCATAAAGTTTTATTGCACCCTC-3′ |
| GATA: | |
| 1a-1012-A sense | 5′-AGGCGAATAGCAATATCTTCCCTGGCTAA-3′ |
| 1a-1012-A antisense | 5′-TTAGCCAGGGAAGATATTGCTATTCGCCT-3′ |
| 1a-1012-G sense | 5′-AGGCGAATAGCAATGTCTTCCCTGGCTAA-3′ |
| 1a-1012-G antisense | 5′-TTAGCCAGGGAAGACATTGCTATTCGCCT-3′ |
| 1a-1521-G sense | 5′-GCTAGCTTTCCCACGATGCTTTGGGCAAG-3′ |
| 1a-1521-G antisense | 5′-CTTGCCCAAAGCATCGTGGGAAAGCTAGC-3′ |
| 1a-1521-C sense | 5′-GCTAGCTTTCCCACCATGCTTTGGGCAAG-3′ |
| 1°-1521-C antisense | 5′-CTTGCCCAAAGCATGGTGGGAAAGCTAGC-3′ |
Results
Genomic Structure of VDR and Homology Analysis
The physical organization of the human VDR gene region on chromosome 12q12-14 (kbp 46950–47350) (fig. 1a) and the VDR gene structure (fig. 1b) is based on PAC clones (PAC228P16 and PAC1057I20), data from two published articles (Miyamoto et al. 1997; Crofts et al. 1998), the Celera database, and our resequencing efforts. VDR encompasses at least 105 kb; the large 5′ region of noncoding exons 1f–1c is 60 kb, with exon 1f located 35 kb upstream of exon 1e, whereas exon 1e is 2 kb upstream of exon 1a. The nearest genes are COL2A1, at a distance of 20 kb upstream (with a small gene-like structure, MGC5576, close to COL2A1), and HDAC7A, at a distance of 10 kb downstream.
The overall percentage of identity between the entire human and mouse VDR genomic sequences is 28.8% (fig. 2). Although coding exons were found in a highly conserved region (86.5%–92.6% identity), except for exon 5 (61.2% identity), we also observed regions of high homology in intronic areas—in total, 14 kb of such regions between exons 1f and 1e, 5 kb of regions between exons 1b and 1c, and a small 500-bp region between exons 2 and 3. Interestingly, there is a lack of homology of human exons 1b, 1e, and 1f with the mouse gene, whereas exons 1a, 1d, and 1c are well conserved. Overall, much of the lack of homology can be explained by the presence of Alu repeats in the human VDR gene. We detected 62 Alu-like sequences, mostly located in introns and between exons 1f and 1e, although one is located in the 3′ UTR (data not shown).
Figure 2.
Conservation of the human and mouse genomic VDR gene sequence. The Y-axis is the homology rate between human and mouse; the X-axis is the physical distance on the human VDR gene. All exons are indicated in purple, the 3′ UTR in light green, and the conserved noncoding region in red. The small black bars on the top of each frame indicate the polymorphisms we observed by resequencing, and the gray arrow on top indicates the transcription direction of VDR.
Resequencing of the VDR Gene
We found 62 polymorphisms (fig. 1c and table 3), including 57 SNPs and 5 VNTR polymorphisms, of which 18 polymorphisms (including 15 SNPs) were not present in the Celera database (chromosome 12, position 47033000–47139000 in the National Center for Biotechnology Information [NCBI] Human Genome [build 35.1, April 2005 freeze]), the NCBI dbSNP (build 123), the Human Gene Mutation Database (HGMD), the HGBASE database, and/or other publications. Of 62 polymorphisms, 9 are located in highly conserved noncoding regions (identity rate >70%) in the 3′ UTR and the promoter region. The average frequency of variation across 22 kb was 1/355 bp, with a range from 1/247 bp (13/3.2 kb) in the 3′ UTR, to 1/315 (13/4.1 kb) in exons 2–9 and surrounding introns, to 1/408 (36/14.7 kb) across the promoter region. Fifty SNPs with an MAF ⩾5% in the 15 subjects were identified as “informative SNPs.”
Table 3.
Sequence Variations of VDR Determined by Sequence Analysis of 30 White Chromosomes
| Location and Polymorphism Nomenclaturea | Variation Number | Minor Allele [MAF %] | Polymorphism and Flanking Sequenceb | Positionc | Characteristic(s)d | Reference(s)c | dbSNP ID(Build 124) |
| Promoter region: | |||||||
| Exon 1f: | |||||||
| 1f-G−1904A | 1 | A [37] | GTGTGTGTGCG/ACCTGTGTGGG | 47136401 | … | Celera (hCV26069889) | ss32465633 |
| 1f-1889 (CATGTGTGTG)2/1 | 2 | (CATGTGTGTG)1 [20] | TGGGTGTG(CATGTGTGTG)2/1TGGATGTGG | 47136219 | USF (−) | Novel | ss32465817 |
| 1f-C−1570T | 3 | T [43] | CATATTTATCC/TTTTTATAC/TTTCTTTCAAG | 47136067 | Evi-1 (−) | Celera (hCV2880884) | ss32465634 |
| 1f-C−1562T | 4 | T [30] | CATATTTATCC/TTTTTATAC/TTTCTTTCAAG | 47136059 | … | Celera (hCV26069888) | ss32465635 |
| 1f-G−1344A | 5 | A [37] | TTAAACAGATG/ATATCATCATC | 47135841 | GATA (+) | Celera (hCV2880883) | ss32465636 |
| 1f-T−1198G | 6 | G [20] | CAGGCTCAGCT/GGCCCTTTGAC | 47135695 | AP4 (+) | dbSNP (ss5759311 and rs4237856); Celera (hCV27909097) | ss32465637 |
| 1f-G−777A | 7 | A [13] | CCAGGTAGGCG/ATGGTGCCCCC | 47135107 | … | ss20420964e | ss32465638 |
| 1f-G−465C | 8 | C [37] | TCCTCCCTGGG/CCAAGCCATCT | 47134962 | … | dbSNP (ss5452989 and rs4073726); Celera (hCV27537612); ss20420962e | ss32465639 |
| 1f-C−217T | 9 | T [30] | CCCCCCTGCTC/TCCTCATGGCC | 47134714 | … | dbSNP (ss6198081 and rs4073729); Celera (hCV27909106); ss20420961e | ss32465640 |
| Exon 1e: | |||||||
| 1e-C−2090T | 10 | T [23] | ACTGGGATTAC/TAGGCCTGAGC | 47100603 | Cdx-A (+) | Novel | ss32465641 |
| 1e-A−2054G | 11 | G [7] | GATTTTTAATA/GCTGTATTTTG | 47100567 | … | Novel | ss32465642 |
| 1e-G−1739A | 12 | A [37] | CTAGGTCACAG/ATAAAAACTTA | 47100296 | Cdx-A (−) | Cdx-2 (Yamamoto et al. 1999); Celera (hCV2880808); ss20420955e | ss32465643 |
| 1e-A−1553G | 13 | G [3] | AGGAAATATAA/GATGAATTAAT | 47100066 | Cdx-A (−) | Novel | ss32465644 |
| 1e-C−1092T | 14 | T [23] | TGAGGACAGGC/TTGCAGTCCTG | 47099649 | … | Celera (hCV2880807) | ss32465645 |
| 1e-C−577A | 15 | A [23] | CTCTGTAGTGC/AAGGAATGGTG | 47099134 | … | Celera (hCV2880806); ss20421064e | ss32465646 |
| 1e-19 (T)9–11 | 16 | … | TGCATGGAAA(T)9–11GTTTTTTAGA | 47098532 | … | Novel | ss32465647 |
| 1e-19 (T)9 | … | (T)9 [20] | … | … | … | … | … |
| 1e-19 (T)10 | … | (T)10 [50] | … | … | … | … | … |
| 1e-19 (T)11 | … | (T)11 [30] | … | … | … | … | … |
| Exon 1a: | |||||||
| 1a-G−1521C | 17 | C [23] | GCTTTCCCACG/CATGCTTTGGG | 47098041 | GATA-1 (+) | ss20420954e | ss32465648 |
| 1a-A−1012G | 18 | G [23] | GAATAGCAATA/GTCTTCCCTGG | 47097576 | GATA-1, -2, -3 (−) | Celera (hCV2880805); ss20420953e | ss32465649 |
| Exon 1d: | |||||||
| 1d+77 (T)23–28 | 19 | … | TTTTAAGTAA(T)23–28ACTTTTCTTT | 47095974 | … | Novel | ss32465818 |
| 1d+77 (T)23 | … | (T)23 [13] | … | … | … | … | … |
| 1d+77 (T)24 | … | (T)24 [40] | … | … | … | … | … |
| 1d+77 (T)25 | … | (T)25 [33] | … | … | … | … | … |
| 1d+77 (T)26 | … | (T)26 [7] | … | … | … | … | … |
| 1d+77 (T)27 | … | (T)27 [3] | … | … | … | … | … |
| 1d+77 (T)28 | … | (T)28 [3] | … | … | … | … | … |
| 1d+425 (TCC)3/2 | 20 | (TCC)2 [13] | TTCTCACAGT(TCC)3/2T(TCC)7/8TCTTGTTCCT | 47095624 | … | Novel | ss32465819 |
| 1d+435 (TCC)7/8 | 21 | (TCC)8 [47] | TTCTCACAGT(TCC)3/2T(TCC)7/8TCTTGTTCCT | 47095612 | … | Novel | ss32465650 |
| Exon 1b: | |||||||
| 1b-T−2746C | 22 | C [23] | AACTCGCTGTT/CGCCTGTGCAG | 47094187 | … | dbSNP (ss6566285 and rs4760658); Celera (hCV27904554) | ss32465651 |
| 1b-G−2528A | 23 | A [37] | GCCCTCTGAGG/AGATAGCAGGA | 47093969 | … | ss20420951e | ss32465652 |
| 1b-C−2481A | 24 | A [13] | GACTCCAAGAC/AAGTTTGAGCC | 47093922 | … | Novel | ss32465653 |
| 1b-A−2225G | 25 | G [37] | GATGCCATGCA/GTGGTTATACC | 47093666 | … | Celera (hCV2880804); ss20421063e | ss32465654 |
| 1b-T−1748A | 26 | A [40] | TACAGCCTCCT/ACATTTTGCTC | 47093189 | … | Celera (hCV2880803) | ss32465655 |
| 1b-G−1641C | 27 | C [3] | GGCAAACCTCG/CCCCACTCTGA | 47093082 | MZF1 (−) | Novel | ss32465656 |
| 1b-G−886A | 28 | A [37] | CAGCTGCACCG/AGCGGGAAAGC | 47092327 | … | Celera (hCV26014291); ss20421061e | ss32465657 |
| 1b-C−673T | 29 | T [30] | CAGCTACTTAC/TTGAGCACTCA | 47092114 | … | Novel | ss32465658 |
| 1b-T−391C | 30 | C [40] | (i) ATTATACACCT/CGCAGTAAATG; (ii) ATTATACACCT/CGCAGTAAATG; (iii) ATTATACACCT/CGCAGTAAATG | 47091832 | (i) E47 (−); (ii) deltaEF1 (+); (iii) USF (+) | ss20421060e | ss32465659 |
| 1b-C25A (Thr/Lys) | 31 | A [23] | CCCAGCTGGAC/AGGAGAAATGG | 47091409 | … | ss20421059e | ss32465660 |
| Exon 1c: | |||||||
| 1c-C−2040T | 32 | T [23] | CCTGCCATCCC/TTTGGCCTGGG | 47076284 | … | ss20421046e | ss32465661 |
| 1c-T−1930C | 33 | C [27] | AAGGTGTCCAT/CGGCTTAGGGT | 47076174 | … | dbSNP (ss4040170 and rs2853564); Celera (hCV15823889); ss20420934e | ss32465662 |
| 1c-G−1633C | 34 | C [27] | GCCTAGCTGTG/CGGACCCTGGG | 47075877 | … | dbSNP (ss3193006 and rs2238135); Celera (hCV16031778); ss20420933e | ss32465663 |
| 1c-C−1453T | 35 | T [27] | (i) TGGTTGTCTAC/TCTGGATGTCA; (ii) TGGTTGTCTAC/TCTGGATGTCA | 47075558 | (i) GATA-1, -2 (−); (ii) AREB6 (+) | dbSNP (ss3193007 and rs1989969); Celera (hCV12060044); ss20420932e | ss32465664 |
| 1c-G−1156A | 36 | A [27] | CCCAGCTTAGG/ATTATCTTGGC | 47075261 | … | dbSNP (ss3193008 and rs2238136); Celera (hCV3290655); ss20421045e | ss32465665 |
| Coding exons and flanking intron region: | |||||||
| E2-G−51A | 37 | A [3] | ATGCCAGCTGG/ACCCTGGCACT | 47070636 | … | Novel | ss32465666 |
| E2-C4T (codon 1) | 38 | T [40] | CTTACAGGGAC/TGGAGGCAATG | 47070443 | … | dbSNP (ss16359158 and rs2228570); FokI (Saijo et al. 1991); HGMD (CM972826); Celera (hCV12060045); ss20420928e | ss32465667 |
| E2-C59T (codon 19) | 39 | T [7] | TTGACCGGAAC/TGTGCCCCGGA | 47070527 | … | Exon 2, C57T (Brown et al. 2000); ss20420927e | ss32465668 |
| E4-C−71T | 40 | T [27] | ACCTTTACCCC/TCAACCGCAA/GGAGGAA | 47049093 | … | Celera (hCV3290629) | ss32465669 |
| E4-A−62G | 41 | G [33] | TTACCCC/TCAACCGCAA/GGAGGAAGGTT | 47049084 | … | Celera (hCV3290628); ss20421022e | ss32465670 |
| E5-A+118T | 42 | T [7] | AACTTACATAA/TATACTGTGCG | 47048463 | … | ss20420902e | ss32465671 |
| E6-G+154A | 43 | A [33] | GTGCAGTGGCG/ACGATCTCGGC | 47046925 | … | Novel | ss32465672 |
| E7-D+75G | 44 | G [13] | TGGGGTTTGG-/GCTCCAATCAG | 47038024 | … | Intron 7, G insertion (Brown et al. 2000) | ss32465673 |
| E8-C2T (codon 303) | 45 | T [3] | CTCTCACAGCC/TGGACACAGCC | 47037891 | … | ss20420899e | ss32465674 |
| E8-G+284A (BsmI) | # | A [41] | ACAGGCCTGCG/ACATTCCCAAT | 47038290 | … | rs1544410 (Morrison et al. 1992) | - |
| E9-G−111C | 46 | C [33] | TAGAGGGGTGG/CCCTAGGGGGT | 47036557 | … | Novel | ss32465675 |
| E9-G−94A | 47 | A [13] | GGGTGCTGCCG/ATTGAGTGTCT | 47036540 | … | Novel | ss32465676 |
| E9-T−48G (ApaI) | 48 | G [33] | AGCAGTGAGGT/GGCCCAGCTGA | 47036493 | … | dbSNP (ss20420896 and rs7975232); ApaI (Morrison et al. 1992); ss20420896e | ss32465677 |
| E9-T32C (codon 352, TaqI) | 49 | C [33] | CCGCGCTGATT/CGAGGCCATCC | 47036308 | … | dbSNP (ss20420895 and rs731236); TaqI (Morrison et al. 1994); HGBASE (SNP000008184); Celera (hCV2404008); ss20420895e | ss32465678 |
| 3′-UTR: | |||||||
| U-A311C | 50 | C [33] | CCACCGCTGCA/CTAAGTGGCTG | 47035772 | … | dbSNP (ss87853 and rs739837); C1588A (Durrin et al. 1999); Celera (hCV2404007); ss20420894e | ss32465679 |
| U-C440G | 51 | G [17] | TCTCCCTCTCC/GTGCCTACTCA | 47035748 | … | ss20420893e | ss32465680 |
| U-G464T | 52 | T [33] | TAAATAATCGG/TCCCACAGCTC | 47035619 | … | dbSNP (ss87854 and rs3847987); Celera (hCV2404006); ss20420892e | ss32465681 |
| U-D796T | 53 | T [30] | GTCCTCCCCC-/TGCCAGTGCCT | 47035391 | … | T2074del (Durrin et al. 1999); ss20421003e | ss32465682 |
| U-T1229C | 54 | C [3] | AAGTGCATGCT/CCTCTGCAGCC | 47034958 | In destabilizing elements | ss20421001e | ss32465683 |
| U-1238 (CCAGC)3/4 | 55 | (CCAGCC)4 [13] | GCATGCT/CCTCTGCAG(CCAGCC)3/4TGGTGGGAAG | 47034947 | In destabilizing elements | AGCCC2517del (Durrin et al. 1999) | ss32465820 |
| U-G1868A | 56 | A [7] | TTCAGTGGGAG/AAAAACACTTG | 47034316 | … | Novel | ss32465684 |
| U-A1909C | 57 | C [30] | TCCCCTCATTA/CAGGAAAACTG | 47034174 | … | C3185A (Durrin et al. 1999); Celera (hCV8716058); HGBASE (SNP000016363); ss20420891e | ss32465685 |
| U-G1982C | 58 | C [7] | AGGACAGGCCG/CGGCGCGGTGG | 47034101 | … | Celera (hCV3290620); ss20420890e | ss32465686 |
| U-2119 (A)13–24 | 59 | … | TTAAAAATAC(A)13–24TAGCCGGGCA | 47033958 | … | Poly(A) (Ingles et al. 1997); Celera (hCV26075087) | ss32465687 |
| U-2119 (A)13 | … | (A)13 [20] | … | … | … | … | … |
| U-2119 (A)16 | … | (A)16 [17] | … | … | … | … | … |
| U-2119 (A)18 | … | (A)18 [13] | … | … | … | … | … |
| U-2119 (A)19 | … | (A)19 [17] | … | … | … | … | … |
| U-2119 (A)23 | … | (A)23 [27] | … | … | … | … | … |
| U-2119 (A)24 | … | (A)24 [7] | … | … | … | … | … |
| U-T2147A | 60 | A [30] | TGGTGGCGCAT/AGCCTGTAATC | 47034035 | … | A3424T (Durrin et al. 1999); ss20420889e | ss32465688 |
| U-G2795A | 61 | A [13] | GGGAGAAAAGG/ATCATCATCGA | 47033289 | In destabilizing elements | dbSNP (ss4040169 and rs2853563); A4107G (Durrin et al. 1999); Celera (hCV15823846); ss20420888e | ss32465689 |
| U-A2978T | 62 | T [13] | AATCCAAGCGA/TGGTCAACAGA | 47033201 | In destabilizing elements | ss20420887e | ss32465690 |
The nomenclature of the polymorphisms is based on resequencing. The first element of the polymorphism ID is the designation of exon (coding exons begin with a capital “E”) or 3′-UTR (“U”); in most IDs, this is followed by a hyphen and the major nucleotide allele (for whites); then, the nucleotide location is designated as plus (+) or minus (−), relative to the first or last base of the nearest exon, or is shown without a sign (+ or −) if it is in the exon; and the last element of the polymorphism ID is the minor nucleotide allele (for whites).
Bold letters indicate the variation, and underlined nucleotides indicate a potential transcription-factor consensus sequence. For multiallelic polymorphisms, we list all observed alleles.
For Celera, dbSNP, HBMD, and HGBASE, accession numbers are in parentheses. The location and accession numbers of SNPs are based on the April 2005 freeze of the Celera database and NCBI dbSNP build 123.
The potential transcription factors are described: (+) indicates the sense recognition sequence and (−) indicates the antisense recognition sequence.
SNPs reported by Nejentsev et al. 2004.
Three informative polymorphisms were located in the so-called destabilizing elements (described elsewhere [Durrin et al. 1999]) in the 3′ UTR of VDR (fig. 1c). Four informative SNPs were observed in coding exons 2–9: a previously identified C→T substitution (E2-C4T, a FokI RFLP), E2-C59T and E8-C2T, which are synonymous substitutions, and a previously identified synonymous SNP (E9-T32C, detected as a TaqI RFLP). In the promoter-region exons 1a–1f, we found 36 sequence variations, including 5 VNTRs and 31 SNPs. One SNP in exon 1b (1b-C25A) would change the predicted amino acid sequence by replacing a threonine with a lysine (1b-Thr8Lys). TRANSFAC analysis of the 5′ upstream regions of promoter exons 1a–1f indicated that 14 polymorphisms change the core recognition sequence of potential transcription factors (see table 3).
LD and Haplotype Analyses
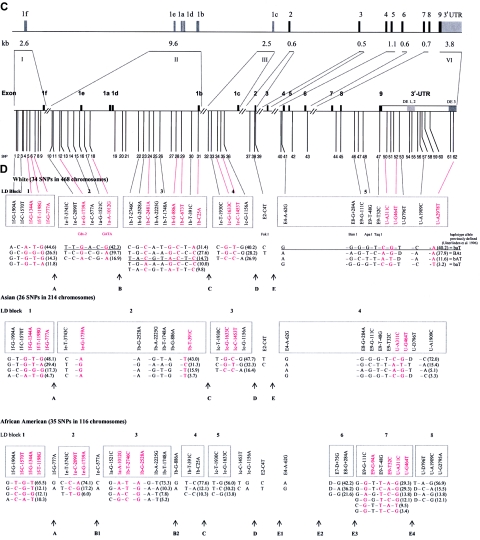
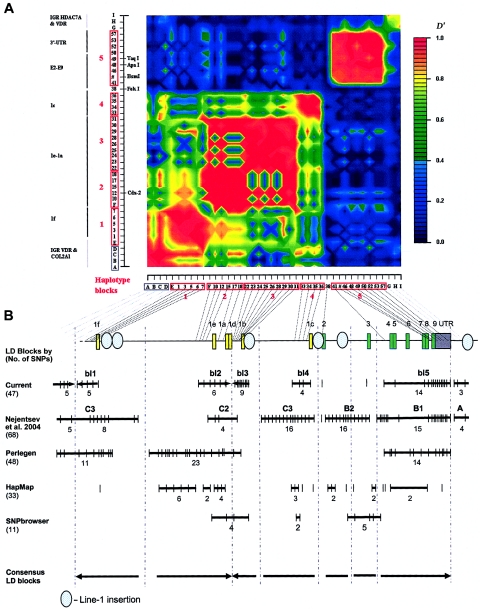
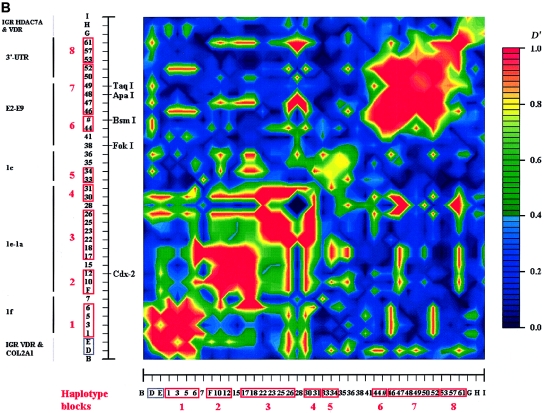
In whites, we identified five blocks with high LD (numbered 1–5 in fig. 3a), which ranged in size from 2 to 17 kb. SNPs in such a block are in strong LD with the other SNPs inside the block but show very little LD with SNPs outside the block. Four blocks (blocks 1–4) were found in the promoter region, whereas the largest block (block 5) encompasses 17 kb and includes exons 4–9 and the 3′ UTR. Blocks 2 and 3, encompassing the 1b–1e promoter region, could not be considered as one LD block in our analyses, even when D′>0.50 is taken as the cutoff to define a block. There are also clear areas of very low (or absent) LD—that is, between exons 1e and 1f (blocks 1 and 2), between exons 2 and 3 (blocks 4 and 5), and at the end of VDR (3′ of block 5). The E2-C4T SNP (the FokI RFLP) has no LD with any of the other SNPs and cannot be assigned to any of the blocks. The most distal 3′ haplotype block (block 5) shows no LD with SNPs after VDR, and the most proximal 5′ (block 1) shows only weak LD with more 5′ SNPs near and within COL2A1. At least seven blocks could be identified when we compared different sources of VDR LD block structures (fig. 3b). It is difficult to define the exact boundaries of these low-LD areas because not all studies analyzed a high density of SNPs across VDR. Interestingly, we observed Line-1 repetitive elements to be localized between the boundaries of the LD blocks of VDR (fig. 3b).
Figure 3.
LD structure of VDR in whites. a, Blocks with pairwise D′>0.8 are numbered 1–5. The analyzed SNPs (table 1) include 39 VDR SNPs, 5 SNPs in the COL2A1 and VDR intergenic region (“IGR VDR & COL2A1”), and 3 SNPs in the VDR and HDAC7A intergenic region (“IGR HDAC7A & VDR”). SNP IDs correspond to those in figure 1 and tables 1 and 3. The red boxes on the X- and Y-axes indicate the high-LD blocks used to define haplotype alleles. The physical organization of VDR is represented with vertical lines on the Y-axis (see also fig. 1). b, Aligned LD analyses from different sources and estimated consensus LD structure of VDR. The total number of SNPs analyzed in each study is indicated in parentheses. Thick lines indicate haplotype blocks, with the number of analyzed SNPs below the line and the name of the block above the line.
The LD map of Han Chinese is similar to that of whites (figs. 1d and 4a), but with only four haplotype blocks and with areas of low LD colocalizing with those in whites. For Asians, we observed a large haplotype block in the promoter region (from 1e-T−3743C to 1b-T−391C), which corresponds to the combined blocks 2 and 3 in whites. This larger promoter block was also observed in whites when we used SNPs with an MAF ⩽5% (data not shown). The LD map of African Americans is more fragmented and is substantially different from those of whites and Asians (figs. 1d and 4b). There are eight small haplotype blocks and seven unlinked SNPs. Again, the E2-C4T SNP (the FokI RFLP) was found to be independent of any other haplotype block. Some areas of low LD are colocalizing in all three ethnic groups, indicated in fig. 1d as A (separating exon 1f from the upstream region to COL2A1), C (separating exon 1c from exons 1e–1b), D (separating exon 2 from exons 3–9/the 3′ UTR), and E (separating exon 4 to the 3′-UTR from the area downstream of VDR), whereas B (separating exon 1f from exons 1e–1b) is shared between whites and African Americans but is absent in Han Chinese. In African Americans, two additional areas of low LD (B1 and B2) can be distinguished.
Figure 4.
LD maps of VDR in different ethnic groups. a, LD map of 33 SNPs in 107 Asians (214 chromosomes). b, LD map of 41 SNPs in 58 African Americans (116 chromosomes).
We reconstructed haplotype alleles and analyzed diversity and frequencies across ethnic groups (fig. 1d; only haplotypes with MAF >3% are shown). The number of haplotype alleles in each of the LD blocks increases from Han Chinese to whites and is largest in African Americans. Only for LD blocks relatively “conserved” across ethnic groups is comparison possible, and this shows substantial differences in haplotype allele frequencies. For example, in block 1 (around exon 1f), the most common allele in whites has a frequency of 44.6%, but this same allele has a frequency of 4.7% in Han Chinese and 10.3% in African Americans. Similarly, the most common allele in block 4 (around exon 1c) has frequency 40.2% in whites, 32.3% in Han Chinese, and 13.8% in African Americans.
Although, for whites, 15 tagSNPs (14 htSNPs and E2-C4T) are required to cover the common genetic diversity across VDR, the required number is only 10 for Han Chinese but increases to 28 for African Americans. When only the tagSNPs were used to genotype a population (6,148 subjects from the Rotterdam Study), the LD pattern obtained was similar to that obtained using the 34 original SNPs with an MAF >5% in the smaller sample of 234 blood-bank donors, which indicates that the tagSNPs effectively predict the other SNPs within the blocks.
We observed that the haplotypes constructed from the well-known BsmI, ApaI, and TaqI RFLPs, as described elsewhere by our group (Uitterlinden et al. 1996), could correctly predict all the common haplotypes at the 3′ UTR in whites, defined using the nine SNPs with an MAF >5% (fig. 1d). For Han Chinese, such correspondence is lower because of the U-D796T polymorphism, which has changed phase, whereas, for African Americans, the haplotype structure of this area is much different.
Study of Association with Osteoporosis
Baseline characteristics of 6,148 white elderly of the Rotterdam Study are presented in table 4. The 15 tagSNPs were in Hardy-Weinberg equilibrium. Haplotype alleles in blocks 2, 3, and 5 had consistent—albeit not always significant—effects on clinical fractures (fig. 5), with similar effects in men and women and for vertebral, hip, and wrist fractures (data not shown).
Table 4.
Characteristics of the Study Population
|
Value for Combined Promoter and 3′-UTR VDR Genotypea |
|||||
| Characteristic | Value for Total Cohort | 0 | 1 | 2 or 3 | P |
| No. (%) of individuals | 6,148 | 1,586 (25.8) | 3,560 (57.9) | 1,002 (16.3) | … |
| No. (%) of females | 3,649 (59.4) | 932 (58.8) | 2,102 (59.0) | 612 (61.1) | .49 |
| Age (years)b | 69.5 ± 9.1 | 69.1 ± 9.1 | 69.5 ± 9.1 | 70.0 ± 9.4 | .03 |
| Height (cm)c | 166.8 ± 9.5 | 167.0 ± 9.5 | 166.7 ± 9.4 | 166.6 ± 9.8 | .16 |
| Weight (kg)d | 73.1 ± 12.0 | 73.3 ± 12.2 | 73.1 ± 12.0 | 72.9 ± 11.7 | .55 |
| Femural neck (FN) BMD (g/cm2)e | .867 ± .142 | .872 ± .143 | .864 ± .142 | .868 ± .145 | .40 |
| Lumbar spine BMD (g/cm2)e | 1.090 ± .198 | 1.098 ± .197 | 1.089 ± .198 | 1.082 ± .199 | .25 |
| FN BMD change (10−3 g/cm2/year)f | −2.1 ± 9.4 | −1.6 ± 8.6 | −1.9 ± 9.5 | −3.2 ± 10.0 | .02 |
The definition of the combined genotypes “0,” “1,” and “2 or 3” are given in table 6.
Age was adjusted for sex.
Height was adjusted for age and sex.
Weight was adjusted for age, sex, and height.
BMD (subset n=5,027) was adjusted for age, sex, height, and weight.
BMD change in 7.4 years follow-up (subset n=2,391) was adjusted for age, sex, and clinical fracture.
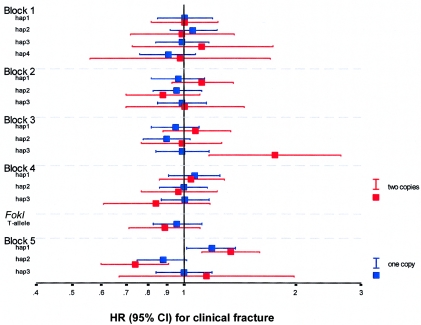
Figure 5.
HR for clinical fracture, by VDR genotypes based on haplotype alleles in five haplotype blocks (1–5) and the FokI RFLP. The HR point estimates and the surrounding 95% CIs are represented with colored squares and lines. The HR for one copy of the test allele versus no copy is in blue; the HR for two copies of the allele versus no copy is in red. The logarithmic HR is plotted for the common haplotype alleles (frequency >3%) in all haplotype blocks (see fig. 1d for whites) and the FokI RFLP.
Subjects homozygous for the block 2–hap1 allele, homozygous for the block 3–hap3 allele, or carrying the block 5–hap1 allele had an increased risk for clinical fracture of 15% (P=.06), 74% (P=.002), and 23% (P=.004), respectively (tables5 and 6). We then investigated the combined effect of promoter and 3′-UTR risk genotypes on fracture risk and found that subjects carrying two or three VDR risk genotypes had a 48% increased risk for clinical fracture (P=.0002). Although age was borderline significantly different by combined VDR genotype, all associations with fracture were independent of age, sex, height, weight, and bone loss (data not shown). By our observations, BMD did not influence the association of individual or combined blocks with fracture risk.
Table 5.
Incidence of Fractures in 6,148 Men and Women from the Rotterdam Study, by VDR Genotype Defined by Haplotype Allele Status in Blocks 2, 3, and 5[Note]
|
Genotype Groupsa |
|||
| Individual LD Blockand Measured Value | Nonhomozygous | Homozygous | P |
| Block 2–hap1: | |||
| No. of cases/total subjects (%) | 695/4,844 (14.3) | 211/1,304 (16.2) | .06 |
| Crude HR (95% CI) | 1 | 1.15 (.97–1.35) | .06 |
| Block 3–hap3: | |||
| No. of cases/total subjects (%) | 876/6,019 (14.6) | 30/129 (23.3) | .006 |
| Crude HR (95% CI) | 1 |
1.74 (1.22–2.50) |
.002 |
| Noncarrier |
Carrier |
||
| Block 5–hap1: | |||
| No. of cases/total subjects (%) | 256/1,988 (12.9) | 650/4,160 (15.6) | .004 |
| Crude HR (95% CI) | 1 | 1.23 (1.07–1.42) | .004 |
Note.— Of 6,148 subjects overall, the total number of cases was 906 (14.7%). The values in bold italics represent significant associations.
“Nonhomozygous” includes the genotype groups without the risk allele and the heterozygotes. “Homozygous” is the genotype group homozygous for the risk allele. “Noncarrier” is the genotype group without the risk allele, and “Carrier” includes the genotype groups heterozygous and homozygous for the risk allele.
Table 6.
Incidence of Fractures in 6,148 Men and Women from the Rotterdam Study, by Combined VDR Genotype[Note]
|
No. of Risk Genotypes in Blocks 2, 3, and 5 |
|||||
| Measured Valueand Fracture Type | Total | 0a | 1b | 2 or 3c | Trend P |
| No. of cases/total subjects (%): | |||||
| Clinical fracture | 906/6,148 (14.7) | 207/1,586 (13.1) | 516/3,560 (14.5) | 183/1,002 (18.3) | .001 |
| Vertebral fractured | 335/3,055 (11.0) | 78/814 (9.6) | 186/1,748 (10.6) | 71/493 (14.4) | .01 |
| Hip fracture | 261/6,148 (4.2) | 61/1,586 (3.8) | 144/3,560 (4.0) | 56/1,002 (5.6) | .05 |
| Wrist fracture | 257/6,148 (4.2) | 58/1,586 (3.7) | 145/3,560 (4.1) | 54/1,002 (5.4) | .04 |
| Crude HR (95% CI): | |||||
| Clinical fracture | … | 1 | 1.14 (.97–1.33) | 1.48 (1.21–1.80) | .0002 |
| Vertebral fracture | … | 1 | 1.12 (.85–1.48) | 1.59 (1.13–2.24) | .01 |
| Hip fracture | … | 1 | 1.06 (.79–1.43) | 1.49 (1.04–2.14) | .04 |
| Wrist fracture | … | 1 | 1.13 (.83–1.53) | 1.53 (1.06–2.21) | .03 |
Note.— The values in bold italics represent significant associations.
“0” indicates the nonhomozygous genotype groups for blocks 2 and 3 (where “nonhomozygous” includes the genotype groups without the risk allele and the heterozygotes) and the noncarrier genotype groups (i.e., the groups without the risk allele) for block 5.
“1” indicates either the homozygous genotype group (i.e., the group homozygous for the risk allele) for block 2 or 3 or the carrier genotype group (i.e., the group with the risk allele) for block 5.
“2 or 3” indicates two or three of the risk genotype groups—the homozygous genotype groups for blocks 2 and 3 and the carrier genotype group for block 5.
Vertebral fracture was diagnosed by x-ray.
The PAR of the VDR genetic markers (table 7) was 1% (frequency of subjects homozygous for the block 3–hap3 allele = 2%) and was 2% for the promoter region (frequency of subjects homozygous for the block 2–hap1 allele = 21%) but increased to 12% for the 3′ region, including the 3′ UTR (frequency of the block 5–hap1 allele carriers = 68%). This was higher than the 1% PAR of the best validated genetic marker for osteoporosis so far (COL1A1 Sp1 [Grant et al. 1996; Uitterlinden et al. 1998a]; in our study population, the frequency of subjects homozygous for the Sp1 T allele was 3%) and was similar to the PARs for smoking and use of a walking aid.
Table 7.
PAR of Independent Risk Factors for Clinical Fracture in 6,148 Men and Women from the Rotterdam Study
| Risk Factor | Frequencyat Baseline(%) | HR (95% CI)a | PAR in % (95% CI)a |
| Age >75 years | 32 | 2.3 (2.1–2.6) | 29 (25–34) |
| T score <−2.5 | 16 | 2.7 (2.3–3.1) | 21 (17–25) |
| Current smoking | 23 | 1.3 (1.1–1.5) | 6 (2–10) |
| Use of walking aid | 12 | 1.3 (1.1–1.6) | 3 (1–4) |
| Genetic markers: | |||
| COL1A1 Sp1 T-allele homozygote | 3 | 1.4 (1.0–1.9) | 1 (0–3) |
| VDR risk genotype: | |||
| Block 2–hap1 | 21 | 1.1 (.9–1.3) | 2 (0–5) |
| Block 3–hap3 | 2 | 1.6 (1.2–2.4) | 1 (0–3) |
| Block 5–hap1 | 68 | 1.2 (1.1–1.4) | 12 (4–21) |
| Carrier of 2 or 3 VDR risk genotypes | 16 | 1.3 (1.1–1.5) | 4 (2–6) |
All HRs and PARs were adjusted for age and sex.
Functionality Studies of Promoter Polymorphisms
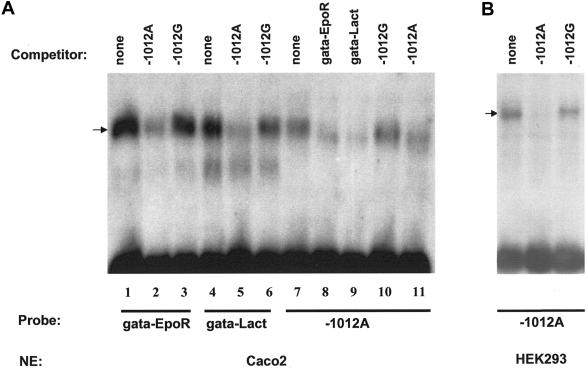
Choosing from among 12 polymorphisms located at potential TFBSs in the VDR promoter region, we performed EMSA for four common (MAF >20%) SNPs (1e-C−2090T, 1e-G1739A, 1a-G1521C, and 1a-A−1012G) in the promoter region of exons 1e and 1a, using Caco2 or HEK293 nuclear extracts (fig. 6). We identified a putative GATA-binding site for the A allele of the 1a-A−1012G SNP in the exon 1a promoter region (AGATAT in reverse orientation) and demonstrated that the G allele has markedly decreased binding to GATA compared with that of the A allele.
Figure 6.
EMSA of 1a-A−1012G for GATA protein. a, GATA binding assay using Caco2 cell-line nuclear extract. The binding of GATA to the 1a−1012A site was analyzed in competition experiments for Caco2 nuclear extract by use of the well-characterized GATA sites of the EpoR and Lactase gene promoters (Zon et al. 1991; Fang et al. 2001). These experiments showed similar binding characteristics of the complexes bound to the 1a−1012A site and to the other GATA sites (compare lane 7 to lanes 1 and 4). In addition, the signal found on the 1a−1012A site was eliminated by a 100-fold excess of unlabeled GATA sites of the EpoR and Lactase genes (see lanes 7–10). Conversely, a 100-fold excess of the 1a−1012A site eliminated the binding of GATA to the EpoR and Lactase GATA sites (lane 1 vs. 2 and lane 4 vs. 5). b, GATA binding using HEK293 cell-line nuclear extract. Competition experiments for HEK293 nuclear extract using the −1012 GATA site as a probe revealed that the 1a−1012G variant was unable to compete with the binding of the 1a−1012A variant.
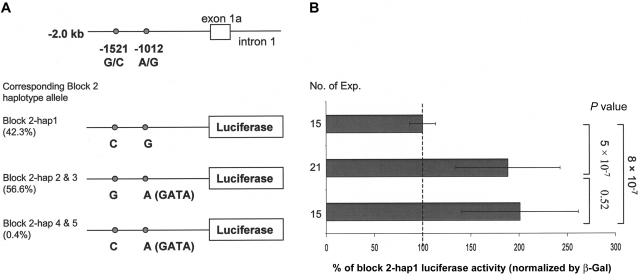
Reporter constructs containing the 2-kb 1a-promoter sequence with the two SNPs 1a-G−1521C and 1a-A−1012G in block 2 (fig. 7) showed that, in HEK293 cells, the normalized luciferase activity of the haplotype allele was decreased by 53% and 50%, compared with that of the hap 2/3 and hap 4/5 alleles, respectively (P=5×10-7 and 8×10-7). This indicates that the G allele of the htSNP 1a-A−1012G has a transcription rate twofold lower than that of the A allele. The same results were observed in COS-7 cells (data not shown).
Figure 7.
Relative luciferase activity in HEK293 cells of VDR exon 1a promoter sequences, including two SNPs. a, The three constructs containing the 2-kb 1a promoter sequence with SNPs 1a-G−1521C and 1a-A−1012G. b, β-galactosidase (beta-Gal)–normalized luciferase activity for the three constructs. The block 2–hap1 allele is set at 100% to be the reference group; P values were calculated by independent t test.
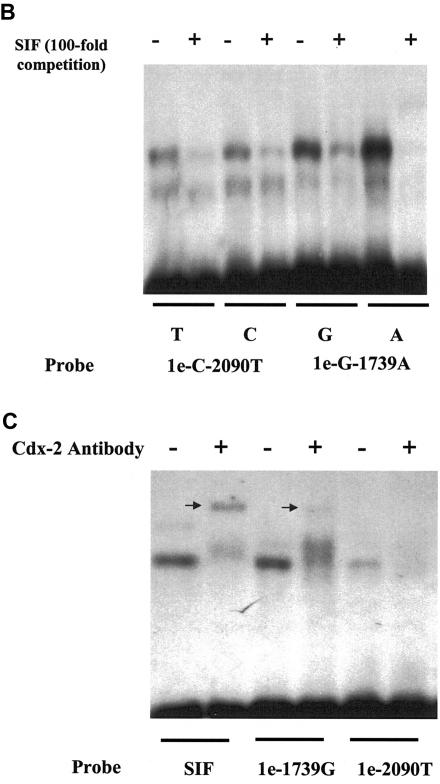
Surrounding sequence analyses showed that the 1e-C−2090T and 1e-G−1739A SNPs are located at potential Cdx-2–binding sites (fig. 8a). EMSA confirmed that the G allele of 1e-G−1739A and the T allele of 1e-C−2090T have a relatively decreased binding to Cdx-2, compared with their allelic counterparts (fig. 8b). The risk allele of the GATA and Cdx-2 promoter SNPs are contained in block 2–hap1, which is the risk allele for fracture.
Figure 8.
Two SNPs in the 1e promoter region of the human VDR gene, located at a Cdx-2 binding site. a, Alignment of 15 well-characterized Cdx-2 sites from mammalian gene promoters. Base usage is summarized in a table and is compared with DNA sequences surrounding SNPs 1e-C−2090T and 1e-G−1739A. DNA bases involved in SNPs are underlined. b, EMSA experiments performed using Caco-2 cell nuclear extracts. Double-strand oligonucleotides containing the sucrose isomaltase (SIF) Cdx2-binding site as a control or sequences encompassing SNPs 1e-C−2090T and 1e-G−1739A were 32P-labeled and were purified on a 10% polyacrylamide gel. The competition experiments were done with an oligonucleotide containing the SIF element. Gel-shift experiments were performed in the absence (−) or presence (+) of a 100-fold excess of cold SIF probe as a competitor, which resulted in relatively more elimination of these specific complexes for the A allele of 1e-G−1739A and the T allele of 1e-C−2090T than for their allelic counterparts. c, Antibody experiments with monoclonal anti–Cdx-2. EMSA was performed with nuclear extract alone (−) or in the presence of monoclonal anti–Cdx-2 antibody (+). Supershift complexes are identified by an arrow. A clear supershift was observed with SIF, but it had a weak signal with 1e−1739G, and a very low intensity of the complex was seen with 1e−2090. Comparison of the surrounding 1e-C−2090T and 1e-G−1739A sequences with the consensus Cdx-2 sequence (panel a) evidenced the presence of a substitution (T→A) in the 1a−2090 sequence, somehow corrupting the Cdx-2 site, which may explain the lower intensity observed in EMSA for this sequence compared with 1e-G−1739A.
Functionality Studies of 3′-UTR Polymorphisms
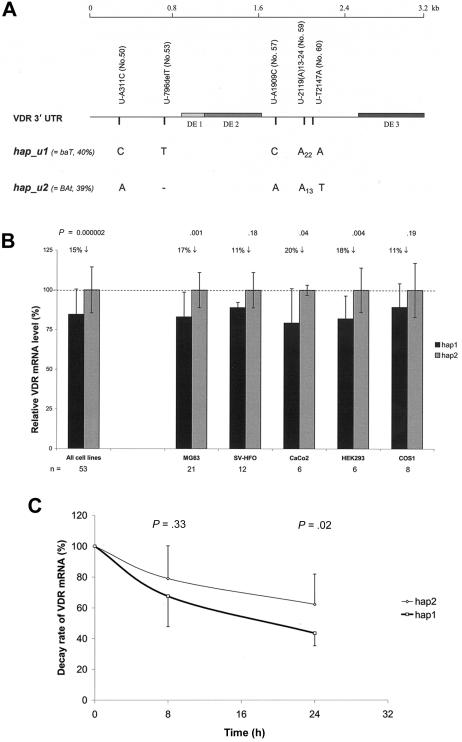
We found that, 24 h after transfection of reporter constructs containing the complete 3.2-kb 3′ UTR of hap1 or hap2, the neomycin-normalized VDR mRNA level of the hap1 transcript was 15% lower than that of the hap2 transcript when results of all tested cell lines were combined (P=2×10-6; n=53), and similar patterns were observed in individual cell lines (fig. 9a and 9b). We then investigated the stability of VDR mRNAs transcribed from hap1 and hap2 and observed that the decay rate of VDR mRNA for hap1 was 30% faster than that for hap2 in the MG63 cell line 24 h after inhibiting transcription (P=.02; n=9) (fig. 9c).
Figure 9.
VDR mRNA expression level and stability analysis by 3′-UTR haplotypes in different cell lines. a, VDR 3′-UTR with sequence variations that distinguish hap1 (corresponding to block 5–hap1) from hap2 (corresponding to block 5–hap2). The SNP variation number (in parentheses) refers to those given in table 3. b, Neomycin-normalized VDR mRNA expression levels (mean ± SD) for VDR 3′-UTR hap1 versus hap2. The level of hap2 is set at 100% to be the reference group; P values were calculated by independent t test; n = number of experiments for each cell line. c, Decay rate of VDR mRNA by hap1 versus hap2, determined in the MG63 cell line. Time point 0 h is defined as 100% mRNA level for both haplotypes; P values for each time point were calculated by independent t test.
Discussion
A major limitation so far of association studies using VDR polymorphisms in relation to complex-disease end points has been the small number of analyzed polymorphisms and, thus, the lack of knowledge about the influence of and relation between other polymorphisms in the gene. In addition, the lack of statistical power of most studies to detect the expected subtle effects as well as the misconceptions about how such small biological effects could be translated into risk of disease have led to a number of controversies in the field. In the current study, we identified 62 polymorphisms in potentially functional areas of VDR, characterized the LD structure, and identified htSNPs. We here combined several studies and databases (Nejentsev et al. 2004; Hinds et al. 2005; Applied Biosystems SNPbrowser Software Web site; HapMap Data Web site; Perlegen Genome Browser Web site) to conclude that there are seven haplotype blocks across the VDR gene in whites. We used 15 tagSNPs to represent the common haplotypes for five of these blocks in potentially functional areas of VDR.
In this study, we also constructed an LD map and identified htSNPs of VDR for three major ethnic populations. There are several further approaches to identify the (set of) responsible functional SNPs in the LD areas where we identified risk haplotype alleles. Since African Americans have, in general, smaller LD blocks, replication of the associations in African Americans could focus on a smaller region with concomitantly fewer SNPs to test functionally. Subsequently, the functionality test we describe here (see figs. 7 and 9) can be repeated, but with constructs of promoter and 3′-UTR variants that differ at only one position.
A large association study with the htSNPs in a population-based cohort (6,148 elderly subjects) identified haplotype alleles in three LD blocks that confer risk for fracture, and we present evidence of intragenic additive effects. These relationships were found in both men and women and for different types of fractures (including vertebral, hip, and wrist fractures) and were independent of age, height, weight, BMD, and BMD change. In our study population (which had a 15% prevalence of fracture among 6,148 subjects), we have 80% statistical power to detect a 25% increased fracture risk for a polymorphism and/or haplotype with a frequency >10%. We analyzed haplotypes instead of individual SNPs, which limited the number of tests. For blocks 2 and 5, we did not adjust the significant P value by the conservative Bonferroni correction for multiple comparisons because, in our previous studies (Uitterlinden et al. 2001; Fang et al. 2003), we observed two fracture-risk alleles in these blocks in a subset of our current study, and we confirmed the association in this study—but this time for the haplotype alleles encompassing the previously observed risk alleles. However, we used P=.005 (.05/11) on the basis of a Bonferroni correction for analyses in the other three blocks and the FokI RFLP; only the association between block 3–hap3 and fracture risk remains borderline significant (P=.002). Thus, we cannot reject the possibility that some of the associations we observed are false positives, even in this very large population.
According to our association analysis, the genetic effect of the VDR polymorphisms on fracture risk is modest (15%–48%), which corresponds to the modest difference (15%) in the VDR mRNA level by genotype in our functionality analysis. In an earlier study (Mann et al. 2001), the “Ss” (or GT) genotype of COL1A1 Sp1 was found to have a 21% increased COL1A1:COL1A2 protein ratio, as measured in osteoblasts, compared with that of the “SS” (or GG) genotype. Another functional study of a 3′-UTR SNP (+1073C/T in the oxidized LDL receptor gene [OLR1]) that is associated with increased risk for Alzheimer disease demonstrated that C-allele carriers had a 41% decreased OLR1 mRNA level compared with that of TT homozygotes (Lambert et al. 2003). These examples indicate that genetic effects of polymorphisms on gene expression and clinical phenotypes are modest.
Elsewhere, we reported that haplotype 1 of the 3′-end variants, defined by only BsmI-ApaI-TaqI RFLPs, was associated with increased fracture risk in 1,004 postmenopausal women (Uitterlinden et al. 2001), which is a subgroup of the current study population. We here confirm this association between BsmI-ApaI-TaqI haplotype 1 and fracture risk in the complete study population (P=.03; data not shown). Yet, in our more detailed haplotype analysis based on more SNPs, we defined a subtype of the BsmI-ApaI-TaqI haplotype 1—that is, of block 5–hap1u (40.2%) (fig. 1d)—that shows a stronger and more significant association with fracture risk. These subtle differences in the exact definition of the risk allele could contribute to heterogeneity in association results observed in different studies. Although we focused here on the more common haplotype alleles in the population, we cannot exclude the idea that (several) less-frequent risk alleles may contribute to the VDR genotype–dependent fracture risk. Further studies will be necessary to assess their contribution. We show that effects are modest (∼20%–70% increased risk), as can be expected for common variants in relation to complex disease. The VDR risk haplotype alleles therefore have a modest influence on individual risk of fracture but make a substantial contribution at the population level (PAR = 4%–12%) in comparison with other genetic markers we previously identified in this population, such as COL1A1 Sp1 (Uitterlinden et al. 1998b) and ESR1 PvuII and XbaI polymorphisms (van Meurs et al. 2003). The most prominent genetic effect on fracture risk, according to the PAR analyses, is the block 5–hap1 risk allele. Many previous conflicting association studies of VDR and BMD as well as fracture usually analyzed (very) small study populations and used the BsmI, ApaI, or TaqI polymorphisms in this block but mostly analyzed them separately. We therefore suppose that the controversy can partly be explained by a lack of statistical power as a result of small sample size and failure to use haplotypes. In addition, population stratification, such as mixed ethnic groups with different allele frequencies; population-specific differences in some environmental factors, such as (dietary) calcium intake, (dietary) vitamin D intake, and sunlight exposure; and other characteristics of the study population pertinent to bone metabolism and fracture risk could result in heterogeneity of association observed across different study populations. On the basis of a meta-analysis of published data on the relationship between the VDR BsmI RFLP and fracture risk, we have some evidence that this could be an important factor (Y. Fang, F. Rivadeneira, J. Ioannidis, and A. Uitterlinden, unpublished data). We have data on serum vitamin D and on dietary calcium intake in a subset of our study population (n=1,312 and 4,747, respectively), but we do not have reliable dietary vitamin D intake data. We note that our study population has a very high dietary calcium intake (1,120 mg/d). For the VDR-fracture relationship, we repeated the analyses but stratified them by the median, tertiles, and quartiles of vitamin D level or dietary calcium intake. However, the association did not differ between these strata, and no interaction was observed (data not shown).
By analyzing functionality of the risk alleles in vitro, we demonstrate that the molecular mechanisms underlying these associations are likely to involve a lower expression of VDR mRNA. Some of the promoter polymorphisms result in altered transcription-factor binding for Cdx-2 and GATA. The previously reported Cdx-2 site at 1e-G−1739A (Yamamoto et al. 1999; Fang et al. 2001) and the GATA site at 1a-A−1012G (Halsall et al. 2005) are encompassed in the block 2–hap1 risk allele. Our functionality experiment and previous ones (Arai et al. 2001) show that these two weak-binding alleles together result in decreased transcription activity of this VDR promoter. Further research is needed to establish, in more detail, in which cells and/or tissues this promoter part is influencing VDR expression. Although GATA is expressed in many tissues, Cdx-2 is expressed predominantly in the intestines. Thus, the hap1 allele might cause relatively lower VDR expression in target cells for vitamin D, including the intestines.
The 3′ UTR of genes is known to be involved in regulation of gene expression, especially through regulation of mRNA stability. The BsmI, ApaI, and TaqI SNPs are anonymous, and block 5 does not include polymorphisms beyond the 3′ UTR of VDR. Therefore, SNPs in the 3′ UTR are the most likely candidates for the truly functional sequence variations that may explain the associations we observed. We identified differences in VDR mRNA expression level and stability between the hap1 and hap2 alleles, which differ at only five positions across the 3.2-kb 3′ UTR. The fracture-risk allele hap1 causes 15% lower levels of mRNA expression than does hap2, in all tested cell lines. This is in line with the 30% faster decay of or lower stability of VDR mRNA we observed in MG63, an osteoblast cell line. This observation also corresponds to other studies performed in vivo and in vitro (Morrison et al. 1992, 1994; Carling et al. 1997; Ogunkolade et al. 2002). This is likely to also result in lower numbers of VDR protein being present in target cells for vitamin D, giving such target cells a decreased response to vitamin D.
VDR as a transcription factor influences the expression of downstream genes, such as TRPV5, TRPV6, and Calbindin, which are involved in calcium absorption and could therefore impact BMD and thus fracture risk. However, we found that the association between VDR genotype and fracture risk was independent of BMD, which suggests that other mechanisms (such as bone microarchitecture, bone quality, and bone strength) determine fracture risk. Alternatively, we can hypothesize that the bone tissue of the subjects carrying a risk allele may have a somewhat lower sensitivity to vitamin D, since the expression of VDR is lower. Therefore, the osteoblast activity could be lower, and the bone formation rate (in the bone-remodeling balance) could be decreased. The age-related expansion of the outer diameter of long bone is associated with a marked increment of bone strength. We have observed recently that the fracture-risk haplotype alleles were also associated with decreased bone size (Y. Fang, J. van Meurs, F. Rivadeneira, N. van Schoor, J. van Leeuwen, P. Lips, H. Pols, and A. Uitterlinden, unpublished data) in the same population. This genotype-related bone geometry difference reflects that the bone gain resulting in outer bone diameter expansion is smaller than the bone loss resulting in inner bone diameter expansion. This bone geometry difference leads to a decrease in bone strength for the risk-allele carriers, possibly increasing the fracture risk.
We demonstrate that polymorphisms within the promoter area and the 3′-UTR area of a gene have effects that can influence VDR gene function in certain cells and/or subjects. Thus, the 5′-promoter and 3′-UTR polymorphisms together can determine how much of a given VDR mRNA level will be expressed in a given target cell. The combined risk genotypes in the promoter region and 3′ UTR represent a moderate genetic effect of the entire VDR gene on fracture risk. The vitamin D endocrine system has been implicated in several other complex diseases, including osteoarthritis, diabetes, and cancer. Whether our findings have relevance for these other diseases needs to be tested in separate association studies using the LD and haplotype information we provide here.
In conclusion, we systematically scanned sequence variations across VDR, identified LD structure and htSNPs for different ethnic populations, and demonstrated that polymorphisms in the 5′ promoter region and the 3′ UTR of VDR contribute to fracture risk in a large population. Functionality experiments in the 5′ promoter region and the 3′ UTR support and provide a possible molecular explanation for the association with fracture risk that we observed.
Acknowledgments
We thank Drs. John Eisman and Linda Crofts for helpful sharing of unpublished sequence information; Dr. Mark Haussler for kindly providing VDR cDNA; and Marco Eijken for initial help with the RT-PCR in the 3′-UTR experiments. We thank all participants of the Rotterdam Study and the general practitioners, the pharmacists, and the many field workers at the research center in Ommoord, Rotterdam, The Netherlands. This project was funded by the Dutch Research Organisation (NWO grants 903-46-178, 925-01-010, 014-90-001, and 911-03-012) and the European Commission (grant QLK6-CT-2002-02629 [“GENOMOS”]).
Appendix A: Supplementary Material and Methods
Sequencing
We sequenced 37 overlapping PCR fragments covering 22 kb of VDR (shown in fig. 1c) in 30 chromosomes, including the 3.2-kb 3′ UTR, coding exons 2–9 and flanking introns (4.1 kb), and the 14.7-kb promoter area containing six exons (1a–1f; shown in fig. 1c). PCR primers were designed in accordance with published sequences (J03258 [Baker et al. 1988], PAC clones, AC004466 [Miyamoto et al. 1997], and AF080454–AF080456 [Crofts et al. 1998; Yamamoto et al. 1999]) and the Celera database. Melting temperatures of primers were calculated using DNAMAN (version 4.0 [Lynnon BioSoft]). Primer sequences and PCR conditions are described in table 3. The 50-μl PCR contained 20–80 ng of genomic DNA, 3–5 pmol primers, and 2 units Taq DNA polymerase (SUPER TAQ [HT Biotechnology] or M1665 [Promega]) and was performed for 25–38 cycles in a Thermal Cycler 480 (Perkin Elmer) or a GeneAmp PCR System 9700 (ABI). Taq PCR Core Kit (5 × Q-Solution [QIAGEN]) was used to amplify some GC-rich PCR fragments in promoter regions (table A1). The sequencing extension reaction system (20 μl) consisted of 11 μl of purified PCR product, 4 μl of Terminator Mixture (ABI PRISM Big-Dye Terminator Cycle Sequencing Ready Reaction Kit), 4 μl of 5 × buffer (400 mM Tris-Cl, pH 9.0; 10 mM MgCl2), and 1 μl of 10 pM primer. We purified PCR and sequencing products by use of Quantum Prep PCR Kleen Spin Columns and Micro Bio-spin Chromatography Columns (BIO-RAD). Sequencing products were analyzed on an ABI PRISM 310 or 3100 automated capillary Genetic Analyzer. All sequencing data were analyzed and aligned using Navigator (Perkin Elmer) and Sequencher (Gene Codes) sequence-analysis software.
Table A1.
Primers and PCR Conditions for the Sequence Analysis of VDR
| Region and Primer Name | Primer Sequence(5′→3′) | MgCl2(nM) | Annealing Temperature(°C) | Size of PCR Product(bp) | GenBanka Accession Number |
| Promoter region: | |||||
| 1f-23h For | GAACCAGGATCATGTTTTGGA | ||||
| 1f-23h Rev | ACAATCCAAGTCTGTGGATAGATG | 1.5b | 57 | 655 | BV210414 |
| 1f-2kb For | CAAGTTTTGCCAGATTCACC | ||||
| 1f-2kb Rev | CCGCAGCACTATACAGAGGT | 1.5b | 58 | 932 | BV210415 |
| 1f-1kb For | CAAGTCCATATCTCCTTCTCCAG | ||||
| 1f-1kb Rev | AAGTGGTTTCCCTACCATCAAT | 1.5b | 59 | 855 | BV210416 |
| 1f-4h For | AGATACCTCTGGTTCCTCTAATGC | ||||
| 1f-4h Rev | CACACTTGTTCACCTCCACAC | 1.5b | 59 | 631 | BV210417 |
| VDR1a-40h For | CTCAAGCATAGTGGCATGATCA | ||||
| VDR1a-40h Rev | TTCCTGATACAAAAGATGTTCTACAATG | 1.5 | 60 | 549 | BV210418 |
| VDR Cdx-2 G For | AGGATAGAGAAAATAATAGAAAACATT | ||||
| VDR Cdx-2 A Rev | ACGTTAAGTTCAGAAAGATTAATTC | 1.5 | 54 | 297 | BV210419 |
| VDR 1a-36h1 For | GAGTCATCCCTGATCCTTTTTGT | BV210420 | |||
| VDR 1a-36h1 Rev | TTCTGAGAAACAGATGAAGTGCC | 1.5 | 60 | 2201 | BV210421 |
| VDR 1a-36h2 For | CTGGCAGAGAGGTCAAGAGAC | BV210422 | |||
| VDR 1a-36h2 Rev | GCTGAGGAGTATCAGAGCTACTG | 1.5 | 60 | 1,119 | BV210423 |
| VDR 1a-36h3 For | GAATGGTGCTTGTCATCGAGG | ||||
| VDR 1a-36h2 Rev | GCTGAGGAGTATCAGAGCTACTG | 1.5 | 60 | 651 | BV210424 |
| VDR 1a-36h4 Forc | AAGACCAGAGAATTGACAGTTCCTT | ||||
| VDR 1a-36h2 Rev | GCTGAGGAGTATCAGAGCTACTG | 1.5 | 56 | 352 | BV210425 |
| VDR 1a-36h2 For | CTGGCAGAGAGGTCAAGAGAC | ||||
| VDR 1a-36h4 Rev | AGGAACTGTCAATTCTCTGGTCTT | 1.5 | 60 | 768 | BV210421 |
| VDR 1a-14h For | GGGGATCCTTCCTATTACTTTCATTACA | ||||
| VDR 1a-14h Rev | CTTAGACTCACTGTGCAGTGGAGATG | 1.5b | 60 | 612 | BV210426 |
| VDR 1a-10h For | GGAGGTCATCGACTGCTGGA | ||||
| VDR 1a-10h Rev | CTAGCTCCGACGAATGGGAAA | 1.5 | 60 | 535 | BV210427 |
| VDR 1a-5h For | ATCTGTGGGATCAGGCTGAGC | ||||
| VDR 1a-5h Rev | CGCCTTTTGACAAGCAGAGACA | 1.5b | 60 | 550 | BV210428 |
| VDR 1ad For | TGGTTGATTCCAAGTCAAGATGG | ||||
| VDR 1ad Rev | CTCCAGCAGTTCTGAGCACCA | 1.5b | 60 | 604 | BV210429 |
| VDR1d+7h For | CGCTAACACAGTGCTTAGCACTT | ||||
| VDR1d+7h Rev | AGTAAAATGCCTGCCCAACTG | 1.5 | 60 | 710 | BV210430 |
| 1gA-For | TCCTGTCTGGGCTCAGTTG | ||||
| 1gA-Rev | TAGCTTGGTAAGGGTCCAAGTC | 1.5b | 60 | 879 | BV210431 |
| 1gB-For | AATTCACAGTCTATGCTCTGGCT | ||||
| 1gB-Rev | GATCCTCTCAGAACTGGACAATAGT | 1.5b | 59 | 896 | BV210432 |
| 1gC-For | TGAGAGATAGCAGGAAGCAGAAC | ||||
| 1gC-Rev | CTGGCTGCTTACCTGCTTTAC | 1.5b | 60 | 967 | BV210433 |
| 1b-2k For | GACCACACTATCCCACAGAAAGT | ||||
| 1b-2k Rev | GAATGTCCCACCTTGCATAAC | 1.5b | 59 | 842 | BV210434 |
| 1b-gap For | AAGCGGAGTCTGTGCAGAC | ||||
| 1b-gap Rev | AGGTGGCTGCATCTCTTTAGA | 1.5b | 59 | 835 | BV210435 |
| 1c-2kb For | AAGGACCGTCCACTATTGGA | ||||
| 1c-2kb Rev | AAGTCCTTCCCAGCTGACC | 1.5b | 59 | 875 | BV210436 |
| 1c-12h For | CTGAGGAATCAATAAGGCCAG | ||||
| 1c-12h Rev | TGGATCTTAGCAGCTGGCT | 1.5b | 59 | 622 | BV210413 |
| 1c-8h For | TTGTCTATGTCTGCAGGTGGA | ||||
| 1c-8h Rev | CTTTTATCTAAGGCGGAGCG | 1.5b | 59 | 816 | BV210437 |
| VDR E1c-For | CACTTCTGTTTGCAGTCACTGA | ||||
| VDR E1c-Rev | GGCTGGATAGGAAACATCAGA | 1.5 | 60 | 489 | BV210438 |
| Exons: | |||||
| VDR E2-For | ATGCTCTGAGCCAGCTATGTAG | ||||
| VDR E2-Rev | GAGAGTCAGAGGAACATCTGGA | 1.5 | 60 | 571 | BV210439 |
| VDR E3-For | TGTCTTCTGTTGGAGAAATGGA | ||||
| VDR E3-Rev | AGTGCATCTGACCCTGGACT | 1.5 | 60 | 495 | BV210440 |
| VDR E4-For | TTTCTTCACACAGTGGAGTGG | ||||
| VDR E4-Rev | GGCTTTGAGGAAGGTCTACAG | 1.5 | 60 | 526 | BV210441 |
| VDR E5-For | ATCCTGAACAGAACTGGGGTA | ||||
| VDR E5-Rev | TGATGCTACACAGCTGGAATC | 1.5 | 60 | 519 | BV210442 |
| VDR E6-For | TACTGCCTTATGCTGCTGAAA | ||||
| VDR E6-Rev | GAGAATCGCTTGAACCTAGGA | 1.5 | 60 | 557 | BV210443 |
| VDR E78-For | CAGCAGGTGTATACCTGTCAAAG | ||||
| VDR E78-Rev | AGCAGGTCTTTGTCCTTCATACT | 1.5 | 60 | 634 | BV210444 |
| VDR E9-For | CTAGGTCTGGATCCTAAATGCA | ||||
| VDR E9-Rev | TTAGGTTGGACAGGAGAGAGAA | 1.5 | 60 | 628 | BV210445 |
| 3′-UTR: | |||||
| VDRU1-For | TGAGTGCAGCATGAAGCTAAC | ||||
| VDRU1-Rev | ATATAACCAGGGCAATGGGAT | 1.5 | 59 | 580 | BV210446 |
| VDRU2-For | TCCTGCCTTACTCACGATAAATAA | ||||
| VDRU2-Rev | CTAGCTCTTAGCCCTGTGGTG | 1.5 | 60 | 612 | BV210447 |
| VDRU3-For | AAGAATTTTCAGACCCCAGC | ||||
| VDRU3-Rev | TTTCCACCTGAAGAATTCTGAG | 1.5 | 60 | 621 | BV210448 |
| VDRU4-For | GAGGAATCAGACTTCACACTGC | ||||
| VDRU4-Rev | AGAGACAGGGTTTCTCCATGTT | 1.5 | 61 | 592 | BV210449 |
| VDRU5-For | GTAGGTGGATCACCTGAGGTC | ||||
| VDRU5-Rev | AGTAACTGATATTTCAGGAGTTCCC | 1.5 | 61 | 656 | BV210450 |
| VDRU5-For | GTAGGTGGATCACCTGAGGTC | ||||
| VDRU5-PoA-Rev | CAGATCCAGACTTGGCTCTTT | 1.5 | 60 | 403 | BV210451 |
| VDRU6-For | ATGCTGTTGCCTCATCTATAACA | ||||
| VDRU6-Rev | ATAATGATTCATCTCCCATAAGGTC | 1.5 | 60 | 680 | BV210452 |
GenBank database accession numbers of the small sequence fragments. The accession numbers of the four long sequence fragments (2.5–9.6 kb; I–IV in fig. 1c) are AY827085–AY827088.
Add 5 × Q-Solution (Qiagen) to the PCR system.
Seminested PCR using the fragment generated with primer set VDR 1a-36h2 For and VDR 1a-36h2 Rev as the template.
EMSA and Transactivation Assay for Promoter Polymorphisms
Cell nuclear extracts from Caco2 (human colonic adenocarcinoma) and HEK293 (human embryonic kidney) cell lines were prepared as described elsewhere (Dame et al. 1985). Annealed oligonucleotides (Addendum 2) were [γ−32P] ATP end-labeled (Amersham Biosciences) by the T3-polynucleotide kinase (Invitrogen Life Technologies) and were purified on a nondenaturing 10% polyacrylamide gel prior to EMSA studies. Nuclear extracts (20 μg) were incubated at 4°C for 30 min in the presence of 1 μg of double-strand poly [dI-dC]–poly [dI-dC] (Amersham Biosciences) and 10 fmoles of purified labeled oligonucleotide in a final buffer containing 10% glycerol, 5 mM Tris (pH 7.5), and 150 mM KCl. Samples were resolved on a 4% nondenaturing polyacrylamide gel in a low-salt buffer (pH 7.5) of Tris (6.7 mM), acetate (3.3 mM), and EDTA (1 mM) . Supershift experiments were done similarly, except for a 1-h incubation at 4°C in the presence of 1 μl of monoclonal anti–Cdx-2 antibody (MU392-UC [Biogenex]) prior to the addition of the oligonucleotide probe.
Three reporter constructs containing the 2-kb 1a promoter sequence with two SNPs (1a-G−1521C and 1a-A−1012G) were created. Site-directed mutagenesis at the SNP locations in the human VDR promoter region was performed using the Gene Editor TM system (Promega). The mutated oligonucleotides used were 5′-AGG CGA ATA GCA ATG TCT TCC CTG GCT AA-3′ for the −1012 SNP and 5′-GCT AGC TTT CCC ACC ATG CTT TGG GCA AG-3′ for the −1521 SNP (the mutated base is underlined). Each site-directed mutant was confirmed by sequencing.
Cells were put in six seeded-well plates (2×105 cells/well) and were grown in 2 ml of medium for 24 h. Then, a mixture of 1 μg of plasmid DNA, containing 50 ng of pCMV-β plasmid (Clontech), 950 ng of luciferase reporter vector, and 2 μl of Fugene-6 (Roche) was added (maintaining the presence of serum and antibiotics). No change of medium occurred during 72 h of culture. Cells were grown for an additional 48 h and were harvested with 200 μl of 1 × reporter lysis buffer (Promega). Luciferase activities were measured in 10 μl of cell extract with a LG Berthold Lumat LB 9507 and were corrected over β-galactosidase activity, determined by a standard colorimetric procedure using o-nitrophenyl-β-D-galactopyranoside as the substrate.
Functionality Experiments for 3′-UTR Polymorphisms
The constructs encompassed VDR exons 2–9, linked to the entire 3′ UTR of hap1 (corresponds to block 5–hap1) or hap2 (corresponds to block 5–hap2), were cloned into a pCI-Neo mammalian expression vector (Promega). The accuracy of all cloning was verified by direct sequencing of constructs.
Before transfection, MG63 (human osteoblast), SV-HFO (human osteoblast), Caco2, HEK293, and Cos1 (green monkey kidney) cells were grown overnight in Dulbecco’s modified Eagle’s medium (Gibco); were supplied with 10% fetal calf serum (FCS [Gibco]), 50 U/ml penicillin, and 50 μg/ml streptomycin (Gibco); and were transfected using Fugene transfection reagent (Roche) at a reagent:DNA ratio of 3:1. After 24 h, cells were harvested and the VDR mRNA level was measured. For the mRNA stability measurements, cells were put on fresh medium with 7.5 μl/ml actinomycin D (Sigma), and cells were harvested at time points 0, 8, and 24 h after addition of actinomycin D.
Harvested cells were washed once with PBS, and total RNA was extracted with High Pure RNA Isolation Kit (Roche). One microgram total RNA was reverse transcribed into cDNA by use of a cDNA synthesis kit in accordance with the protocol of the manufacture (MBI Fermentas). Quantitative real-time PCR was performed using an ABI PRISM 7700 sequence detector. A 25-μl reaction system with qPCR Core kit (Eurogentec) contained 20 ng cDNA, 5 mM MgCl2, 200 μM dNTPs, and 0.025 U/μl Hot GoldStar enzyme. Primers and probes were designed using the Primer Express program (version 1.5 [ABI]). The cDNA primers VDR For (5′-CCTCCAGTTCGTGTGAATGATG-3′), VDR Rev (5′-TCATGTCTGAAGAGGTGATACA-3′), NEO For (5′-GCGCCCGGTTCTTTTTG-3′), and NEO Rev (5′-CCTCGTCCTGCAGTTCATTCA-3′) were used to amplify the VDR and neomycin cDNA, respectively. Neomycin was used as an internal control at the 0 h time point. Reaction conditions of real time PCR were followed: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. All transfection experiments were performed 6–21 times. The mRNA levels were calculated by the equation 2(20-Ct), where Ct is the cycle threshold of real-time PCR.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Applied Biosystems SNPbrowser Software, http://events-na.appliedbiosystems.com/mk/get/snpb_landing?isource=fr_E_RD_www_allsnps_com_snpbrowser
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for detected polymorphisms [accession numbers ss32465817–ss32465820 and ss32465633–ss32465690])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for reference sequences [accession numbers BV210413–BV210452 and AY827085–AY827088])
- Genomatix, http://www.genomatix.de/cgi-bin/./matinspector_prof/mat_fam.pl
- GOLD, http://www.sph.umich.edu/csg/abecasis/GOLD
- HaploBlockFinder V0.7, http://cgi.uc.edu/cgi-bin/kzhang/haploBlockFinder.cgi [DOI] [PubMed]
- Haploxt, http://archimedes.well.ox.ac.uk/
- HapMap Data, http://www.hapmap.org/cgi-perl/gbrowse/gbrowse/hapmap
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for VDR) [PubMed]
- Perlegen Genotype Browser, http://genome.perlegen.com/browser/index.html
- Phase, https://depts.washington.edu/ventures/clickthru/ReleaseAgreement.php?raf=PHASEV2
- TFSEARCH: Searching Transcription Factor Binding Sites, http://www.cbrc.jp/research/db/TFSEARCH.html
- Vista Tools, http://www-gsd.lbl.gov/VISTA/index.shtml
References
- Arai H, Miyamoto KI, Yoshida M, Yamamoto H, Taketani Y, Morita K, Kubota M, Yoshida S, Ikeda M, Watabe F, Kanemasa Y, Takeda E (2001) The polymorphism in the caudal-related homeodomain protein Cdx-2 binding element in the human vitamin D receptor gene. J Bone Miner Res 16:1256–1264 [DOI] [PubMed] [Google Scholar]
- Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, Pike JW, Shine J, O’Malley BW (1988) Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci USA 85:3294–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SB, Brierley TT, Palanisamy N, Salusky IB, Goodman W, Brandi ML, Drueke TB, Sarfati E, Urena P, Chaganti RS, Pike JW, Arnold A (2000) Vitamin D receptor as a candidate tumor-suppressor gene in severe hyperparathyroidism of uremia. J Clin Endocrinol Metab 85:868–872 [DOI] [PubMed] [Google Scholar]
- Burger H, van Daele PL, Algra D, van den Ouweland FA, Grobbee DE, Hofman A, van Kuijk C, Schutte HE, Birkenhager JC, Pols HA (1994) The association between age and bone mineral density in men and women aged 55 years and over: the Rotterdam Study. Bone Miner 25:1–13 [DOI] [PubMed] [Google Scholar]
- Carling T, Ridefelt P, Hellman P, Rastad J, Akerstrom G (1997) Vitamin D receptor polymorphisms correlate to parathyroid cell function in primary hyperparathyroidism. J Clin Endocrinol Metab 82:1772–1775 [DOI] [PubMed] [Google Scholar]
- Crofts LA, Hancock MS, Morrison NA, Eisman JA (1998) Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc Natl Acad Sci USA 95:10529–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame MC, Pierce EA, DeLuca HF (1985) Identification of the porcine intestinal 1,25-dihydroxyvitamin D3 receptor on sodium dodecyl sulfate/polyacrylamide gels by renaturation and immunoblotting. Proc Natl Acad Sci USA 82:7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin LK, Haile RW, Ingles SA, Coetzee GA (1999) Vitamin D receptor 3′-untranslated region polymorphisms: lack of effect on mRNA stability. Biochim Biophys Acta 1453:311–320 [DOI] [PubMed] [Google Scholar]
- Fang R, Olds LC, Santiago NA, Sibley E (2001) GATA family transcription factors activate lactase gene promoter in intestinal Caco-2 cells. Am J Physiol Gastrointest Liver Physiol 280:G58–G67 [DOI] [PubMed] [Google Scholar]
- Fang Y, van Meurs JB, Bergink AP, Hofman A, van Duijn CM, van Leeuwen JP, Pols HA, Uitterlinden AG (2003) Cdx-2 polymorphism in the promoter region of the human vitamin D receptor gene determines susceptibility to fracture in the elderly. J Bone Miner Res 18:1632–1641 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Grant SF, Reid DM, Blake G, Herd R, Fogelman I, Ralston SH (1996) Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I α 1 gene. Nat Genet 14:203–205 [DOI] [PubMed] [Google Scholar]
- Halsall JA, Osborne JE, Pringle JH, Hutchinson PE (2005) Vitamin D receptor gene polymorphisms, particularly the novel A-1012G promoter polymorphism, are associated with vitamin D3 responsiveness and non-familial susceptibility in psoriasis. Pharmacogenet Genomics 15:349–355 [DOI] [PubMed] [Google Scholar]
- Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, Jurutka PW (1998) The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res 13:325–349 [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079 [DOI] [PubMed] [Google Scholar]
- Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA (1991) Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol 7:403–422 [DOI] [PubMed] [Google Scholar]
- Ingles SA, Haile RW, Henderson BE, Kolonel LN, Nakaichi G, Shi CY, Yu MC, Ross RK, Coetzee GA (1997) Strength of linkage disequilibrium between two vitamin D receptor markers in five ethnic groups: implications for association studies. Cancer Epidemiol Biomarkers Prev 6:93–98 [PubMed] [Google Scholar]
- Lambert JC, Luedecking-Zimmer E, Merrot S, Hayes A, Thaker U, Desai P, Houzet A, Hermant X, Cottel D, Pritchard A, Iwatsubo T, Pasquier F, Frigard B, Conneally PM, Chartier-Harlin MC, DeKosky ST, Lendon C, Mann D, Kamboh MI, Amouyel P (2003) Association of 3′-UTR polymorphisms of the oxidised LDL receptor 1 (OLR1) gene with Alzheimer’s disease. J Med Genet 40:424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker AC, Wahner HW, Dunn WL, Calvo MS, Harris TB, Heyse SP, Johnston CC Jr, Lindsay R (1998) Updated data on proximal femur bone mineral levels of US adults. Osteoporos Int 8:468–489 [DOI] [PubMed] [Google Scholar]
- Mann V, Hobson EE, Li B, Stewart TL, Grant SF, Robins SP, Aspden RM, Ralston SH (2001) A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest 107:899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey EV, Spector TD, Eyres KS, Fern ED, O’Rourke N, Vasikaran S, Kanis JA (1993) The assessment of vertebral deformity: a method for use in population studies and clinical trials. Osteoporos Int 3:138–147 [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Kesterson RA, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S, Inoue Y, Morita K, Takeda E, Pike JW (1997) Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol 11:1165–1179 [DOI] [PubMed] [Google Scholar]
- Morrison NA, Qi JC, Tokita A, Kelly PJ, Crofts L, Nguyen TV, Sambrook PN, Eisman JA (1994) Prediction of bone density from vitamin D receptor alleles. Nature 367:284–287 [DOI] [PubMed] [Google Scholar]
- Morrison NA, Yeoman R, Kelly PJ, Eisman JA (1992) Contribution of trans-acting factor alleles to normal physiological variability: vitamin D receptor gene polymorphism and circulating osteocalcin. Proc Natl Acad Sci USA 89:6665–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S, Godfrey L, Snook H, Rance H, Nutland S, Walker NM, Lam AC, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Tuomilehto-Wolf E, Tuomilehto J, Newport MJ, Clayton DG, Todd JA (2004) Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum Mol Genet 13:1633–1639 [DOI] [PubMed] [Google Scholar]
- Ogunkolade BW, Boucher BJ, Prahl JM, Bustin SA, Burrin JM, Noonan K, North BV, Mannan N, McDermott MF, DeLuca HF, Hitman GA (2002) Vitamin D receptor (VDR) mRNA and VDR protein levels in relation to vitamin D status, insulin secretory capacity, and VDR genotype in Bangladeshi Asians. Diabetes 51:2294–2300 [DOI] [PubMed] [Google Scholar]
- Saijo T, Ito M, Takeda E, Huq AH, Naito E, Yokota I, Sone T, Pike JW, Kuroda Y (1991) A unique mutation in the vitamin D receptor gene in three Japanese patients with vitamin D-dependent rickets type II: utility of single-strand conformation polymorphism analysis for heterozygous carrier detection. Am J Hum Genet 49:668–673 [PMC free article] [PubMed] [Google Scholar]
- Schuit SC, van der Klift M, Weel AE, de Laet CE, Burger H, Seeman E, Hofman A, Uitterlinden AG, van Leeuwen JP, Pols HA (2004) Fracture incidence and association with bone mineral density in elderly men and women: the Rotterdam Study. Bone 34:195–202 [DOI] [PubMed] [Google Scholar]
- Uitterlinden AG, Burger H, Huang Q, Fang Y, McGuigan FE, Grant SF, Hofman A, van Leeuwen JP, Pols HA, Ralston SH (1998a) Relation of alleles of the collagen type Iα1 gene to bone density and the risk of osteoporotic fractures in postmenopausal women. N Engl J Med 338:1016–1021 [DOI] [PubMed] [Google Scholar]
- Uitterlinden AG, Burger H, Huang Q, Yue F, McGuigan FE, Grant SF, Hofman A, van Leeuwen JP, Pols HA, Ralston SH (1998b) Relation of alleles of the collagen type 1A1 gene to bone density and the risk of osteoporotic fractures in postmenopausal women. N Engl J Med 338:1016–1021 [DOI] [PubMed] [Google Scholar]
- Uitterlinden AG, Pols HA, Burger H, Huang Q, Van Daele PL, Van Duijn CM, Hofman A, Birkenhager JC, Van Leeuwen JP (1996) A large-scale population-based study of the association of vitamin D receptor gene polymorphisms with bone mineral density. J Bone Miner Res 11:1241–1248 [DOI] [PubMed] [Google Scholar]
- Uitterlinden AG, Weel AE, Burger H, Fang Y, van Duijn CM, Hofman A, van Leeuwen JP, Pols HA (2001) Interaction between the vitamin D receptor gene and collagen type Iα1 gene in susceptibility for fracture. J Bone Miner Res 16:379–385 [DOI] [PubMed] [Google Scholar]
- Van der Klift M, De Laet CE, McCloskey EV, Hofman A, Pols HA (2002) The incidence of vertebral fractures in men and women: the Rotterdam Study. J Bone Miner Res 17:1051–1056 [DOI] [PubMed] [Google Scholar]
- van Meurs JB, Schuit SC, Weel AE, van der Klift M, Bergink AP, Arp PP, Colin EM, Fang Y, Hofman A, van Duijn CM, van Leeuwen JP, Pols HA, Uitterlinden AG (2003) Association of 5′ estrogen receptor alpha gene polymorphisms with bone mineral density, vertebral bone area and fracture risk. Hum Mol Genet 12:1745–1754 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Miyamoto K, Li B, Taketani Y, Kitano M, Inoue Y, Morita K, Pike JW, Takeda E (1999) The caudal-related homeodomain protein Cdx-2 regulates vitamin D receptor gene expression in the small intestine. J Bone Miner Res 14:240–247 [DOI] [PubMed] [Google Scholar]
- Young RP, Lau EM, Birjandi Z, Critchley JA, Woo J (1996) Interethnic differences in hip fracture rate and the vitamin D receptor polymorphism. Lancet 348:688–689 [DOI] [PubMed] [Google Scholar]
- Zon LI, Youssoufian H, Mather C, Lodish HF, Orkin SH (1991) Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc Natl Acad Sci USA 88:10638–10641 [DOI] [PMC free article] [PubMed] [Google Scholar]