Abstract
The resistance of Klebsiella pneumoniae BM4493, isolated in Ho Chi Minh City, Vietnam, to cefotaxime and aztreonam was due to production of a novel β-lactamase, CTX-M-17. The blaCTX-M-17 gene was borne by 7,086-bp plasmid pIP843, which was entirely sequenced and which was found to belong to the ColE1 family. The 876-bp blaCTX-M-17 gene differed from blaCTX-M-14 by 2 nucleotides, which led to the single amino acid substitution Glu289→Lys. blaCTX-M-17 was flanked upstream by an ISEcp1-like element and downstream by an insertion sequence (IS) IS903 variant designated IS903-C. The transcriptional start site of blaCTX-M-17 was located 109 nucleotides upstream from the initiation codon in the ISEcp1-like element, which also provided the promoter sequences. Plasmid pIP843, which was non-self-transferable and nonmobilizable, contained five open reading frames transcribed in the same orientation. Regions homologous to sequences coding for putative RNA II and RNA I transcripts, a rom gene, which is involved in initiation of replication, and a cer-like gene, which is responsible for the stability of ColE1-like plasmids, were identified. Consensus sequences for putative replication (oriV) and transfer (oriT) origins were present. Results of primer extension experiments indicated that ISEcp1 provides the promoter for expression of blaCTX-M-17 and may contribute to dissemination of this gene.
The CTX-M enzymes represent a new family of class A extended-spectrum β-lactamases. The first member, MEN-1 (CTX-M-1), was reported at the beginning of the 1990s (3, 4). In contrast to TEM- and SHV-type cefotaxime-hydrolyzing extended-spectrum β-lactamases, CTX-M β-lactamases are much more active against cefotaxime as a substrate than against ceftazidime. This probably results from changes in amino acids critical for extended-spectrum activity (3, 17, 20, 26).
To date, the fast-growing CTX-M family comprises 19 members, isolated from various enterobacterial species in different geographic areas. The strains producing CTX-M enzymes were mainly found during nosocomial outbreaks that occurred in Japan (21, 26), Europe (9, 18), and South America (7, 31, 35). More recently, they have been isolated in Africa (23), China (GenBank accession nos. AF252621 to AF252623), and Korea (32). On the basis of their amino acid sequence similarities, the CTX-M enzymes have been classified into four subgroups (7, 8). The first one contains CTX-M-1 (MEN-1) (3, 4), CTX-M-3 (18), CTX-M-10 (31), CTX-M-12 (23), and CTX-M-15 (22); the second subgroup includes CTX-M-2 (5), CTX-M-4 (15), CTX-M-5, CTX-M-6, and CTX-M-7 (16), and Toho-1 (21); the third subgroup consists of CTX-M-8 (7); and the fourth subgroup contains Toho-2 (26), CTX-M-9 (36), CTX-M-13 (GenBank accession no. AF252621), CTX-M-14 (32), and CTX-M-16 (8). The divergence in amino acid sequences among the subgroups (between 70 and 99% sequence identity) (8), as well as the geographical dispersion of the host strains, renders prediction of the origin of CTX-M enzymes difficult. The blaCTX-M-2-like gene is closely related to the chromosomal kluA-1 β-lactamase gene of Kluyvera ascorbata (GenBank accession no. AJ272538), and it has thus been proposed that the KluA-1 enzyme could be the ancestor of the CTX-M β-lactamases belonging to the first subgroup (A. Philippon, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr 2044a, 1999). Little is known about the environment of the structural genes for CTX-M-type enzymes, and their promoters have not been identified experimentally.
Numerous bla genes are located on plasmids, and some of them are part of transposons or constitute cassettes in integrons (27, 34). The aim of this work was to characterize the gene coding for the CTX-M-type β-lactamase of Klebsiella pneumoniae BM4493 and BM4494, which were isolated in Vietnam and which are resistant to cefotaxime and apparently susceptible to ceftazidime, and to identify its genetic basis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The origins and properties of the strains used in this study are shown in Table 1. The strains were grown in brain heart infusion (BHI) broth and agar (Difco Laboratories, Detroit, Mich.) at 37°C. Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France). The MICs of the β-lactams, alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml), were determined by agar dilution with 104 CFU per spot on Mueller-Hinton medium.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| JM83 | araΔ(lac-proAB)rpsL(Strr)[F80dlacΔ(lacZ)M15] | 38 |
| C600Rif | e14-(mcrA)thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21 | 24 |
| HB101 | hsdS20 Δ(gpt-proA)62 leu supE44-ara-14 galK2 lacY1 Δ(merC-mrr) rpsL20 (Strr)xyl-5 mtl-1 recA13 | 37 |
| DH5α | (φ80ΔlacZΔM15) endA1 recA1 hsdR17 supE44 thi-1 λ gyrA96 relA1 Δ(lacZYA-argF)U169 | 43 |
| K. pneumoniae | ||
| BM4493 | Ctxr Cmr Gmr Ntr Smr Sur Tmr Tpr | Wild-type strain |
| BM4494 | Ctxr Cmr Gmr Rifr Smr Sur Tmr | Wild-type strain |
| Plasmids | ||
| pBGS18 | Tra Mob Kmr | 39 |
| pIP843 | Tra Mob+ CtxrblaCTX-M-17 | From BM4493 |
| pIP843-1 | Tra Mob CtxrblaCTX-M-17 (IS903-CΩ4bp) | This work |
| pIP844 | Tra Mob CtxrblaCTX-M-17 | From BM4494 |
| pAT797 | pBGS18Ω 4-kb HindIII from pIP844, Ctxr Kmr | This work |
| pOX38 | Tra+ IncF1 Gmr | 11 |
Abbreviations: Tra+, self-transferable; Tra−, non-self-transferable; Mob+, mobilizable; Mob−, nonmobilizable; Apr, ampicillin resistance; Ctxr, cefotaxime resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Ntr, netilmicin resistance; Rifr, rifampin resistance; Smr, streptomycin resistance; Sur, sulfonamide resistance; Tmr, tobramycin resistance; Tpr, trimethoprim resistance.
Transfer of resistance to β-lactams.
Transfer of resistance from the K. pneumoniae strains by conjugation (45) or by transformation with purified plasmid DNA (37) was carried out as described previously with selection on 4 μg of cefotaxime per ml by using Escherichia coli C600Rif and E. coli JM83 as recipients, respectively.
DNA manipulations.
Total DNA and plasmid DNA were prepared as described previously (37). Plasmid DNA was purified with a Wizard minipreps DNA kit (Promega, Madison, Wis.). Restriction with endonucleases was done according to the recommendations of the supplier (Life Technologies Inc., Gaithersburg, Md.). Plasmid DNA was analyzed by electrophoresis on a 1.2% agarose gel (Gibco-BRL-Life Technologies, Eragny, France) containing 0.15 μg of ethidium bromide per ml (Pharmacia-Biotech, Orsay, France).
Detection of blaCTX-M-like genes was performed by amplifying a ca. 550-bp internal fragment with degenerate primers CTX-MA-1 (5′-SCSATGTGCAGYACCAGT-3′) and CTX-MA-2 5′-CCGCRATATGRTTGGTGGTG-3′) (where S is G or C, Y is C or T, and R is A or G), whose sequences are complementary to a conserved portion of the sequences of the blaCTX-M genes (for CTX-MA-1, complementary from positions 270 to 287 of blaCTX-M-1; for CTX-MA-2, complementary from positions 794 to 813 of blaCTX-M-1). The primers were prepared at the Unité de Chimie Organique, Institut Pasteur, Paris, France. PCRs were performed with 100-μl reaction mixtures consisting of 1× Pfu DNA polymerase buffer, 2 IU of Pfu DNA polymerase (Stratagene, La Jolla, Calif.), 1.5 mM MgCl2, 50 μM (each) deoxynucleoside triphosphates, 50 pmol of each primer, and 25 ng of DNA in a GeneAmp PCR system 2400 (Perkin-Elmer Cetus, Norwalk, Conn.). The PCR mixture was submitted to a denaturation step (2 min at 94°C), followed by 30 cycles of amplification (45 s of denaturation at 94°C, 1 min of annealing at 52°C, and 1 min of elongation at 72°C) and 10 min at 72°C as the last step.
Plasmid DNA of strain BM4494 and pBGS18 DNA were digested with HindIII, mixed, ligated with T4 ligase (Pharmacia Biotech, Saint-Quentin en Yvelines, France), and introduced by transformation (37) into E. coli JM83 with selection on BHI medium containing 4 μg of cefotaxime per ml. The transformants were screened for plasmids by agarose gel electrophoresis.
Sequencing of both strands of pIP843 DNA was performed directly with a CEQ 2000 DNA Analysis System automatic sequencer (Beckman Instruments, Inc., Palo Alto, Calif.). Primers CTX-MA-1 and CTX-MA-2 were used for initial sequencing, and the primers used for further sequencing were deduced from the sequence determined from the initial sequencing. Sequencing of the insert in pAT797 was determined similarly.
Disruption of the putative transposase of IS903-C.
Insertion sequence (IS) IS903-C of pIP843 contained a single MluI site. Plasmid pIP843 DNA was digested with MluI, and the ends were blunted with Klenow polymerase (37); IS903-C was then self-ligated with T4 DNA ligase (Pharmacia) and introduced by transformation into E. coli. This inserted a 4-bp CGCG sequence, which introduced a frame shift, and the plasmid carrying IS903-CΩbp was designated pIP843-1.
Assay for transposition of ISEcp1-like element by plasmid conduction.
Plasmid pIP843 (Tra− Mob− Ctxr) carrying the ISEcp1-like element and IS903-CΩbp was introduced by transformation into E. coli HB101 (recA Strr) harboring pOX38 (Tra+ Gmr). The resulting strain, HB101(pOX38, pIP843-1), was mated with E. coli DH5α (recA Nalr), and transconjugants were selected on BHI medium containing nalidixic acid (40 μg/ml) and ampicillin (100 μg/ml) to test for the presence of cointegrated plasmids that could have been generated by the replicative transposition of the ISEcp1-like element.
Southern hybridization.
pOX38 DNA and cointegrated plasmid DNA were purified with a plasmid purification system (Qiagen Inc., Valencia, Calif.) and digested with EcoRI (Life Technologies Inc.), which is one of the enzymes that cuts ISEcpI at a single site. The DNA fragments were separated by electrophoresis on 0.9% agarose gels and transferred to a Hybond-N+ membrane (Amersham International, Little Chalfont, England) by vacuum with a Trans Vac TE80 apparatus (Hoefer Scientific Instruments, San Francisco, Calif.). The 1,656-bp ISEcp1 fragment was amplified by PCR, labeled with [α-32P]dCTP (3,000 Ci/mM; Amersham Radiochemical Center, Amersham, England) with the Megaprime DNA Labeling System (Amersham Pharmacia Biotech, Little Chalfont, England), and used as a probe for Southern hybridization (37).
RNA manipulation and primer extension.
Total RNA was extracted from 10 ml each of exponentially growing cultures of BM4493 and E. coli JM83/pIP843 with an optical density at 600 nm of ca. 0.7, as described previously (2). Oligonucleotide PE (5′-GCTGCACCGCACTCGTCTGCGCATAAAGCG-3′), complementary to a sequence 74 to 103 bp downstream from the site of initiation of the blaCTX-M-17 gene, was 5′ labeled with [γ-32]ATP (6,000 Ci/mM; Amersham Radiochemical Center) and T4 nucleotide kinase (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), 105 cpm was incubated with 50 μg of RNA overnight at 30°C, and extension was performed with 40 U of avian myeloblastosis virus reverse transcriptase (Boehringer, Mannheim, Germany) for 90 min at 42°C, as described previously (2). Primer elongation products were analyzed by electrophoresis on 6% denaturating polyacrylamide gels.
Computer analysis of sequence data.
Nucleotide and amino acid sequence data were analyzed with the GCG sequence analysis software package (version 7; Genetics Computer Group, Madison, Wis.). The sequences in the GenBank and SwissProt databases were screened for similarity to the sequences obtained in the present study.
Nucleotide sequence accession numbers.
The sequences of pIP843 and pIP844 have been deposited in the GenBank (Los Alamos, N.M.) data library under accession nos. AY033516 and AF454633, respectively.
RESULTS AND DISCUSSION
Characterization of K. pneumoniae clinical isolates.
K. pneumoniae BM4493 was isolated in Ho Chi Minh City, Vietnam, in 1996 from a urine specimen of an 8-month-old child hospitalized for acute pneumonia. The strain was resistant to cefotaxime and aztreonam but was susceptible to cefoxitin and ceftazidime (Table 2). Synergy between cefotaxime and clavulanic acid was observed by double-disk diffusion. A PCR amplification product from total DNA of the strain was obtained with degenerate primers specific for the blaCTX-M-like genes. K. pneumoniae BM4494 had a phenotype of resistance to β-lactams, similar to that of BM4493, and was isolated in Ho Chi Minh City during the same period. Strains BM4493 and BM4494 were also resistant to chloramphenicol, trimethoprim-sulfamethoxazole, and gentamicin (data not shown).
TABLE 2.
MICs of various β-lactams with and without clavulanic acid for the strains tested
| Antibiotic | MIC (μg/ml)
|
||
|---|---|---|---|
| K. pneumoniae BM4493 | E. coli JM83/ pIP843 | E. coli JM83 | |
| Amoxicillin | >256 | >256 | 4 |
| Piperacillin | 256 | 256 | 1 |
| Piperacillin-CLAa | 4 | 2 | 1 |
| Cefalothin | >256 | >256 | 2 |
| Cefuroxime | >256 | 256 | 2 |
| Cefoxitin | 4 | 4 | 1 |
| Cefoxitin-CLA | 2 | 2 | 1-2 |
| Cefotaxime | 64 | 128 | <0.125 |
| Cefotaxime-CLA | 0.5 | 0.5 | <0.125 |
| Ceftazidime | 8 | 4 | <0.125 |
| Ceftazidime-CLA | 0.25 | 0.25 | <0.125 |
| Aztreonam | 16 | 16 | <0.125 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml.
Transfer of resistance to antibiotics.
All attempts to transfer resistance to cefotaxime by conjugation from K. pneumoniae BM4493 and BM4494 to E. coli C600Rif were unsuccessful. The plasmid DNA contents of BM4493 and BM4494 were used to transform E. coli JM83, and analysis of the transformants resistant to cefotaxime and aztreonam indicated acquisition of plasmids of ca. 7 and 11 kb, respectively, designated plasmids pIP843 and pIP844, respectively (data not shown).
β-Lactam susceptibility of K. pneumoniae.
The MICs of β-lactams for BM4493, E. coli JM83/pIP843, and E. coli JM83 are listed in Table 2. K. pneumoniae BM4493 and E. coli JM83/pIP843 were resistant to high levels of amoxicillin, piperacillin, cephalothin, cefuroxime, and cefotaxime and to low levels of aztreonam. The MICs of cefoxitin and ceftazidime were only slightly increased. The activities of the penicillins and cefotaxime against both strains were restored in the presence of clavulanic acid. The MICs of β-lactams, with and without clavulanic acid, for K. pneumoniae BM4494 and E. coli JM83/pIP844 were similar (data not shown).
Genetic organization of plasmid pIP843.
Analysis of the sequence of pIP843 indicated that it was a 7,086-bp ColE1-like replicon, which is known to replicate under the control of proteins encoded by the host chromosome (14). The plasmid contained five open reading frames (ORFs) that were transcribed in the same orientation and that were preceded by putative Shine-Dalgarno sequences. A consensus sequence for an oriV origin of replication was located from positions 6527 to 6529 in an AT-rich region (30, 38, 41). There were two domains in pIP843 (Fig. 1): the first was responsible for replication and its regulation, and the second contained four adjacent ORFs.
FIG. 1.
Genetic organization of plasmid pIP843. Heavy bar, regulatory region for replication; thin line, noncoding region; bar consisting of multiple rectangles, AT-rich region. The putative functional regions are as follows: RNA II transcript (nt 6024 to 6676); RNA I transcript (complementary to nt 6031 to 6128); oriV replication origin (nt 6527 to 6529); rom-like genes (ORF1; nt 6717 to 6908); oriT transfer origin (nt 6934 to 300); cer-like region (nt 346 to 611); ISEcp1-like element (nt 1138 to 2973); ISEcp1-like ORF2 transposase (nt 1324 to 2586); CTX-M-17 ORF3 (nt 2836 to 3711); IS903 (nt 3714 to 4770); IS903-C ORF4 transposase (nt 1324 to 2586); and outer membrane lipoprotein ORF5 (nt 4918 to 5706).
The first domain on both sides of the oriV origin of replication contained sequences which could control the initiation of DNA replication and the plasmid copy number. ColE1-like plasmids replicate by a common mechanism of initiation (13, 30, 38). On the basis of sequence similarity, the first region, 503 bp upstream from oriV, would code for an RNA II-like element, the primer for initiation of replication, on one strand (positions 6024 to 6676) and would code for an RNA I-like element, an antisense RNA molecule that acts as a negative regulator of replication initiation, on the other strand (positions 6128 to 6031). By hybridizing to RNA II, RNA I inhibits hybrid formation between the preprimer RNA and its DNA template (24). The second region, 191 bp downstream from oriV (positions 6717 to 6908), corresponded to a 192-bp rom-like ORF (ORF1) encoding a putative Rom (RNA I modulator) protein (42), an additional negative regulator of plasmid replication produced by many ColE1-type plasmids. Rom, also called Rop, enhances the interaction of the RNA I inhibitor with its target, resulting in a reduction in the frequency of replication initiation (11). The pIP843 Rom-like protein consisted of 63 amino acids which displayed 58.1% identity to the Rop protein of ColE1. Despite the sequence heterogeneity of Rom and Rop proteins, Rom proteins have been shown to be functional (28).
A second AT-rich portion of ca. 1,300 bp (nucleotides [nt] 6908 to 1137) was located downstream from the rom-like gene and contained two important regions. The 452-bp sequence adjacent to rom (positions 6934 to 300) displayed 61% identity with the oriT transfer origin of plasmids R46 and pCU1 (12, 33). The lack of a mob gene in pIP843 is consistent with the fact that it could not be mobilized. The 265-bp segment, from nt 346 to 611, displayed 90 and 64% identities with the cer (ColE1 resolution) recombination regions of pJHCMW1 (13) and ColE1 (25), respectively. The cer region plays a role in plasmid stability through plasmid monomerization (25, 40).
The second domain in pIP843 contained four ORFs. The first was a 1,656-bp insertion element from nt 1138 to 2973 that was nearly identical to ISEcp1, recently detected in plasmid pST01 from E. coli (P. D. Stapleton, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457, 2000; GenBank accession no. AJ242809). The ends of the ISEcp1-like element were composed of two imperfect inverted repeats with five nucleotide mismatches. The left end was 22 bp (positions 1138 to 1159), and the right one was 24 bp (nt 2950 to 2973). ORF2 (nt 1324 to 2586) encoded a 420-amino-acid putative transposase that differed from that of ISEcp1by 4 amino acids: Asn100→Ile, Ser126→Ile, Trp347→Gly, and Phe366→Val. It displayed 24% identity with the IS492 transposase from Bacteroides fragilis.
A 876-bp ORF (ORF3; nt 2836 to 3711), which could encode a 291-amino-acid protein closely related to CTX-M-type β-lactamases, was located 43 bp downstream from ISEcp1. The consensus sequences 70SXXK73, 130SDN132, E166, and 234KTG236 (Ambler numbering [1]), typical of class A serine-active β-lactamases, were present. The sequence of the bla gene of BM4493, designated blaCTX-M-17, was 98% identical to that of blaCTX-M-9 (36) and was closest to that of blaCTX-M-14 (32) with only two base pair changes that led to the single Glu289→Lys substitution. These observations indicate that CTX-M-17 belongs to the fourth group of CTX-M β-lactamases mentioned above.
Two nucleotides from the stop codon, the blaCTX-M-17 gene was flanked by a 1,057-bp IS903-like element (locations 3714 to 4770) designated IS903-C. The IS903-C copy possessed 18-bp terminal inverted repeats and an ORF (ORF4) coding for a 308-amino-acid putative transposase. The IS903-C copy was 95 and 96.5% identical to IS903 (19) and IS903-B (29), respectively. It differed from IS903 by 42 mutations, leading to 10 amino acid changes, including six isofunctional substitutions, and differed from IS903-B by 30 mutations, leading to 7 amino acid changes, including five isofunctional substitutions. IS903-B and IS903 differ by 34 nucleotide substitutions, which lead to eight amino acid alterations that do not affect the activity of the transposase (29).
The 789-bp ORF5 (positions 4918 to 5706) could encode a 261-amino-acid truncated version of the TonB-dependent protein, an outer membrane lipoprotein involved in iron uptake.
In summary, blaCTX-M-17 was flanked upstream by the ISEcp1-like element and downstream by IS903-C, a genetic organization that could promote mobility of the blaCTX-M-17 gene. However, target duplications were not detected at the left end of ISEcp1 or at the right end of IS903-C.
Transcriptional analysis of blaCTX-M-17 gene.
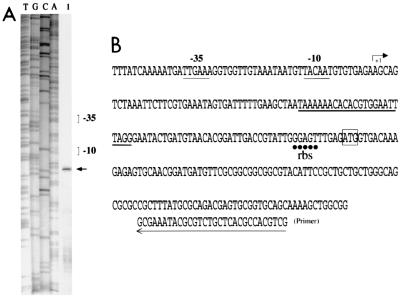
Primer extension with oligodeoxynucleotide PE (complementary to a sequence 74 to 103 bp downstream from the initiation site of blaCTX-M-17) indicated that the transcriptional start site was located in ISEcp1 109 nucleotides from the start codon of the blaCTX-M-17 gene (Fig. 2). The −35 (TTGAAA) and −10 (TACAAT) sequences of the blaCTX-M-17 promoter were separated by 18 bp and were closely related to the consensus motifs for the σ70 factor of E. coli RNA polymerase (10). These results are not in agreement with those suggesting that the putative promoter is upstream from the blaCMY-2 gene, as has been proposed previously (6).
FIG. 2.
Identification of the transcriptional start site for blaCTX-M-17 in strain BM4493 by primer extension analysis. (A) Lane 1, primer elongation product obtained with oligodeoxynucleotide PE and 50 μg of total RNA of BM4493 (arrow); lanes T, G, C, and A, results of sequencing reactions performed with pIP843 DNA as a template and the PE primer. (B) Sequence from nt 2676 to 2950 (the numbering is that for the sequence of the strain with GenBank accession no. AY033516); +1, transcriptional start site for blaCTX-M-17 in BM4493, indicated by an arrow; the −35 and −10 promoter sequences upstream from the transcriptional start site are underlined with a thin line; the right terminal repeat of the ISEcp1-like element is underlined with a thick line; the ATG start codon of blaCTX-M-17 is boxed; and the ribosome-binding site (rbs) is underlined with dots. The location of the PE primer is indicated with an arrow at the bottom.
Transposition of ISEcp1-like element.
Transposition of the ISEcp1-like element was assayed by plasmid conduction in E. coli. E. coli HB101 harboring plasmids pOX38 (Tra+ Gmr) and pIP843-1 (Tra− Mob− Ctxr) (in which the putative transposase of IS903-C has been disrupted, as described above) was mated with E. coli DH5α (Nalr). Transconjugants were obtained on medium containing nalidixic acid (40 μg/ml) and cefotaxime (4 μg/ml) at a frequency of 10−6 per donor. A transconjugant that was resistant to cefotaxime and gentamicin and that may have been generated by cointegrate formation between pIP843 (Tra− Mob− Ctxr) carrying the ISEcp1-like element and pOX38 (Tra+ Gmr) was selected for further study. The plasmid content of the transconjugant was purified, digested with EcoRI (an enzyme that cuts the ISEcp1-like element at a single site), and analyzed by Southern hybridization with an ISEcp1-specific probe for putative duplication of ISEcp1. The results obtained indicate the presence of a single copy of the ISEcp1-like element (data not shown), in agreement with the one-ended model for transposition of ISEcp1 proposed by Stapleton (40th ICAAC; GenBank accession no. AJ242809).
Genomic environment of blaCTX-M-17 in K. pneumoniae BM4494.
Study of K. pneumoniae BM4494, which was isolated in Vietnam during the same period that strain BM4493 was isolated, permitted detection of a blaCTX-M-17 gene borne on nonconjugative 11-kb plasmid pIP844. A part of the pIP844 HindIII insert in pAT797, consisting of 2,625 bp, was sequenced. Interestingly, this sequence revealed the presence of an identical blaCTX-M-17 sequence flanked immediately downstream by IS903-C and upstream by a truncated ISEcp1 element in which the end containing the blaCTX-M-17 promoter was conserved.
A data bank search showed that ISEcp1was also present upstream from various genes coding for other broad-spectrum β-lactamases and AmpC cephalosporinases, such as blaCTX-M-15 (22), blaCMY-5 (44; GenBank accession no. Y17716), blaCMY-2 (GenBank accession no. X91840), and blaACC-1 (GenBank accession no. AJ133121). In plasmid pST01, blaCMY-4 is flanked upstream by ISEcp1 and downstream by part of a gene coding for an outer membrane lipoprotein. Similar to blaCTX-M-17 in pIP843, blaCTX-M-14 is found in association with ISEcp1and IS903 32; GenBank accession no. AF252622). Our data suggest that the −35 and −10 sequences located in the ISEcp1-like element could direct transcription not only of blaCTX-M-17 but also of other bla genes following a transposition event. The fact that the same IS903-C copy was found to be present downstream from the blaCTX-M-17 gene in two different plasmids, whereas an entire or a truncated ISEcp1-like element was present upstream, suggests that IS903-C plays a role in the dissemination of the blaCTX-M-17 gene. An epidemiological study to establish the prevalence of this mechanism of resistance to cefotaxime in Vietnam is being carried out.
Acknowledgments
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases. V.C. was a recipient of a fellowship from the Réseau International des Instituts Pasteur et Instituts Associés.
We thank S. Magnet and G. Gerbaud for technical advice and G. Arlet for helpful discussions.
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J.-M. Frere, J.-M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Barthélémy, M., J. Peduzzi, H. Bernard, C. Tancrède, and R. Labia. 1992. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15-22. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., H. Grimm, and S. Schweighart. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294-298. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilheilm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 6.Bauernfeind, A., I. Stemplinger, R. Jungwirth, and H. Giamarellou. 1996. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, R., J. L. M. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet, R., C. Dutour, J. L. M. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 11.Cesarini, G., M. Helmer-Citterich, and L. Castagnoli. 1991. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 7:230-235. [DOI] [PubMed] [Google Scholar]
- 12.Coupland, G. M., A. M. C. Brown, and N. S. Willetts. 1987. The origin of transfer (oriT) of the conjugative plasmid R46: characterization by deletion analysis and DNA sequencing. Mol. Gen. Genet. 208:219-225. [DOI] [PubMed] [Google Scholar]
- 13.Dery, K. J., R. Chavideh, V. Waters, R. Chamorro, L. S. Tolmaky, and M. E. Tolmasky. 1997. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid 38:97-105. [DOI] [PubMed] [Google Scholar]
- 14.Donoghue, D. J., and P. A. Sharp. 1978. Replication of colicin E1 plasmid DNA in vivo requires no plasmid-encoded proteins. J. Bacteriol. 133:1287-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazouli, M., E. Tzelepi, S. V. Sidorenko, and L. S. Tzouvelekis. 1998. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob. Agents Chemother. 42:1259-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazouli, M., E. Tzelepi, A. Markogiannakis, N. J. Legakis, and L. S. Tzouvelekis. 1998. Two novel plasmid-mediated cefotaxime-hydrolysing β-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol. Lett. 165:289-293. [DOI] [PubMed] [Google Scholar]
- 17.Gazouli, M., N. J. Legakis, and L. S. Tzouvelekis. 1998. Effect of substitution of Asn for Arg-276 in the cefotaxime-hydrolyzing class A β-lactamase CTX-M-4. FEMS Microbiol. Lett. 169:289-293. [DOI] [PubMed] [Google Scholar]
- 18.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grindley, N., and C. M. Joyce. 1981. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harbor Symp. Quant. Biol. 45:125-133. [DOI] [PubMed] [Google Scholar]
- 20.Ibuka, A., A. Taguchi, M. Ishiguro, S. Fushinobu, Y. Ishii, S. Kamitori, K. Okuyama, K. Yamaguchi, M. Konno, and H. Matsuzawa. 1999. Crystal structure of the E166A mutant of extended-spectrum β-lactamase Toho-1 at 1.8 Å resolution. J. Mol. Biol. 285:2079-2087. [DOI] [PubMed] [Google Scholar]
- 21.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karim, A., L. Poirel, S. Nagarajan, and P. Nordman. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 23.Kariuki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacatena, R. M., and G. Cesareni. 1981. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature 294:623-626. [DOI] [PubMed] [Google Scholar]
- 25.Leung, D. W., E. Chen, G. Cachianes, and D. V. Goeddel. 1985. Nucleotide sequence of the partition function of Escherichia coli plasmid ColE1. DNA 4:351-355. [DOI] [PubMed] [Google Scholar]
- 26.Ma, L., Y. Yshii, M. Ishiguro, H. Matsuzawa, and K. Yamaguchi. 1998. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob. Agents Chemother. 42:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mabilat, C., J. Lourencao-Vital, S. Goussard, and P. Courvalin. 1992. A new example of physical linkage between Tn1 and Tn21: the antibiotic multiple resistance region of plasmid pCFF04 encoding extended spectrum β-lactamase TEM-3. Mol. Gen. Genet. 235:113-121. [DOI] [PubMed] [Google Scholar]
- 28.Mikiewicz, D., B. Wro'bel, G. Wegrzyn, and A. Plucienniczak. 1997. Isolation and characterization of a ColE1-like plasmid from Enterobacter agglomerans with a novel variant of rom gene. Plasmid 38:210-219. [DOI] [PubMed] [Google Scholar]
- 29.Mollet, B., S. Iida, and W. Arber. 1985. An active variant of the procaryotic transposable element IS903 carries an Ambler stop codon in the middle of an open reading frame. Mol. Gen. Genet. 199:534-536. [DOI] [PubMed] [Google Scholar]
- 30.Nomura, N., and Y. Murooka. 1994. Characterization and sequencing of the region required for replication of a non-self-transmissible plasmid pEC3 isolated from Erwinia carotovora subsp. carotovora. J. Ferment. Bioeng. 78:250-254. [DOI] [PubMed] [Google Scholar]
- 31.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pai, H., E.-H. Choi, H.-J. Lee, J. Y. Hong, and G. A. Jacoby. 2001. Identification of CTX-M-14 extended-spectrum beta-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39:3747-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson, E. S., and V. N. Iyer. 1992. The oriT region of the conjugative transfer system of plasmid pCU1 and specificity between it and the mob region of other N tra plasmids. J. Bacteriol. 174:499-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., T. Nass, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynaud, A., J. Peduzzi, M. Barthélémy, and R. Labia. 1991. Cefotaxime-hydrolyzing activity of the β-lactamase of Klebsiella oxytoca D488 could be related to a threonine residue at position 140. FEMS Microbiol. Lett. 81:185-192. [DOI] [PubMed] [Google Scholar]
- 36.Sabate, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Selzer, G., T. Som, T. Itoh, and J. Tomizawa. 1983. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell 32:119-129. [DOI] [PubMed] [Google Scholar]
- 39.Spratt, B. G., P. J. Hedge, S. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 40.Summers, D. K., and D. J. Sherratt. 1984. Multimerization of high copy number plasmids causes instability: ColE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097-1103. [DOI] [PubMed] [Google Scholar]
- 41.Tomizawa, J.-I., H. Ohmori, and R. E. Bird. 1977. Origin of replication of colicin E1 plasmid DNA. Proc. Natl. Acad. Sci. USA 74:1865-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Twigg, A. J., and D. J. Sherratt. 1980. trans-Complementable copy-number mutants of plasmid ColE1. Nature 238:216-218. [DOI] [PubMed] [Google Scholar]
- 43.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization of nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]