Abstract
It is not uncommon to see amphotericin B treatment failure in patients with systemic infection caused by Candida lusitaniae. We report a patient with stage IV ovarian carcinoma and C. lusitaniae sepsis whose treatment with amphotericin B failed. The initial blood isolate was susceptible to amphotericin B in vitro; however, the MIC for a blood isolate recovered 7 weeks after treatment began showed a fourfold increase. Direct subculture of two positive blood samples obtained within a week of the patient's death showed the coexistence of two distinct colony color variants on CHROMagar Candida (CAC). One variant was susceptible to amphotericin B, and one was resistant. These results emphasize the importance of repeat amphotericin B susceptibility testing for patients with persistent C. lusitaniae infection. The presence of colony variants on CAC may signal the emergence of amphotericin B resistance in C. lusitaniae and should be investigated.
Candida lusitaniae is considered an opportunistic pathogen, causing infection primarily in immunocompromised patients (1-5, 17). Recent studies have shown that the incidence of serious infection caused by C. lusitaniae is increasing. Clinical management of systemic infection by this organism is challenging because of innate amphotericin B resistance in some isolates (3, 10, 16). Moreover, some isolates of C. lusitaniae may develop amphotericin B resistance in vivo, a finding which is supported by in vitro studies (14). Recently, Yoon et al. reported high-frequency, reversible, in vitro switching of isolates from being amphotericin B susceptible to amphotericin B resistant after exposure to the drug (18). Here we report a case of fatal systemic infection caused by C. lusitaniae with amphotericin B treatment failure. Blood cultures obtained during therapy yielded colonies with distinct differences in color on CHROMagar Candida (CAC) between amphotericin B-susceptible and amphotericin B-resistant strains.
CASE REPORT
A 69-year-old woman with stage IV metastatic ovarian carcinoma had fever for 5 days, respiratory distress, and metabolic acidosis despite metronidazole and imipenem therapy for 2 weeks. The culture of a central venous pressure catheter tip and three Isolator (Wampole Laboratories, Cranbury, N.J.) blood cultures drawn 2 days later were positive for C. lusitaniae. The catheter line was removed, and empiric treatment with fluconazole was started on day 1 (Table 1). The treatment was switched to amphotericin B at 35 mg (0.7 mg/kg of body weight) four times a day on day 6 and then to lipid complex amphotericin B (Abelcet) at 350 mg (7 mg/kg) four times a day on day 10 because of persistent fungemia, which was confirmed by another positive blood culture. The patient's symptoms improved, and after approximately 5 weeks of hospitalization, she was discharged on continuing treatment with lipid complex amphotericin B. Seven days after discharge, she experienced shortness of breath and was readmitted. She denied having fever, chills, or nausea. A pleural effusion prompted a left pleurocentesis, which produced 1,300 ml of fluid. The admission blood culture and later blood cultures were positive for C. lusitaniae despite continued amphotericin therapy. The patient's condition deteriorated with the development of hypotension and profound sepsis of an apparently fungal etiology. She expired 1 week after readmission and 8 weeks after the initiation of amphotericin therapy.
TABLE 1.
Summary of fungal culture results
| Day of culture | Sample source | Presence of C. lusitaniae |
|---|---|---|
| 18 days prior to first positive culture | Blood | No |
| 11 days prior to first positive culture | Blood | No |
| Day 1 (empiric fluconazole treatment started) | Central venous pressure catheter tip | Yes |
| Day 3 | Blooda | Yes |
| Day 6 (amphotericin B treatment started) | Blood | Yes |
| Day 10 (lipid complex amphotericin B started) | Blood | Yes |
| Day 13 | Urine | No |
| Day 15 | Blood | Yes |
| Day 20 | Urine | Yes (10,000 CFU/ml) |
| Day 36 | Urine | No |
| Day 55 | Bloodb | Yes (two colony types) |
| Day 56 | Pleural fluid | No |
| Day 57 | PICCc catheter tip | No |
| Day 59 | Blood | Yes |
| Day 61 | Blood | Yes (two colony types) |
| Day 61 | Endotracheal aspirate | No |
| Day 61 | Urine | Yes (80,000 CFU/ml) |
| Day 62 (patient expired) |
First isolate subculture sent to reference laboratory.
Second isolate subculture sent to reference laboratory.
PICC, peripherally inserted central catheter.
MATERIALS AND METHODS
Isolates.
C. lusitaniae was identified by pigment production on CAC (Hardy Diagnostics, Santa Maria, Calif.), a negative germ tube test, morphological characteristics on cornmeal agar with polysorbate 80, and the API 20C AUX test (bioMerieux/Vitek, St. Louis, Mo.).
In-house antifungal susceptibility tests.
The isolates were retrieved from a −70°C freezer, and two samples were serially subcultured to Sabouraud dextrose agar (Emmons modification) before testing.
(i) Etest.
Etests (AB Biodisk, Skolne, Sweden) were performed and the results were interpreted according to Etest technical guide 4. Isolates from a 24-h-old culture on Sabouraud dextrose agar (Emmons modification) were suspended in saline to achieve a 0.5 McFarland turbidity. Four hundred microliters of the inoculum was dispensed to the center of a 150-mm-diameter plate containing RPMI 1640, 2% glucose, MOPS (morpholinepropanesulfonic acid), and 1.5% Bacto Agar. The inoculum was swabbed in three directions over the entire agar surface and then allowed to dry for 15 min before Etest strips were applied. The plates were incubated at 35°C in a moist incubator for 24 h. The amphotericin MIC was read from the scale as the lowest drug concentration at which there was 100% inhibition of the organism. Fluconazole and itraconazole MICs were read as the lowest drug concentrations at which there was 80% inhibition of the organism.
(ii) Sensititre YeastOne.
Sensititre YeastOne tests (Trek Diagnostics, Westlake, Ohio) were performed and the results were interpreted according to the manufacturer's guidelines. Isolates from a 24-h-old culture on Sabouraud dextrose agar (Emmons modification) were suspended in autoclaved, demineralized water to achieve a 0.5 McFarland turbidity. A working suspension was made by adding 20 μl of the suspension to 11 ml of inoculum broth. Then, 100 μl of inoculum was added to each well of the Sensititre panel. The panels were incubated for 24 h at 35°C in a non-CO2 incubator. The MIC was read as the lowest drug concentration that prevented a color change of the medium from blue or purple to red.
Molecular typing.
The two color variants of C. lusitaniae isolated from our patient and a blood culture isolate of C. lusitaniae recovered from another Stanford University Medical Center patient (control) were compared by use of SfiI and NotI restriction endonuclease digestion of genomic DNA, followed by pulsed-field gel electrophoresis (PFGE) and karyotyping of whole-genomic DNA by PFGE. The lysis enzyme used was lyticase (Sigma, St. Louis, Mo.). Digest PFGE was performed at 6 V/cm, with a voltage ramp of 10 to 90 s for 24 h in 1% SeaKem agarose. Karyotype PFGE used Fastlane agarose and was performed at 4.5 V/cm, with a ramp of 120 to 280 s for 48 h.
RESULTS
Microbiology culture results.
Table 1 summarizes the results of fungal cultures performed during all episodes of C. lusitaniae infection in the patient. Isolation of C. lusitaniae from the catheter tip 2 days prior to the first positive blood culture suggested the catheter tip as the source of candidemia (Table 1). A urine culture, collected 17 days after the first positive blood culture, also grew C. lusitaniae. After 8 weeks of continued amphotericin B treatment, the organism was recovered from multiple blood cultures and a urine culture obtained 1 day prior to the patient's death on day 62.
In vitro antifungal susceptibility results.
Subcultures of C. lusitaniae isolated from two blood cultures were sent for antifungal susceptibility testing to a reference laboratory which uses the NCCLS M-27A macrobroth method. The first subculture represented the growth from blood obtained before amphotericin B treatment was initiated. The second subculture was from blood cultures obtained over 7 weeks later. An unusual colony color on CAC was not noted for colonies from either of the two blood cultures. Although both cultures were interpreted by the reference laboratory as being susceptible to amphotericin B, fluconazole, and itraconazole, the amphotericin B MIC for the second isolate was 1.0 μg/ml (Table 2) whereas an MIC of 0.25 μg/ml was recorded for the initial isolate. The fluconazole and itraconazole MICs were the same for both isolates.
TABLE 2.
Comparison of antifungal susceptibilities of blood isolates of C. lusitaniae prior to and during amphotericin B treatmenta
| Drug | Prior to amphotericin B treatment
|
During amphotericin B treatment
|
||
|---|---|---|---|---|
| MIC (μg/ml) | Interpretation | MIC (μg/ml) | Interpretation | |
| Amphotericin B | 0.25 | Sb | 1.0 | S |
| Fluconazole | 1.0 | S | 1.0 | S |
| Itraconazole | <0.5 | S | <0.5 | S |
These results were reported by a reference laboratory using the NCCLS macrodilution reference standard methodology, which was different from the in-house testing procedure used for the comparison represented in Table 3. The inoculum used by the reference laboratory originated from a random selection of colonies without regard to colony color on CAC.
S, susceptible.
Identification of an emerging amphotericin B-resistant strain.
Two distinct colors were seen on the CAC subculture from the blood culture (BACTEC; BD Microbiology Systems, Sparks, Md.) drawn 7 weeks after amphotericin B treatment was started. One colony type was described as blue, and the other was described as purple. Both were subsequently identified as C. lusitaniae. The expected color range of C. lusitaniae on CAC is pink to grayish purple (13). No differences in the colors, textures, or sizes, of the colonies on Sabouraud agar were noted. The clinical significance of mixed colony morphologies on CAC was not appreciated at the time that subcultures were sent to the reference laboratory for MIC testing. Both colony types were included by chance in a retrospective susceptibility study performed months later in our laboratory. To our surprise, the blue colonies were susceptible to amphotericin B while the purple colonies were resistant (Table 3). Based on the interpretive guidelines in NCCLS document M-27A (12), both colony types were deemed susceptible to fluconazole and itraconazole (Table 3). We then attempted to further characterize the two color-variant strains.
TABLE 3.
Comparison of antifungal susceptibilities of two color variants of C. lusitaniae isolated on CACa
| Drug | Test method | Blue colonies
|
Purple colonies
|
||
|---|---|---|---|---|---|
| MIC (μg/ml) | Interpre- tation | MIC (μg/ml) | Interpre- tation | ||
| Amphotericin B | Sensititre | 0.5 | S | 2 | R |
| Etest | 0.25 | S | 4 | R | |
| Fluconazole | Sensititre | 1.0 | S | 0.38 | S |
| Etest | 0.25 | S | 0.094 | S | |
| Itraconazole | Sensititre | 0.03 | S | 0.016 | S |
| Etest | 0.032 | S | 0.016 | S | |
The interpretations were based on results obtained with NCCLS document M-27A. S, susceptible; R, resistant.
Molecular typing.
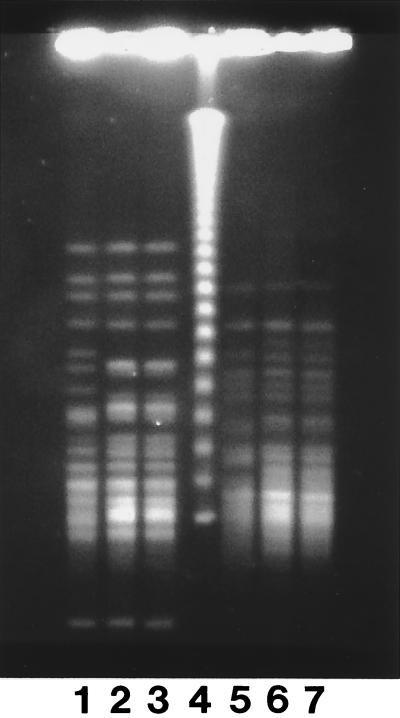
The PFGE patterns of both the SfiI and the NotI restriction digests of the two color variants (Fig. 1, lanes 2 and 6 and lanes 3 and 7) were indistinguishable from each other but were different from that of the control. The single band variation between the karyotypes of the case isolates (data not shown) was insufficient to make us consider the isolates different.
FIG. 1.
Results of PFGE of NotI (lanes 1 to 3) and SfiI (lanes 5 to 7) digests of a control isolate of C. lusitaniae (lanes 1 and 5), of the amphotericin B-susceptible isolate from our patient (blue colony on CAC) (lanes 2 and 6), and of the amphotericin B-resistant isolate from our patient (purple colony on CAC) (lanes 3 and 7). Lane 4, lambda ladder.
DISCUSSION
C. lusitaniae is considered an opportunistic organism whose association with invasive infection has increased in recent years (1, 2, 3, 5, 16). Unlike Candida albicans, which is rarely resistant to amphotericin B, C. lusitaniae has consistently been associated with the failure of amphotericin B treatment in patients with invasive disease (3, 10, 14, 15). Resistance may be innate or may develop during treatment.
Here we report the first clinical case of C. lusitaniae developing amphotericin B resistance during amphotericin B treatment and exhibiting an accompanying difference in colony color on CAC between susceptible and resistant strains. The antifungal susceptibility tests performed according to the NCCLS M-27A macrobroth method by a reference laboratory showed a fourfold increase of the amphotericin B MIC for the isolate recovered after 7 weeks of amphotericin B treatment compared to that for an isolate recovered prior to amphotericin B treatment. The MIC of 1.0 μg/ml reported for the isolate recovered after 7 weeks of treatment may reflect the testing of a mixture of susceptible and resistant clones. Our laboratory probably picked a random sample of colonies from Sabouraud agar, on which morphotype differences were not seen. The reference laboratory did not interpret either strain as being resistant to amphotericin B, and the patient was continued on amphotericin B therapy despite the rise in the amphotericin B MIC for the second isolate.
The discernible difference in colony color on CAC between the susceptible and resistant strains of C. lusitaniae isolated from the blood culture on day 55 (Table 1) was not seen with colonies on standard fungal agars. Previously published molecular epidemiology studies suggest that repeat isolates of C. lusitaniae from the same individual are generally from the same strain (6-9, 11, 15). The PFGE restriction digest patterns of the two case variants reported here were indeed indistinguishable by PFGE.
In their in vitro studies, Yoon et al. found a change in the cellular morphology of C. lusitaniae when it switched from being amphotericin B sensitive to amphotericin B resistant in the presence of the drug (18). It is possible that cellular changes associated with drug resistance are visualized on CAC as differences in colony color. A significant challenge is to identify the amphotericin B-resistant strains of C. lusitaniae when patients are infected concurrently with resistant and sensitive strains. For this reason, we have reported a potential visual clue. Only laboratory awareness and further experience will show whether this is a reliable indicator of amphotericin B resistance in C. lusitaniae or a unique occurrence. Meanwhile, we emphasize the importance of repeat testing of amphotericin B resistance in patients with persistent infection.
Acknowledgments
Some supplies were provided by Trek Diagnostic Systems, Inc., and AB Biodisk.
REFERENCES
- 1.Blinkhorn, R. J., D. Adelstein, and P. J. Spagnuolo. 1989. Emergence of a new opportunistic pathogen, Candida lusitaniae. J. Clin. Microbiol. 27:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christenson, J. C., A. Guruswamy, G. Mukwaya, and P. Rettig. 1987. Candida lusitaniae: an emerging human pathogen. Pediatr. Infect. Dis. J. 6:755-757. [PubMed] [Google Scholar]
- 3.Guinet, R., J. Chanas, A. Goullier, G. Bonnefoy, and P. Ambroise-Thomas. 1983. Fetal septicemia due to amphotericin B-resistant Candida lusitaniae. J. Clin. Microbiol. 18:443-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadfield, T. L., M. B. Smith, R. E. Winn, M. G. Rinaldi, and C. Guerra. 1987. Mycosis caused by Candida lusitaniae. Rev. Infect. Dis. 9:1006-1012. [DOI] [PubMed] [Google Scholar]
- 5.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King, D., J. Rhine-Chalberg, M. A. Pfaller, S. A. Moser, and W. G. Merz. 1995. Comparison of four DNA-based methods for strain delineation of Candida lusitaniae. J. Clin. Microbiol. 33:1467-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathaba, L. T., G. Davis, and J. R. Warminton. 1995. The genotypic relationship of Candida albicans strains isolated from the oral cavity of patients with denture stomatitis. J. Med. Microbiol. 42:372-379. [DOI] [PubMed] [Google Scholar]
- 8.McCullough, M. J., B. C. Ross, B. D. Dwyer, and P. C. Reade. 1994. Genotype and phenotype of oral Candida albicans from patients infected with the human immunodeficiency virus. Microbiology 140:1195-1202. [DOI] [PubMed] [Google Scholar]
- 9.Mercure, S., S. Poirier, G. Lemay, P. Auger, S. Montplaisir, and L. deRepentigny. 1993. Application of biotyping and DNA typing of Candida albicans to the epidemiology of recurrent vulvovaginal candidiasis. J. Infect. Dis. 168:502-507. [DOI] [PubMed] [Google Scholar]
- 10.Merz, W. G. 1984. Candida lusitaniae: frequency of recovery, colonization, infection, and amphotericin B resistance. J. Clin. Microbiol. 20:1194-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merz, W. G., U. Khazan, M. A. Jabra-Rizk, L.-C. Wu, G. J. Osterhout, and P. F. Lehmann. 1992. Strain delineation and epidemiology of Candida (Clavispora) lusitaniae. J. Clin. Microbiol. 30:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. M-27A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Odds, F. C., and R. Bernaerts. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pappagianis, D., M. S. Collins, R. Hector, and J. Remington. 1979. Development of resistance to amphotericin B in Candida lusitaniae infecting a human. Antimicrob. Agents Chemother. 16:123-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller, M. A., S. A. Messer, and R. J. Hollis. 1994. Strain delineation and antifungal susceptibilities of epidemiologically related and unrelated isolates of Candida lusitaniae. Diagn. Microbiol. Infect. Dis. 20:127-133. [DOI] [PubMed] [Google Scholar]
- 16.Powderly, W. G., G. S. Kobayashi, G. P. Herzig, and G. Medoff. 1988. Amphotericin B-resistant yeast infection in severely immunocompromised patients. Am. J. Med. 84:826-832. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez, V., J. A. Vazquez, D. Barth-Jones, L. Dembry, J. D. Sobel, and M. J. Zervos. 1992. Epidemiology of nosocomial acquisition of Candida lusitaniae. J. Clin. Microbiol. 30:3005-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon, S. A., J. A. Vazquez, P. E. Steffan, J. D. Sobel, and R. A. Akins. 1999. High-frequency, in vitro reversible switching of Candida lusitaniae clinical isolates from amphotericin B susceptibility to resistance. Antimicrob. Agents Chemother. 43:836-845. [DOI] [PMC free article] [PubMed] [Google Scholar]