Abstract
Since 1992 there have been seven major outbreaks of cholera in Argentina. Susceptibility analysis of 1,947 isolates (40% of reported cases) of Vibrio cholerae O1 biotype El Tor suggested the presence of extended-spectrum β-lactamases (ESBLs) in 28 isolates. Because of their different susceptibility profiles, V. cholerae isolates M1502, M1516, M1573, and M3030 (all of which are of the Ogawa serotype) were selected for the present study. By susceptibility analysis, isoelectric focusing, and PCR-based restriction fragment length polymorphism analysis, CTX-M-type enzymes were identified in three isolates, whereas a PER-2-type enzyme, in addition to a TEM-1-like enzyme, was identified in the other isolate. The presence of these ESBLs in V. cholerae isolates resulted in MICs well below those commonly observed for members of the family Enterobacteriaceae. Genes that encode both ESBLs were transferred to Escherichia coli by conjugation, together with all determinants of resistance to non-β-lactam antibiotics (gentamicin, kanamycin, and sulfamethoxazole for all isolates; amikacin and streptomycin for three isolates; trimethoprim, tetracycline, and chloramphenicol for two isolates). Plasmid profile analysis and Southern blotting revealed the presence of single plasmids of about 150 kb in the four V. cholerae isolates and their respective transconjugants and revealed that the plasmids harbored genes encoding CTX-M-type or PER-2-type ESBLs. These results strongly suggest the broad spread of these ESBLs among genera belong to families other than the Enterobacteriaceae.
Extended-spectrum β-lactamases (ESBLs) have been classified by Bush et al. (8) in functional group 2be or in Ambler molecular class A. These enzymes include in their spectra of activity penicillins, most cephalosporins, and aztreonam; they do not hydrolyze cephamycins (cefoxitin, cefotetan, and moxalactam) and carbapenems (imipenem and meropenem); and they are inhibited by clavulanic acid, tazobactam, and sulbactam (21). Since 1983, plasmidic ESBLs have been extensively reported worldwide, mainly among species of the family Enterobacteriaceae (17, 21; see http://www.lahey.org/studies/webt.htm). Most plasmidic ESBLs belong to the TEM or SHV type (21). Two novel sets of ESBLs that are unrelated to the TEM and SHV types and that belong to group 2be of Bush et al. (8) have been observed. First, there are the PER-type enzymes (3, 25, 26), which are more active against ceftazidime than cefotaxime. PER-1 was isolated from Pseudomonas aeruginosa (26), Salmonella enterica serovar Typhimurium (46), and Acinetobacter spp. (47), whereas PER-2 was reported in members of the family Enterobacteriaceae from Argentina (3; M. F. Galas, F. G. Pasterán, R. G. Melano, A. E. Petroni, G. Lopez, A. C. Corso, M. A. Rossi, and WHONET Collaborative Group, 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E109, 1998; M. F. Galas, M. J. Rapoport, F. G. Pasterán, R. G. Melano, A. E. Petroni, P. G. Ceriana, WHONET-Argentina Collaborative Group, and M. A. Rossi, 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1474, 1999; F. G. Pasteran, R. G. Melano, M. F. Galas, M. M. Rodriguez, WHONET-Argentina Collaborative Group, and M. A. Rossi, 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1475, 1999). Second, there are the CTX-M β-lactamases, which are more active against cefotaxime than ceftazidime (for a recent review, see reference 45). These enzymes have been observed in members of the family Enterobacteriaceae since 1990, and 14 members of the family have been reported to date (5, 6, 12, 18, 27, 36, 45).
Nowadays, CTX-M-producing enterobacteria appear to be widely spread over very distant geographic regions including Japan and Taiwan (45, 49), several European countries (9, 12, 27, 36, 38, 45), South America (5, 6, 45), and very recently, Kenya (18). In particular, CTX-M-2 was broadly detected mainly in Argentina and also in neighboring countries (31, 35, 45; Galas et al., 38th ICAAC)). Comparisons of the amino acid sequences of the CTX-M family showed four clusters or groups. The first group includes CTX-M-1, CTX-M-3, CTX-M-10, and CTX-M-12; the second group comprises CTX-M-2, CTX-M-4 to CTX-M-7, and Toho-1; the third group contains Toho-2, CTX-M-9, and CTX-M-16; and the fourth group consists of CTX-M-8 (5, 6, 18).
The broad spread of these enzymes among human pathogenic bacteria which have never been shown to harbor ESBLs could constitute a major health concern and an epidemiologic problem. Since 1991 Vibrio cholerae O1 biotype El Tor has become one of the most clinically important reemerging pathogens in Latin America because it has caused cholera outbreaks and endemic diarrhea. From 1992 to 1998, during seven cholera seasons in Argentina, 1,947 isolates of V. cholerae O1 biotype El Tor (40.3% of isolates from reported cases) were received at the National Reference Laboratory of Argentina. Ampicillin resistance was detected in 34 of 1,947 isolates. Surprisingly, 28 of the ampicillin-resistant isolates showed strong reductions in their susceptibilities to extended-spectrum cephalosporins (ESCs) (cefotaxime and/or ceftazidime), cefoperazone, and aztreonam. ESC resistance was reversed by clavulanic acid, suggesting the presence of ESBLs. In this work, we report on the first identification in V. cholerae of two plasmid-mediated ESBLs: CTX-M-type and PER-2-type ESBLs. These facts strongly suggest the broad spread of these enzymes among genera belonging to families other than the Enterobacteriaceae.
(This study was presented in part at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, 24 to 27 September 1998, San Diego, Calif. [M. Galas, A. Petroni, R. Melano, A. Corso, M. Rodriguez, M. L. Cacace, A. M. Bru, and A. Rossi, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C174, 1998].)
MATERIALS AND METHODS
Bacterial strains.
From February 1992 to June 1998, a total of 1,947 V. cholerae isolates were received at the National Reference Laboratory (Table 1). Susceptibility analysis was carried out by the disk diffusion method according to the specifications of NCCLS (24) (adopting the criteria for the family Enterobacteriaceae for antimicrobial agents other than ampicillin, tetracycline, trimethoprim-sulfamethoxazole, chloramphenicol, and sulfonamides), and 28 isolates showed resistance to cefotaxime and/or ceftazidime, cefoperazone, and aztreonam. These V. cholerae isolates (all isolates were of the Ogawa serotype) were grouped according to their susceptibility profiles, and one isolate from each group was selected to carry out this study, namely, isolates M1502, M1516, M1573, and M3030 (Table 1). V. cholerae M1502 has already been described (34). V. cholerae M1516 was isolated from a 43-year-old female outpatient with vomiting and moderate dehydration in Tartagal, Argentina (north of Salta Province), in March 1993. This isolate was one of three isolates that had identical susceptibility profiles and that had been detected in members of the same family. V. cholerae M1573 was isolated from a hospitalized newborn male with diarrhea who had been treated with ampicillin plus gentamicin in Tartagal (April 1993). ESC-resistant V. cholerae isolates were not detected during the following three cholera seasons (a 4-year period). Toward the end of the sixth cholera season (March to April 1997) a small outbreak due to an ESC-resistant strain of V. cholerae O1 occurred in Irigoyen and Pichanal, Argentina (north of Salta Province). The outbreak began with a 1-year-old male patient who acquired the illness during a hospitalization and rapidly spread into the community, resulting in 24 cases in seven families. The representative isolate selected for this study (isolate M3030) was isolated from a 48-year-old woman in the first family affected; she had initially been treated with a single dose of doxycycline.
TABLE 1.
Susceptibility profile distributions for ESC-resistant V. cholerae isolates from the seven Argentinean cholera seasons
| Cholera season (time period)a | No. of reported casesb | No. of NRL casesc | Relevant susceptibility profile of ESC-resistant isolates (no. of isolates)d | Isolate selected |
|---|---|---|---|---|
| 1 (1992) | 446 | 122 | NDe | |
| 2 (1992-1993) | 1,633 | 646 | AMP-CFP-CTX-ATM-GEN-KAN-SFI (1) | M1502 |
| AMP-CFP-CTX-ATM-AMK-GEN-KAN-STR- CHL-ERY-SXT-TET-O129 (3) | M1516 | |||
| AMP-CFP-CTX-CAZ-ATM-GEN-STR-SFI (1) | M1573 | |||
| 3 (1993-1994) | 1,442 | 497 | ND | |
| 4 (1994-1995) | 185 | 117 | ND | |
| 5 (1995-1996) | 427 | 265 | ND | |
| 6 (1996-1997) | 688 | 291 | AMP-CFP-CTX-ATM-AMK-GEN-KAN-STR- CHL-NIT-SXT-TET-O129 (23) | M3030 |
| 7 (1998) | 12 | 9 | ND |
Cholera seasons comprised time periods which generally extended from October to May.
Reported cases were recorded by the Sistema Nacional de Vigilancia Epidemiológica, Ministerio Nacional de Salud y Acción Social.
Number of isolates analyzed at the National Reference Laboratory (NRL).
Resistance or reduced susceptibility are indicated. Abbreviations: AMK, amikacin; AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CFP, cefoperazone; CHL, chloramphenicol; CTX, cefotaxime; ERY; erythromycin; GEN, gentamicin; KAN, kanamycin; NIT, nitrofurantoin; O129, vibriostatic agent; SFI, sulfisoxazole; STR, streptomycin; SXT; trimethoprim-sulfamethoxazole; TET, tetracycline.
ND, no ESC-resistant isolates were detected.
A susceptible V. cholerae 2717 O1 biotype El Tor isolate used in the susceptibility profile analysis was isolated (February 1993) from a 16-month-old female patient with diarrhea, vomiting, and dehydration symptoms.
Spontaneous mutants of Escherichia coli C600 and ER1793 (New England Biolabs, Beverly, Mass.) resistant to nalidixic acid and rifampin were obtained as described previously (37) and were used as recipient strains for bacterial conjugation experiments.
Media and chemicals.
Mueller-Hinton broth and agar, tryptic soy broth, and Luria-Bertani (LB) broth and agar were from Difco (Detroit, Mich.). Restriction enzymes were from New England Biolabs. Routine chemicals were from Merck (Darmstadt, Germany).
Antimicrobial agents and susceptibility testing.
The following antimicrobial agents were obtained from standard laboratory powders and the indicated sources: amikacin, aztreonam, and cefepime, Bristol-Myers Squibb; amoxicillin, Roemmers; clavulanic acid, SmithKline Beecham; ampicillin, Temis Lostaló; chloramphenicol, Parke-Davis; cefotaxime, Argentia; cefoxitin, imipenem, and norfloxacin, Merck Sharp & Dohme; ceftazidime, Glaxo; ceftibuten, gentamicin, nitrofurantoin, sulfamethoxazole, and trimethoprim, Schering Plough; cephalothin, erythromycin, piperacillin, streptomycin, and tazobactam, Wyeth-Ayerst; kanamycin, Armstrong; sulfisoxazole, Abbott; tetracycline, Microsules y Bernabó; and vibriostatic agent O129, Sigma.
MICs were determined by the agar dilution technique according to the specifications of NCCLS (23) by adoption of the criteria for the family Enterobacteriaceae for antimicrobial agents other than ampicillin, tetracycline, trimethoprim-sulfamethoxazole, chloramphenicol, and sulfonamides. Clavulanic acid was used at a ratio of 1 to 2 in combination with amoxicillin or at a constant concentration of 4 μg/ml in combinations with other β-lactams. Tazobactam was used at a constant concentration of 4 μg/ml.
The following reference strains were used for quality control of dilution tests: P. aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and E. coli ATCC 25922 and ATCC 35218. Since the MICs of antimicrobial agents for V. cholerae 2717 were in the range of those previously reported for susceptible isolates (34, 48), this isolate was used as a control strain for analysis of the V. cholerae O1 biotype El Tor susceptibility profile.
Conjugation assays.
Biparental conjugations were performed as follows. Cells of both the donor and the recipient strains were mixed on LB agar at a ratio of 5 to 1, and the mixture was incubated for 18 h at 35°C. Transconjugants were selected on Mueller-Hinton agar supplemented with nalidixic acid (50 μg/ml) and cefotaxime (8 μg/ml) or ceftazidime (0.5 μg/ml), as indicated.
β-Lactamase preparation.
Bacterial cultures were grown overnight at 35°C in 10 ml of brain heart infusion broth. Cells were harvested by centrifugation, washed, and resuspended in 0.5 ml of 10 mM phosphate buffer (pH 7.0). Cell suspensions were disrupted at 4°C by sonication (two pulses of 45 s at 20 Hz, separated by an interval of 30 s, on a Vibra Cell Sonifier [Sonics and Materials Inc., Danbury, Conn.]) and centrifuged (17,000 × g for 10 min at 4°C) for debris removal. The supernatants were used for isoelectric focusing (IEF) assays.
IEF of β-lactamases.
Crude β-lactamase extracts were focused on broad-range precast polyacrylamide gels (pH 3.5 to 9.5; Ampholine PAGplate; Pharmacia Biotech, Uppsala, Sweden) with a Multiphor II apparatus (Pharmacia LKB, Uppsala, Sweden), according to the instructions of the manufacturer. β-Lactamase bands were visualized by the iodometric method described by Labia and Barthélémy (20), with minor modifications. To differentiate between bands produced by broad-spectrum β-lactamase or ESBL activities, after IEF the gel was covered with an agar overlay containing cephaloridine (1 mg/ml) plus penicillin (0.1 mg/ml) or ceftazidime (1 mg/ml) plus ceftriaxone (1 mg/ml) as developing substrates, respectively. The gel was incubated at room temperature until the bands could be visualized. For β-lactamase inhibition procedures, the gel was covered with a piece of filter paper impregnated with 1 mM clavulanic acid or cefoxitin, as indicated, and incubated for 10 min at room temperature before development, as described above. The following β-lactamases with known pIs were used as standards: TEM-1 (pI 5.4), PER-2 (pI 5.4), SHV-2 (pI 7.6), P99 (pI 7.8), CTX-M-2 (pI 7.9), and SHV-5 (pI 8.2).
PCR amplifications and PCR-RFLP analysis.
Primers specific for the TEM and SHV β-lactamase gene families (primers PTEM and PSHV, respectively) (1) were used for PCRs. For amplification of complete structural genes highly related to that for the CTX-M-2 enzyme (blaCTX-M-2), the following primers were designed from its reported DNA sequence (2): 5′-CGGAATTCATGATGACTCAGAGCATTCG-3′ and 5′-GCTCTAGATTATTGCATCAGAAACCGTG-3′ (primers PCTX-M-2 forward and PCTX-M-2 reverse, respectively). For the identification of the gene encoding PER-2-type enzymes (blaPER-2-type), primers 5′-GTAGTATCAGCCCAATCCCC-3′ and 5′-CCAATAAAGGCCGTCCATCA-3′ (primers PPER forward and PPER reverse, respectively) were designed from the reported DNA sequence (3). PCR amplifications were performed with a Perkin-Elmer Cetus thermal cycler in a final volume of 50 μl containing 20 pmol of each primer, 25 μM each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 2.5 U of Taq polymerase (Promega, Madison, Wis.). DNA templates were prepared by lysing one or two colonies of V. cholerae isolates or E. coli transconjugants in 50 μl of boiling water, as described previously (28, 29). Two microliters was used for the PCR. The PCR program was 10 min of denaturation at 94°C; 25 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 30 s of extension at 72°C; and a final extension step of 5 min at 72°C. The resulting amplification products were run in a 1.5% agarose gel and purified by using a Wizard PCR Preps DNA Purification System (Promega), according to the recommendations of the manufacturer. Purified products were used for PCR-based restriction fragment length polymorphism (PCR-RFLP) analysis, as described previously (37), or for construction of the labeled probes used in the hybridization assays.
Plasmid profile analysis and hybridization.
Plasmid DNA was purified from V. cholerae or E. coli transconjugant cells as described previously (22). Plasmidic DNAs (about 250 ng) were analyzed by electrophoresis in 0.7% agarose gels (Tris-acetate buffer), by using plasmids of known size as standards, and were then transferred onto and immobilized on a nylon transfer membrane (Zeta-Probe GT; Bio-Rad, Richmond, Calif.). DNA-DNA hybridizations were performed by the method described by Southern (39). Labeling of purified PCR products used as probes and stripping and reprobing of the membranes were performed with a Dig DNA Labeling and Detection kit (Boehringer Mannheim, Mannheim, Germany) by the procedures recommended by the manufacturer.
RESULTS
Antibiotic susceptibilities of V. cholerae isolates and conjugational transfer of resistance.
The susceptibility profiles of V. cholerae M1502, M1516, M1573, and M3030 are given in Table 2. With the exception of the MICs of cefoxitin and imipenem, the MICs of all β-lactams tested were higher for these four isolates than for susceptible isolate 2717. Two different susceptibility profiles for β-lactams were recognized, and the four isolates were grouped into groups A (V. cholerae M1502, M1516, and M3030) and B (isolate M1573). The MICs of cephalothin, cefotaxime, and cefepime were eight, four, and two times higher, respectively, for the group A isolates than for the group B isolate, whereas the MICs of ceftazidime and ceftibuten for the group A isolates were essentially the same as those for V. cholerae 2717. The MICs of ceftazidime, ceftibuten, and aztreonam were 64, 64, and 16 times higher, respectively, for the group B isolate than for the group A isolates.
TABLE 2.
Antibiotic susceptibilities of V. cholerae isolates and E. coli transconjugants
| Agent(s)a | MIC (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
V. cholerae isolates
|
E. coli transconjugants
|
Susceptible strainsb
|
|||||||||
| Group A
|
Group B, M1573 | Group A
|
Group B, M1561 | ||||||||
| M1502 | M1516 | M3030 | M1538 | M3099 | M3033 | 2717 | C600 | ER1793 | |||
| AMPc | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 | 4 | 2 | 4 |
| AMC | 8 | 4 | 8 | 16 | 16 | 16 | 16 | 16 | 4 | 4 | 4 |
| PIP | 128 | 128 | 128 | 128 | 1,024 | 1,024 | 1,024 | 256 | <0.25 | 0.5 | 1 |
| TZP | 0.25 | <0.25 | <0.25 | 1 | 1 | 1 | 1 | 4 | 0.25 | 0.5 | 1 |
| CEF | 512 | 128 | 256 | 32 | 1,024 | >1,024 | >1,024 | 128 | 2 | 4 | 8 |
| FOX | 8 | 4 | 8 | 4 | 4 | 4 | 4 | 4 | 8 | 2 | 4 |
| CTX | 4 | 2 | 2 | 0.5 | 64 | 128 | 128 | 8 | <0.03 | <0.03 | <0.03 |
| CTX-CLA | <0.03 | <0.03 | <0.03 | <0.03 | 0.06 | 0.13 | 0.13 | <0.03 | <0.03 | <0.03 | <0.03 |
| CAZ | 0.25 | 0.13 | 0.13 | 8 | 2 | 8 | 4 | 128 | 0.13 | 0.13 | 0.13 |
| CAZ-CLA | 0.13 | 0.06 | 0.06 | 0.13 | 0.13 | 0.25 | 0.25 | 0.13 | 0.13 | 0.13 | 0.06 |
| FEP | 2 | 2 | 2 | 1 | 16 | 32 | 64 | 8 | <0.5 | <0.5 | <0.5 |
| FEP-CLA | <0.50 | <0.50 | <0.50 | <0.50 | <0.50 | <0.50 | <0.50 | <0.50 | <0.5 | <0.5 | <0.5 |
| CTB | 0.50 | 1 | 1 | 64 | 2 | 4 | 4 | 64 | 0.25 | 0.25 | 0.25 |
| CTB-CLA | 0.25 | 0.25 | 0.25 | 0.25 | 0.50 | 0.25 | 0.50 | 0.50 | 0.25 | 0.25 | 0.25 |
| ATM | 8 | 8 | 8 | 128 | 16 | 32 | 64 | 128 | 1 | 0.06 | 0.06 |
| IPM | 1 | 1 | 1 | 1 | 0.13 | 0.25 | 0.25 | 0.13 | 1 | 0.13 | 0.13 |
| AMK | 16 | 16 | 16 | 4 | 4 | 8 | 4 | 0.25 | 4 | 0.5 | 0.25 |
| GEN | 256 | 128 | 256 | 256 | 64 | 256 | 256 | 32 | 1 | 0.25 | <0.13 |
| KAN | 256 | 256 | 256 | 8 | 128 | 128 | 64 | 8 | 4 | 0.50 | 0.50 |
| STR | 8 | 256 | 256 | 128 | 1 | >2,048 | >2,048 | 32 | 8 | 1 | 2,048 |
| CHLc | <1 | 4 | 2 | 1 | 4 | 128 | 32 | 4 | <1 | 8 | 4 |
| ERYc | 0.50 | 32 | 0.50 | 0.50 | 16 | 256 | 32 | 32 | 0.50 | 16 | 32 |
| TMP | <0.12 | 512 | 256 | <0.12 | <0.12 | 512 | 512 | 1 | <0.12 | <0.12 | <0.12 |
| SFI | 1,024 | 1,024 | 1,024 | 1,024 | >1,024 | >1,024 | >1,024 | >1,024 | 16 | 8 | 4 |
| SXTc | 0.25 | 32 | 32 | <0.13 | <0.13 | 64 | 64 | <0.13 | <0.13 | <0.13 | <0.13 |
| TETa | 0.25 | 2 | 4 | 0.25 | 1 | 64 | 64 | 1 | 0.25 | 2 | 1 |
| NORc | <0.004 | <0.004 | <0.004 | <0.004 | 0.25 | 0.50 | 0.50 | 0.50 | <0.004 | 0.25 | 0.50 |
| NITc, d | 2 | 4 | 32 | 4 | 8 | 8 | 8 | 8 | 2 | 8 | 8 |
| O129 | 4 | >256 | 256 | 2 | 32 | >256 | 256 | 128 | 2 | 32 | 16 |
Abbreviations: AMC, amoxicillin-clavulanic acid; CLA, clavulanic acid; CTB, ceftibuten; CEF, cephalothin; FOX, cefoxitin; FEP, cefepime; IPM, imipenem; NOR, norfloxacin; PIP, piperacillin; TZP, piperacillin-tazobactam; TMP; trimethoprim. The abbreviations for the remaining agents are given in footnote d of Table 1.
V. cholerae 2717 is a susceptible isolate. E. coli C600 and ER1793 (STRr) were used as recipient strains for bacterial conjugations.
First-line antibiotics for treatment of V. cholerae infections (4).
The use of nitrofurantoin is equivalent to the use of furazolidone, the MICs of which for susceptible isolates from Argentina were in the range 0.5 to 0.12 μg/ml.
In order to analyze if the V. cholerae resistance determinants were harbored in conjugative plasmids, isolates M1502 and M1573 were mated with E. coli C600 (selections were carried out with nalidixic acid plus cefotaxime for M1502 or with nalidixic acid plus ceftazidime for M1573), rendering transconjugants M1538 and M1561, respectively. In addition, V. cholerae M1516 and M3030 were conjugated with E. coli ER1793 (selections were achieved with nalidixic acid plus cefotaxime), rendering transconjugants M3099 and M3033, respectively. The susceptibility profile of each E. coli transconjugant matched that of the respective donor V. cholerae isolate (Table 2). For the group A transconjugant strains, the MICs of cefotaxime (64 to 128 μg/ml) and cefepime (16 to 64 μg/ml) were the highest and the MICs of ceftazidime (2 to 8 μg/ml), ceftibuten (0.5 to 1 μg/ml), and aztreonam (16 to 64 μg/ml) were the lowest. Otherwise, for the group B transconjugant strain, the MICs of ceftazidime (128 μg/ml), ceftibuten (32 μg/ml), and aztreonam (128 μg/ml) were the highest and the MICs of cefotaxime (8 μg/ml) and cefepime (8 μg/ml) were the lowest. For both groups A and B, for either V. cholerae isolates or E. coli transconjugants, clavulanate reduced the MICs nearly to the MICs for the susceptible strains.
Even though selection of transconjugants was achieved only with cefotaxime or ceftazidime, determinants of resistance to all non-β-lactam antibiotics, with the exception of reduced susceptibility to nitrofurantoin, shown by V. cholerae M3030, were also transferred to E. coli: gentamicin, kanamycin, and sulfisoxazole for four isolates; amikacin and streptomycin for three isolates; and trimethoprim, tetracycline, and chloramphenicol for M1516 and M3030, which were also resistant to vibriostatic agent O129.
IEF of β-lactamases.
IEF of crude homogenates of group A V. cholerae isolates (M1502, M1516, and M3030) and their respective E. coli transconjugants (M1538, M3099, and M3033) showed both broad-spectrum β-lactamase activities at pI 5.4 and ESBL activities at the same pI as that for the CTX-M-2 standard (pI 7.9). All these activities were inhibited by clavulanic acid. IEF of crude extracts of V. cholerae M1573 and E. coli M1561 (group B) revealed the presence of ESBL activities at the same pI as that for the PER-2 standard (pI 5.4), and the activities were also susceptible to clavulanic acid inhibition. The presence of a broad-spectrum β-lactamase activity at pI 5.4 in M1573 and M1561 could not be investigated by the standard IEF assay because of the overlapping ESBL band at the same pI. Therefore, in situ inhibition with cefoxitin was performed on the IEF gel. The ESBL activity at pI 5.4, like the activity of the PER-2 standard, was inhibited, whereas the broad-spectrum β-lactamase activity at pI 5.4 and the ESBL activity at pI 7.9 were not.
Identification of β-lactamases by PCR-RFLP analysis.
Both β-lactam susceptibility profiles and IEF analysis suggested the presence of a CTX-M-type enzyme and a PER-like enzyme in the transconjugant strains of groups A and B, respectively. Therefore, PCR assays were carried out with total DNA from the four V. cholerae isolates and their respective E. coli transconjugants as templates and with primers PCTX-M-2, PPER, PTEM, or PSHV. When primers PCTX-M-2 were used, only templates from the group A V. cholerae isolates and their respective transconjugants produced amplification products which were of the same size (900 bp), as expected from the computational analysis of the reported DNA sequences of the genes highly related to blaCTX-M-2. The group of highly related genes comprised genes encoding CTX-M-2 (2), CTX-M-4 (13), CTX-M-5 (7), CTX-M-6 and CTX-M-7 (12), and Toho-1 (16). To differentiate among these possibilities, we designed a PCR-RFLP analysis with restriction enzyme BsaHI, which includes in its recognition sequence the single point mutation between the CTX-M-2 and the Toho-1 structural genes (Table 3). The amplification products obtained were not digested by BsaHI and, in addition, showed the same restriction map for EcoRV, HincII, PstI, and SphI compared to that for the blaCTX-M-2 gene (Table 3).
TABLE 3.
PCR-RFLP analysis to differentiate among genes highly related to blaCTX-M-2a
| Substrate | Length(s) (bp) of DNA fragment(s) obtained after digestion with:
|
||||||
|---|---|---|---|---|---|---|---|
| BsaHI | BsaHI + EcoRV | EcoRV | BsaHI + PstI | PstI | SphI + PstI | SphI | |
| Group A | 900 | 750, 150 | 750, 150 | 500, 400 | 500, 400 | 500, 310, 90 | 810, 90 |
| CTX-M-2 | 900 | 752, 148 | 752, 148 | 500, 400 | 500, 400 | 500, 310, 90 | 810, 90 |
| Toho-1 | 833, 67 | 752, 81, 67 | 752, 148 | 433, 400, 67 | 500, 400 | 500, 310, 90 | 810, 90 |
| CTX-M-4 | 509, 391 | 391, 361, 148 | 752, 148 | 500, 391, 9 | 500, 400 | 500, 310, 90 | 810, 90 |
| CTX-M-5 | 900 | 752, 148 | 752, 148 | 500, 400 | 500, 400 | 500, 400 | 900 |
| CTX-M-6 | 509, 385, 6 | 385, 361, 148, 6 | 752, 148 | 500, 385, 9, 6 | 500, 400 | 500, 400 | 900 |
| CTX-M-7 | 509, 391 | 391, 361, 148 | 752, 148 | 500, 391, 9 | 500, 400 | 500, 310, 90 | 810, 90 |
The percent identities between blaCTX-M-2 and Toho-1, CTX-M-4, CTX-M-5, CTX-M-6, and CTX-M-7 were 99.9, 98.1, 99.2, 97.5, and 98.4% respectively (NALIGN program; PCGene; IntelliGenetics). PCR products from amplifications with PCTX-M-2 and whole DNA extracted from group A V. cholerae isolates and their derivative transconjugants were incubated with restriction enzymes, and the fragment sizes (boldface type) were estimated after electrophoresis on a 1.8% agarose gel. The sizes of the restriction fragments from the theoretical PCR products from amplifications with PCTX-M-2 and genes highly related to blaCTX-M-2 were deduced from the reported DNA sequences.
The PPER primers generated amplification products (740 bp) only with the templates from the group B V. cholerae isolate and its transconjugant. Because of the high degree of DNA homology between the blaPER-1 and blaPER-2 genes, the PCR assay did not allow differentiation between such genes. A PCR-RFLP analysis carried out with both group B amplification products and HindIII, PstI, and SphI showed a unique restriction fragment profile which was identical to that expected for the blaPER-2 gene and different from that expected for the blaPER-1 gene (data not shown). The use of PTEM primers with all DNA templates produced amplification products of 500 bp, which is of the expected size of products from a gene encoding a TEM-like enzyme, while the use of PSHV primers produced no amplification product.
Plasmid profiles and locations of genes encoding ESBLs.
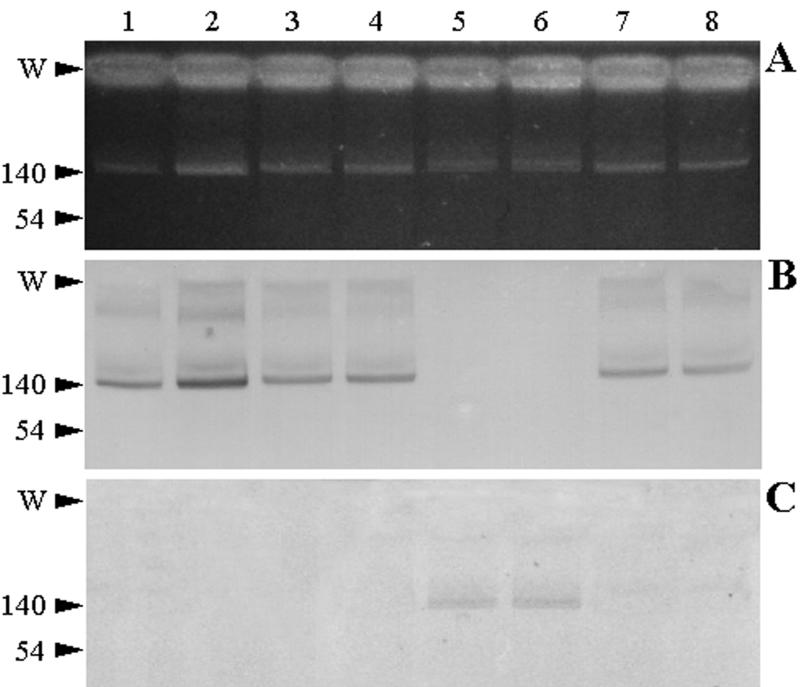
Analysis of the plasmid profiles of V. cholerae M1502, M1516, M1573, and M3030 and their respective transconjugants (M1538, M3099, M1561, and M3033) showed the presence of single plasmids of about 150 kb in each strain (Fig. 1A). The DNA hybridization analysis with the 900-bp PCR product from group A isolates as a probe located the gene encoding the CTX-M-type enzyme on the megaplasmids of group A isolates and their respective transconjugants (Fig. 1B). Reprobing of the same membrane with the 740-bp PCR product from the group B isolate as a probe showed that the megaplasmid of this isolate and its transconjugant harbored the gene encoding the PER-2-type ESBL (Fig. 1C).
FIG. 1.
Plasmid profiles and Southern blots of V. cholerae isolates and their derivative transconjugants. Lanes: 1, 3, 5, and 7, V. cholerae isolates M1502, M1516, M1573, and M3030, respectively; 2, 4, 6, and 8, E. coli transconjugants M1538, M3099, M1561, and M3033, respectively. (A) Plasmid DNA was extracted as indicated in Materials and Methods and analyzed on a 0.7% agarose gel. (B) The gel was transferred and blotted onto a nylon membrane and hybridized against a 900-bp PCR product from group A isolates as a probe. (C) The membrane was stripped and reprobed with the 740-bp PCR product from the group B isolate. The locations of the loading wells (W) and reference plasmids (numbers indicate sizes [in kilobases]) are shown on the left.
DISCUSSION
The most common mechanism of resistance to β-lactam antibiotics, including recent cephalosporins, is the production of a β-lactamase enzyme. Since 1971 transferable ampicillin resistance have been described in V. cholerae isolates from Africa, Asia, and Latin America (10, 19, 32, 33, 42, 43, 44). However, only two plasmidic β-lactamases have been identified to date. First, TEM-1 a broad-spectrum β-lactamase was identified in isolates from Asia and Africa (10, 33). Second, SAR-1, a carbenicillinase with ampicillin-hydrolyzing activity lower than that of TEM-1, was described in only two isolates from Tanzania (33). In previous work (34), we reported for the first time on the isolation of an ESBL-producing V. cholerae strain (i.e., isolate M1502). In the present work, in addition to a TEM-1-type β-lactamase, we have identified two extended-spectrum plasmid-borne enzymes not related to TEM- or SHV-type enzymes in V. cholerae O1 isolates recovered during several cholera seasons in Argentina. For group A isolates, the β-lactam susceptibility profiles of the transconjugant strains, for which the cefotaxime/ceftazidime MIC ratio was 32, were almost identical to those reported for strains producing several CTX-M enzymes (16, 45), even though different E. coli recipient strains (C600 and ER1793) were used in the mating experiments. The results of IEF and PCR-RFLP assays suggest the presence of either CTX-M-2 or a highly related enzyme different from those grouped in the CTX-M-2 cluster already reported (5, 45). Otherwise, for the group B isolate, the β-lactam susceptibility profile of the transconjugant strain, for which the cefotaxime/ceftazidime MIC ratio was 0.06, was very similar to those reported for strains producing PER enzymes (26; A. Bauernfeind et al., Abstr. 34th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C74, 1994). These data, in conjunction with the results of IEF and PCR-RFLP analysis, are consistent with the presence of a PER-2-type enzyme in V. cholerae M1561 and its transconjugant, M1573. Cefoxitin inhibition of this enzyme, performed on the IEF gel before the iodometric method was carried out, allowed the identification of TEM-1-like activity.
The expression of CTX-M-type and PER-2-type enzymes in V. cholerae isolates resulted in MICs of β-lactams lower than those commonly observed for CTX-M- or PER-2-producing strains that belong to the family Enterobacteriaceae (with the exception of the MICs for β-lactams which are poor substrates for these enzymes, such as cefoxitin and imipenem). This fact was more evident with ESCs, such as cefotaxime, ceftazidime, and cefepime, for which the MICs were 32, 32, and 16 times higher, respectively, for the E. coli transconjugants than for the V. cholerae isolates. Therefore, differences between V. cholerae and enterobacteria would exist in terms of the levels of expression of ESBLs, the susceptibilities of penicillin-binding proteins to such β-lactam agents, or the periplasmic concentrations of the drugs (see below).
Genetic determinants conferring resistance to non-β-lactam antimicrobial agents were also encoded by the 150-kb plasmid harbored in each V. cholerae isolate. Multidrug resistance in V. cholerae O1 isolates has so far been linked to plasmids, and the presence of self-transmissible multidrug-resistant plasmids from 100 to 200 kb has been reported in V. cholerae O1 isolates from Asia, Africa, and Ecuador (10, 14, 19, 33, 40, 41, 43) and, more recently, in isolates from Albania, Italy, and Brazil (11, 15). However, plasmids such those harbored by isolates M1516 and M3030, which encode resistance to gentamicin, kanamycin, amikacin, streptomycin, trimethoprim-sulfamethoxazole, tetracycline, and chloramphenicol and, in the case of M1516, to erythromycin, in addition to β-lactam antibiotic resistance, have not previously been reported. Moreover, to the best of our knowledge, gentamicin resistance was reported only in V. cholerae O1 isolates with two different resistance patterns (R types) from two small outbreaks that occurred in Dacca, Bangladesh, in 1981 (41) and in a single nosocomial pediatric ward in Samutsakorn, Thailand, in 1982 (40). In this work we have reported on gentamicin resistance in V. cholerae O1 isolates with four different R types from two distantly occurring cholera seasons (4 years between the outbreaks). In addition, all the ESBL-producing V. cholerae isolates were coresistant to gentamicin, as has been observed for ESBL-producing enterobacterial isolates from Argentina (Galas et al., 38th ICAAC; Galas et al., 39th ICAAC; Pasterán et al., 39th ICAAC). This fact suggests that both resistance determinants may be easily cosegregated and stably maintained.
The MICs of tetracycline and chloramphenicol for both V. cholerae isolates resistant to such drugs (M1516 and M3030) were 32- to 16-fold lower than those for their respective E. coli transconjugant strains (M3099 and M3033, respectively), whereas no differences in susceptibilities to aminoglycosides, trimethoprim, and sulfisoxazole were observed between the original hosts and the derived transconjugants. This differential expression of tetracycline and chloramphenicol resistance mechanisms between V. cholerae and E. coli has been reported previously (30). This fact, in addition to the low levels of resistance to β-lactams discussed above, may be due to differences between the E. coli envelope and that of V. cholerae, which could lack a permeability barrier or an active multidrug efflux mechanism that enhances antibiotic input into the cell.
Although replacement of the fluid lost in the stool remains the crucial element for the treatment of patients with cholera, antimicrobial therapy is also important, since this can reduce the volume of stool purged during illness, as well as shorten the duration of symptoms and the excretion of vibrios in the feces (4). In addition, antimicrobial drugs are also used as chemoprophylaxis in close human contacts in attempts to control the epidemic spread from patients found to have cholera. To date, seven drugs have been recommended for the treatment of cholera: ampicillin, doxycycline (a long-acting form of tetracycline), co-trimoxazole, erythromycin, furazolidone, chloramphenicol, and norfloxacin (4). Multidrug-resistant plasmids harbored by isolate M1516 or M3030 carried genetic determinants that conferred resistance to four (isolate M3030) or five (isolate M1516) of such first-line antimicrobial agents. The clinical relevance of these facts has not been established and deserves special attention when one is using prophylaxis measures.
In this work we have also reported on the first cholera outbreak due to an ESBL-producing isolate. The outbreak comprised 24 reported cases in a community of sugarcane workers in Irigoyen and Pichanal, Argentina, which are north of Salta Province. Interestingly, the V. cholerae O1 strain isolated from the first cholera patient in the outbreak (a 1-year-old male who acquired the illness during a hospitalization) showed reduced susceptibility to nitrofurantoin, which was a common characteristic of isolates from the sixth cholera epidemic in Argentina, but was susceptible to all other antibiotics tested. Furthermore, an E. coli strain isolated from the urine of the second case patient in the outbreak (the first one from whom an ESC-resistant V. cholerae strain had been isolated) showed the same resistance phenotype as that for transconjugant strain M3033. These data suggest that multidrug-resistant V. cholerae isolates could have resulted from the acquisition of the 150-kb plasmid encoding multidrug resistance in a conjugation event with E. coli. The fact that CTX-M-2 is the most frequent ESBL observed in E. coli and members of the family Enterobacteriaceae in Argentina (31, 35; Galas et al., 38th ICAAC; Galas et al., 39th ICAAC; Pasterán et al., 39th ICAAC; A. C. Gales, T. M. Lewis, J. M. Casellas, V. Prado, J. Smayevsky, and R. N. Jones, 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1487, 1999) supports this hypothesis. In addition, the risks for the potential spread of antimicrobial resistance genes from V. cholerae to other bacteria have been pointed out (19). Whatever the case, the findings of this work point out the broad spread of ESBLs among genera belonging to families other than the Enterobacteriaceae.
Acknowledgments
We thank Norma Binztein and Marta Rivas for providing epidemiologic data from the Argentinean cholera seasons; María Inés Caffer for serotyping the V. cholerae isolates; and Marisa Rodríguez, Gustavo López, and Ezequiel Tuduri Franco for technical assistance.
Footnotes
Dedicated to the memory of Alicia Rossi.
REFERENCES
- 1.Arlet, G., and A. Philippon. 1989. Construction by polymerase chain reaction and intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 82:19-26. [DOI] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, Ö. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennish, M. L. 1994. Cholera: pathophysiology, clinical features and treatment, p. 229-255. In I. K. Wachsmuth, P. A. Blake, and Ø. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 5.Bonnet, R., C. Dutour, J. L. M. Sampaio, C. Chanal, D. Sirot, R. Labia, C. de Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R., J. L. Sampaio, R. Labia, C. de Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and The French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupont, M.-J., M. Jouvenot, G. Couetdic, and Y. Michel-Briand. 1985. Development of plasmid-mediated resistance in Vibrio cholerae during treatment with trimethoprim-sulfamethoxazole. Antimicrob. Agents Chemother. 27:280-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falbo, V., A. Carattoli, F. Tosini, C. Pezzella, A. M. Dionisi, and I. Luzzi. 1999. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob. Agents Chemother. 43:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazouli, M., E. Tzelepi, A. Markogiannakis, N. J. Legakis, and L. S. Tzouvelekis. 1998. Two novel plasmid-mediated cefotaxime-hydrolyzing β-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol. Lett. 165:289-293. [DOI] [PubMed] [Google Scholar]
- 13.Gazouli, M., E. Tzelepi, S. V. Sidorenko, and L. S. Tzouvelekis. 1998. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob. Agents Chemother. 42:1259-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass, R. I., M. I. Huq, J. V. Lee, E. J. Threlfall, M. R. Khan, A. R. M. A. Alim, B. Rowe, and R. J. Gross. 1983. Plasmid-borne multiple drug resistance in Vibrio cholerae serogroup O1, biotype El Tor: evidence for a point-source outbreak in Bangladesh. J. Infect. Dis. 147:204-209. [DOI] [PubMed] [Google Scholar]
- 15.Hofer, E., B. R. Quintaes, E. M. dos Reis, D. dos P. Rodrigues, L. M. Seki, I. S. Feitosa, L. H. Ribeiro, and M. R. Ferreira. 1999. Emergencia da multipla resistencia a antimicrobianos em Vibrio cholerae isolados de pacientes com gastroenterite no Ceara, Brasil. Rev. Soc. Bras. Med. Trop. 32:151-156. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby, G. A., and A. A. Medeiros. 1991. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:1697-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariuki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse, H., H. Sørum, F. C. Tenover, and Ø. Olsvik. 1995. A transferable multiple drug resistance plasmid from Vibrio cholerae O1. Microb. Drug Resist. 1:203-210. [DOI] [PubMed] [Google Scholar]
- 20.Labia, R., and M. Barthélémy. 1979. L'enzymogramme des beta-lactamases: adaptation en cel de la methode iodometrique. Ann. Microbiol. (Paris) 130B:295-304. [PubMed] [Google Scholar]
- 21.Medeiros, A. A. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24(Suppl. 1):19-45. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, M., S. Sato, T. Ohya, S. Suzuki, and S. Ikeda. 1986. Plasmid profile analysis in epidemiological studies of animal Salmonella typhimurium infection in Japan. J. Clin. Microbiol. 23:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., vol. 20, no. 2. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed., vol. 20, no. 1. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Nordmann, P., and T. Naas. 1994. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob. Agents Chemother. 38:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordmann, P., E. Ronco, T. Naas, C. Duport, Y. Michel-Briand, and R. Labia. 1993. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsvik, Ø., T. Popovic, and P. I. Fields. 1993. PCR detection of toxin genes in strains of Vibrio cholerae O1, p. 266-270. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. American Society for Microbiology, Washington, D.C.
- 29.Olsvik, Ø., and N. A. Strockbine. 1993. PCR detection of heat-stable, heat-labile, and shiga-like toxin genes in Escherichia coli, p. 271-276. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. American Society for Microbiology, Washington, D.C.
- 30.Ouellette, M., G. Gerbaud, and P. Courvalin. 1988. Genetic, biochemical and molecular characterization of strains of Vibrio cholerae multiresistant to antibiotics. Ann. Inst. Pasteur Microbiol. 139:105-113. [PubMed] [Google Scholar]
- 31.Power, P., M. Radice, C. Barberis, C. de Mier, M. Mollerach, M. Maltagliatti, C. Vay, A. Famiglietti, and G. Gutkind. 1999. Cefotaxime-hydrolysing beta-lactamases in Morganella morganii. Eur. J. Clin. Microbiol. Infect. Dis. 18:743-747. [DOI] [PubMed] [Google Scholar]
- 32.Rahal, K., G. R. Gerbaud, and Y. A. Chabbert. 1973. Caractérisation d'un facteur de résistance transférable de Vibrio cholerae biotype eltor. Ann. Microbiol. (Inst. Pasteur) 124B:283-294. [PubMed] [Google Scholar]
- 33.Reid, A. J., and S. G. B. Amyes. 1986. Plasmid penicillin resistance in Vibrio cholerae: identification of new β-lactamase SAR-1. Antimicrob. Agents Chemother. 30:245-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi, A., M. Galas, N. Binztein, M. Rivas, M. I. Caffer, A. Corso, M. Radice, and G. Gutkind. 1993. Unusual multiresistant Vibrio cholerae O1 El Tor in Argentina. Lancet 342:1172-1173. [DOI] [PubMed] [Google Scholar]
- 35.Rossi, A., H. Lopardo, M. Woloj, A. M. Picandet, M. Mariño, M. Galas, M. Radice, and G. Gutkind. 1995. Non-typhoid Salmonella spp. resistant to cefotaxime. J. Antimicrob. Chemother. 36:697-702. [DOI] [PubMed] [Google Scholar]
- 36.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Simarro, E., F. Navarro, J. Ruiz, E. Miro, J. Gómez, and B. Mirelis. 2000. Salmonella enterica serovar Virchow with CTX-M-like β-lactamase in Spain. J. Clin. Microbiol. 38:4676-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 40.Tabtieng, R., S. Wattanasri, P. Echeverria, J. Seriwatana, L. Bodhidatta, A. Chatkaeomorakot, and B. Rowe,. 1989. An epidemic of Vibrio cholerae El Tor Inaba resistant to several antibiotics with a conjugative group C plasmid coding for type II dihydrofolate reductase in Thailand. Am. J. Trop. Med. Hyg. 41:680-686. [DOI] [PubMed] [Google Scholar]
- 41.Threlfall, E. J., and B. Rowe. 1982. Vibrio cholerae El Tor acquires plasmid-encoded resistance to gentamicin. Lancet i:42. [DOI] [PubMed] [Google Scholar]
- 42.Threlfall, E. J., B. Rowe, and I. Huq. 1980. Plasmid-encoded multiple antibiotic resistance in Vibrio cholerae El Tor from Bangladesh. Lancet i:1247-1248. [DOI] [PubMed] [Google Scholar]
- 43.Threlfall, E. J., B. Said, B. Rowe, and A. Dávalos-Pérez. 1993. Emergence of multiple drug resistance in Vibrio cholerae O1 El Tor from Ecuador. Lancet 342:1173.. [DOI] [PubMed] [Google Scholar]
- 44.Towner, K. J., N. J. Pearson, and F. O'Grady. 1979. Resistant Vibrio cholerae El Tor in Tanzania. Lancet ii:147-148. [DOI] [PubMed] [Google Scholar]
- 45.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 46.Vahaboglu, H., L. M. C. Hall, L. Mulazimoglu, S. Dodanli, I. Yildirim, and D. M. Livermore. 1995. Resistance to extended-spectrum cephalosporins, caused by PER-1 β-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J. Med. Microbiol. 43:294-299. [DOI] [PubMed] [Google Scholar]
- 47.Vahaboglu, H., R. Öztürk, G. Aygün, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1 type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto, T., G. B. Nair, M. J. Albert, C. C. Parodi, and Y. Takeda. 1995. Survey of in vitro susceptibilities of Vibrio cholerae O1 and O139 to antimicrobial agents. Antimicrob. Agents Chemother. 39:241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. Antimicrob. Agents Chemother. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]