Abstract
A total of 204 isoniazid (INH)-resistant strains of Mycobacterium tuberculosis isolated from different patients in the northwestern region of Russia from 1996 to 2001 were screened by a PCR-restriction fragment length polymorphism (RFLP) assay. This assay uses HapII cleavage of an amplified fragment of the katG gene to detect the transversion 315AGC→ACC (Ser→Thr), which is associated with INH resistance. This analysis revealed a 93.6% prevalence of the katG S315T mutation in strains from patients with both newly and previously diagnosed cases of tuberculosis (TB). This mutation was not found in any of 57 INH-susceptible isolates included in the study. The specificity of the assay was 100%; all isolates that contained the S315T mutation were classified as resistant by a culture-based susceptibility testing method. The Beijing genotype, defined by IS6110-RFLP analysis and the spacer oligonucleotide typing (spoligotyping) method, was found in 60.3% of the INH-resistant strains studied. The katG S315T shift was more prevalent among Beijing genotype strains than among non-Beijing genotype strains: 97.8 versus 84.6%, respectively, for all isolates, including those from patients with new and previously diagnosed cases, isolated from 1999 to 2001 and 100.0 versus 86.5%, respectively, for isolates from patients with new cases isolated from 1996 to 2001. The design of this PCR-RFLP assay allows the rapid and unambiguous identification of the katG 315ACC mutant allele. The simplicity of the assay permits its implementation into routine practice in clinical microbiology laboratories in regions with a high incidence of TB where this mutation is predominant, including northwestern Russia.
The last decade of the 20th century was marked by the reemergence and epidemic spread of tuberculosis (TB) in the Russian Federation. In 1999, the prevalence of tuberculosis in the northwestern region of Russia, including St. Petersburg, with a total population of about 14 million, was estimated to be 278.2 per 100,000 population, the incidence was estimated to be 68.1 per 100,000 population, and the mortality rate was estimated to be 9.4 per 100,000 population. Coincident with the reemergence of TB has been the emergence of multiple-drug-resistant (MDR) Mycobacterium tuberculosis strains. MDR M. tuberculosis strains are generally considered those resistant to at least rifampin (RIF) and isoniazid (INH). These drugs are recommended by the World Health Organization DOTS (directly observed therapy, short course) regimen (40) and are used in the standard treatment protocol officially adopted by the Russian Ministry of Health (18).
Resistance to RIF in M. tuberculosis has been associated with mutations in rpoB, the gene coding for the β subunit of RNA polymerase, the main target of RIF (28). It has been demonstrated that 95 to 98% of resistant isolates have mutations in rpoB. Ninety-five percent of the mutations are located in a core region of rpoB (22). This implies that rather elaborate and expensive methods like sequencing, the line probe assay, and assays that use microarrays are required to identify these mutations (3). Unlike RIF resistance, INH resistance is apparently controlled by a more complex genetic system that involves several genes (22, 25). However, extensive studies have demonstrated that INH resistance is most frequently associated with a single mutation in katG, a gene that encodes the catalase-peroxidase enzyme in M. tuberculosis. INH is a prodrug and requires catalytic activation to be converted into its active form. The M. tuberculosis catalase-peroxidase enzyme has been shown to accomplish this function (4). Early studies showed that complete deletion of the gene is rare, likely due to the importance of its peroxidase component for cell viability (7, 19, 28). For this reason the predominant mode of acquisition of resistance via katG alterations is the selection of particular mutations that decrease the catalase activity but that maintain a certain level of the peroxidase activity of the enzyme in viable INH-resistant (INHr) organisms. Such mutations were found in up to 90% of the INHr strains. One particular substitution in codon 315, AGC→ACC (Ser→Thr), was reported to be the most frequent. This mutation appears to provide the optimal balance between decreased catalase activity and a sufficiently high level of peroxidase activity in KatG (22). The phenotypic level of resistance of such strains is typically in the intermediate range (1 to 2 μg/ml) (25). Also, InhA (enoyl-ACP-reductase), a protein involved in mycolic acid and subsequent cell wall biosyntheses, was identified as a main target of INH, and the mutations linked to the INH resistance phenotype were described in two regions of the inhA locus. The mutations in the putative promoter region upstream of orf1-inhA are thought to increase the level of InhA protein expression, thereby elevating the drug target levels and producing INH resistance by a drug titration mechanism (15). The inhA mutations which change the NADH binding site of InhA most affected by INH were also described in resistant isolates (23). Two other genes related to INH resistance have recently been suggested: the kasA gene, which encodes another INH target, β-ketoacyl-ACP-synthase, and the regulatory region of the ahpC gene, which encodes alkylhydroperoxidase. Mutations in these genes seem to provide a supplementary mechanism of resistance and need further investigation (10, 11, 22, 27).
In a previous study (12), we showed a high prevalence of the katG S315T substitution among 24 INHr M. tuberculosis strains recovered from November 1993 to March 1995 in the St. Petersburg area of Russia. For the present study we extended the surveillance to the years 1996 to 2001. We screened a representative selection of INHr M. tuberculosis isolates recovered from patients in northwestern Russia from 1996 to 2001 for the presence of the katG S315T mutation. We also analyzed the frequency of the S315T shift in different patient groups and isolates of different M. tuberculosis genotypes (strains).
MATERIALS AND METHODS
M. tuberculosis isolates and susceptibility testing.
A total of 261 strains recovered from 261 different adult patients (age range, 15 to 63 years) were studied. Some of these patients (n = 139) had newly diagnosed pulmonary TB, and others had previously been diagnosed with pulmonary TB and had active TB during the surveillance period for the present study. These patients originated from St. Petersburg and three neighboring provinces of northwestern Russia (Leningrad Oblast, Novgorod, and Pskov) and were admitted to the hospital of the St. Petersburg Institute of Phthisiopulmonology and the City Anti-Tuberculosis Dispensary of St. Petersburg between 1996 and 2001. According to the World Health Organization definition, TB in a patient who had not received antituberculous treatment for more than 1 month was considered a new case of TB (40).
For each patient, only the first available isolate was included in the study. Löwenstein-Jensen medium was used for cultivation of isolates, and susceptibility testing was performed by the absolute concentration method, as recommended by the Russian Ministry of Health (Order No. 558 of 28 June 1978) and as has been described previously (38). A microbial suspension containing 5 × 108 organisms/ml was prepared according to McFarland turbidity standards and was diluted 1:10; then, 0.2 ml of the dilution was added to Löwenstein-Jensen medium with or without a drug. The culture tubes were incubated at 37°C, and growth was monitored after 3 weeks of incubation and assessed as described previously (39). An isolate was considered resistant when bacterial growth occurred in the presence of a concentration of 1 μg of INH per ml, 20 μg of RIF per ml, 5 μg of streptomycin (STR) per ml, 2 μg of ethambutol per ml, and 100 μg of pyrazinamide (pH 5.6) per ml. The method of absolute concentration was previously shown in a comparative study with the National Mycobacterial Reference Laboratory in Turku, Finland, to give results concordant with those generated by the proportion method in our setting (38).
PCR-RFLP analysis.
DNA preparations were obtained as described by van Embden et al. (32) or Mazars et al. (14). Amplification of the fragment with katG codon 315 (the fragment in katG from positions 904 to 1103; http://genolist.pasteur.fr/TubercuList) was performed in a PTC-100 thermal controller (MJ Research, Inc.) with primers katG1F (5′-AGCTCGTATGGCACCGGAAC) and katG4RB (5′-AACGGGTCCGGGATGGTG) in 30 μl of a PCR mixture (15 pmol of each primer, 1.5 mM MgCl2, 1 U of recombinant Taq DNA polymerase [Amersham Pharmacia Biotech], 200 μM each deoxynucleoside triphosphate) under the following conditions: initial denaturation at 95°C for 4 min; 30 cycles of 94°C for 1 min, 61°C for 1 min, and 72°C for 1 min; and a final elongation at 72°C for 4 min. The amplified fragment was assessed by electrophoresis in a 1.5% agarose gel and was cleaved with HapII (Amersham Pharmacia Biotech) according to the instructions of the manufacturer. The restriction fragments obtained were electrophoresed in a 1.5% agarose gel and were visualized under UV light on a transilluminator.
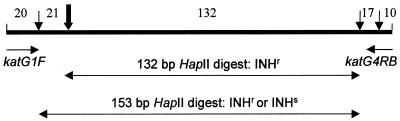
This PCR-restriction fragment length polymorphism (RFLP) assay was designed to detect the katG codon mutation AGC(Ser)→ ACC(Thr), which leads to the INH resistance phenotype. This mutation creates an additional HapII site (CCGG) and thus can be detected by use of this restriction endonuclease. The primers were selected to amplify a rather short 200-bp katG fragment spanning codon 315 in order to avoid interference with other HapII sites situated in the proximity of this region that would otherwise have produced additional screening fragments (Fig. 1). Time-consuming excision of the product from the gel and purification procedures were omitted in our study, and the PCR product was directly subjected to HapII cleavage. As a result, the longest RFLP product obtained was 132 bp for the INHr strain with a mutated 315ACC allele and 153 bp for the katG codon 315 wild type or differently mutated allele. These indicative bands could be clearly discriminated by 1.5% agarose minigel electrophoresis: they were the only visible bands, while the shorter 10- to 21-bp bands (Fig. 1) ran out of the gel. Figure 2 presents the different typical profiles generated by this PCR-RFLP assay. Theoretically, by this assay one could also detect heteroresistance (a mixed population of INHr strains with 315ACC and INH-susceptible [INHs] M. tuberculosis strains), as both bands (bands of 132 and 153 bp) are clearly distinguishable in a single lane (Fig. 2, lane 8).
FIG. 1.
Schematic illustration of the katG 200-bp fragment amplified with primers katG1F and katG4RB. Vertical thin arrows indicate HapII restriction sites (CCGG); the bold arrow marks codon 315, where the AGC→ACC transversion creates an additional HapII site in INHr M. tuberculosis strains. Horizontal double-headed arrows represent the principal HapII digestion products resulting from different katG codon 315 alleles.
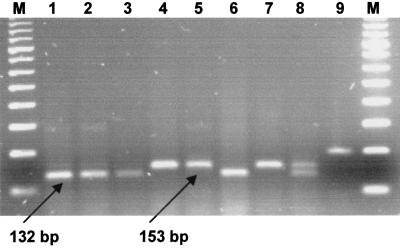
FIG. 2.
Gel electrophoresis of the amplified katG fragment and the products of its digestion by HapII. Lanes: 1, 2, 3, and 6, products obtained by HapII digestion of katG of INHr strains with mutated katG (315ACC); 4, 5, and 7, products obtained by HapII digestion of katG of strains not harboring the katG AGC→ACC mutation at codon 315; 8, artificially mixed digests of PCR products from both wild-type and ACC alleles of katG codon 315; 9, undigested amplified 200-bp katG fragment; M, 100-bp DNA ladder (Amersham Pharmacia Biotech).
Control reactions for the detection of false-positive results due to possible contamination with previously amplified amplicons were performed as follows. A negative control sample (distilled water) was included in each PCR run; no contamination was detected.
DNA fingerprinting.
The DNA of the strains studied was also subjected to IS6110 RFLP typing (32) and spacer oligonucleotide typing (spoligotyping) (8), as described previously. The IS6110 RFLP assay patterns were compared, and a dendrogram was constructed with the GelCompar (version 4.1) package (BVBI Applied Maths, Kortrijk, Belgium) by the unweighted pair group method of arithmetic averages by using the Dice coefficient.
RESULTS
A total of 204 INHr isolates and 57 INHs isolates from 261 different patients were examined. Of the INHr strains, 112 were from patients who had active TB during the surveillance period from 1996 to 2001 but who had previously (from 1974 to 1995) been diagnosed with TB and 92 were from patients with newly diagnosed cases.
The distributions of the susceptibility profiles of the INHr strains among the different patient groups are presented in Table 1. Interestingly, 200 of the 204 (98.0%) INHr strains were also resistant to streptomycin; four strains were INH monoresistant (3 of these INH-monoresistant strains were isolated from patients with new cases [Table 1]).
TABLE 1.
Susceptibility profiles of INHr M. tuberculosis strains
| Patient group | No. of strainsa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H | SH | SR | HR | SHR | SHE | SHRE | SHRZ | SHREZ | Total | |
| Patients with newly diagnosed TB | 3 | 22 | 0 | 0 | 45 | 1 | 8 | 8 | 5 | 92 |
| Patients with previously diagnosed TB | 1 | 9 | 1 | 1 | 75 | 0 | 6 | 15 | 4 | 112 |
| Total | 4 | 31 | 1 | 1 | 120 | 1 | 14 | 23 | 9 | 204 |
Drug resistance abbreviations: S, STR; R, RIF; H, INH; E, ethambutol, Z, pyrazinamide.
The 57 randomly selected INHs strains included in the study showed the following resistance profiles: 39 were susceptible to all drugs tested, 10 were resistant to streptomycin alone, 1 was resistant to pyrazinamide, 1 was resistant to RIF only, and 6 were resistant to both streptomycin and RIF. None of these isolates exhibited the katG S315T mutation. Since other rarely described mutations in codon 315 confer INH resistance (22, 25), we assume that these INHs strains had a wild-type allele at codon 315 (AGC).
A discrepancy between the results of phenotypic and genotypic drug resistance testing was found for three strains that were phenotypically identified as INH susceptible but that had the S315T mutation. These DNA samples were retested by PCR-RFLP analysis, and the prior results were confirmed. Phenotypic susceptibility testing was repeated for the same isolates, and they were proved to be resistant. For further analysis, the final phenotypic test result was considered correct for these three strains.
The results of the PCR-RFLP assay of INHr isolates are summarized in Table 2. Analysis of the distribution of the katG 315ACC allele showed that it was highly prevalent in strains from patients with both previously diagnosed and new cases: 92.9 and 94.6%, respectively. As MDR is defined as resistance to at least RIF and INH and because almost all the isolates (200 of 204) were STR resistant, we subdivided the strains into two groups: INHr and RIFs strains and INHr and RIFr strains. There was essentially no difference in the frequency of this mutation between MDR (INHr and RIFr, 94.6%) and INHr and RIFs (89.7%) strains. Taken together, these data indicate that in our setting the specificity and sensitivity of this PCR-RFLP assay for the detection of INH resistance were 100 and 93.6%, respectively.
TABLE 2.
Frequency of katG 315ACC allele among 204 INH-resistant M. tuberculosis strains revealed by PCR-RFLP analysis
| Phenotype and patient group | No. of strains with 315ACC allele/total no. of strains tested
|
% Strains with 315ACC allele | ||||
|---|---|---|---|---|---|---|
| 1996-1997 | 1998 | 1999 | 2000-2001 | Total | ||
| INHr | 29/30 | 28/30 | 84/92 | 50/52 | 191/204 | 93.6 |
| Previously diagnosed cases | 23/24 | 21/22 | 31/37 | 29/29 | 104/112 | 92.9 |
| INHr and RIFs | 0/0 | 7/7 | 2/3 | 2/2 | 11/12 | |
| INHr and RIFr | 23/24 | 14/15 | 29/34 | 27/27 | 93/100 | |
| New cases | 6/6 | 7/8 | 53/55 | 21/23 | 87/92 | 94.6 |
| INHr and RIFs | 1/1 | 2/2 | 15/16 | 6/8 | 24/27 | |
| INHr and RIFr | 5/5 | 5/6 | 38/39 | 15/15 | 63/65 | |
| Beijing genotype | 11/12 | 17/18 | 54/56 | 36/36 | 118/122 | 96.7 |
| Other genotypes | 18/18 | 11/12 | 30/36 | 14/16 | 73/82 | 89.0 |
| New cases, Beijing genotype | 3/3 | 4/4 | 34/34 | 14/14 | 55/55 | 100.0 |
| New cases, other genotypes | 3/3 | 3/4 | 19/21 | 7/9 | 32/37 | 86.5 |
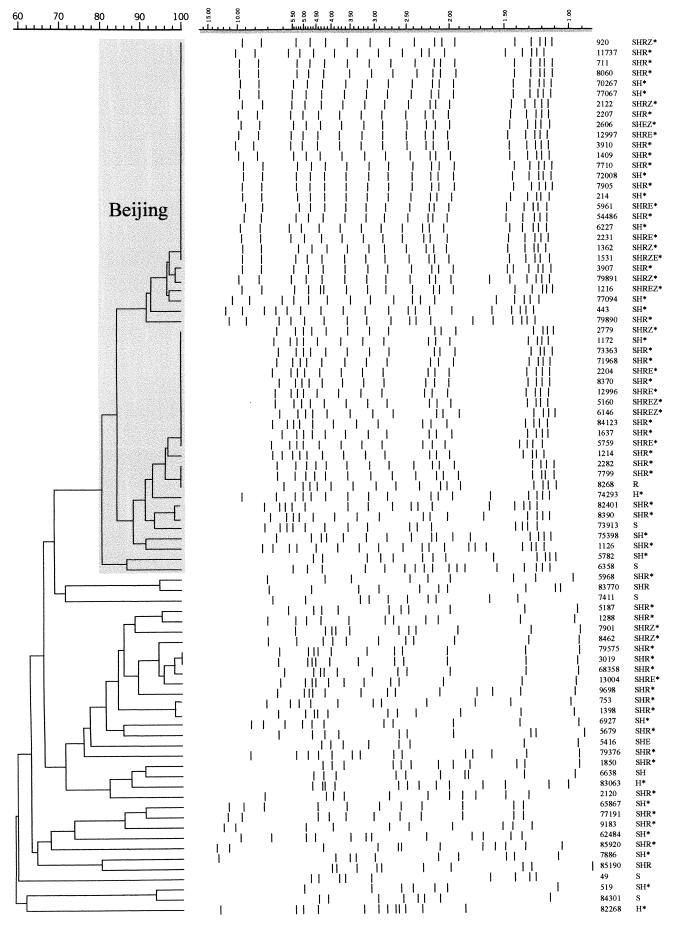
Two standardized DNA fingerpinting techniques, spoligotyping (8) and IS6110 RFLP typing (32), were used to differentiate the strains in order to assess their genetic relatedness. Forty-one different profiles were obtained by spoligotyping (Table 3), and the profiles were compared with those in the spoligotype database of Sola et al. (26). A distinctive pattern that consisted of signals 35 to 43 was shared by the majority (121 of 204) of strains. This nine-signature-signal spoligotyping profile (spoligoprofile) (Table 3, type R0 and 1) is typical of the Beijing family genotype (26, 35). Two strains showed incomplete Beijing profiles that lacked single signals 37 and 40 (Table 3, type R33 and types R34 and 190, respectively). Other than Beijing types, the 38 particular spoligotypes included from 1 to 11 strains (Table 3). The 85 drug-resistant isolates from patients with newly diagnosed TB were subjected to IS6110 RFLP fingerprinting; the profiles obtained were used to construct a dendrogram (Fig. 3). A total of 18 different IS6110 RFLP profiles were identified among the 52 strains of the Beijing family (Fig. 3, spoligotypes R0 and 1); these profiles clustered at a level of 80% similarity (Dice coefficient). In particular, two distinct profiles within the Beijing cluster were shared: one by 22 strains and the other by 12 strains (Fig. 3). Thirty-two different IS6110 RFLP profiles were identified among 33 strains of genotypes other than the Beijing genotype. The number of copies of IS6110 per isolate ranged from 14 to 19 for the Beijing family strains and from 7 to 14 for strains of the other genotypes (Fig. 3).
TABLE 3.
Schematic representation of spoligoprofiles of the M. tuberculosis strains studieda
| Spoligoprofile | Typeb | Reference no.c | No. of strains |
|---|---|---|---|
| R0 | 1 | 121 | |
| R2 | 53 | 4 | |
| R8 | 253 | 2 | |
| R9 | 50 | 1 | |
| R11 | 42 | 8 | |
| R12 | 161 | 1 | |
| R14 | 252 | 11 | |
| R16 | 35 | 2 | |
| R19 | 4 | ||
| R21 | 1 | ||
| R22 | 1 | ||
| R23 | 237 | 1 | |
| R24 | 251 | 2 | |
| R26 | 1 | ||
| R27 | 2 | ||
| R28 | 2 | ||
| R29 | 1 | ||
| R30 | 254 | 6 | |
| R31 | 1 | ||
| R32 | 1 | ||
| R33 | 1 | ||
| R34 | 190 | 1 | |
| R38 | 1 | ||
| R39 | 191 | 2 | |
| R40 | 1 | ||
| R41 | 1 | ||
| R42 | 47 | 3 | |
| R43 | 1 | ||
| R44 | 2 | 1 | |
| R45 | 1 | ||
| R46 | 2 | ||
| R48 | 1 | ||
| R49 | 1 | ||
| R50 | 2 | ||
| R51 | 2 | ||
| R52 | 2 | ||
| R53 | 1 | ||
| R54 | 1 | ||
| R55 | 1 | ||
| R56 | 1 | ||
| R57 | 1 |
The boldface numbers represent the spoligotypes of the Beijing family.
Arbitrary code accepted in our laboratory.
Spoligotype from the database of Sola et al. (26).
FIG. 3.
Dendrogram obtained by the IS6110 RFLP assay-based unweighted pair group method of arithmetic averages for the drug-resistant M. tuberculosis clinical isolates from patients with new cases of TB. The Beijing cluster is in the shaded area. The positions of the bands in each lane are adjusted (normalized) so that the band positions for all strains are comparable. The scale on the left depicts similarity coefficients, which are defined elsewhere (32); the scale on the right (shaded) shows band sizes (in kilobase pairs) obtained by the IS6110 RFLP assay. The presence of the mutation at katG codon 315 (AGC→ACC) is indicated by an asterisk. Drug resistance abbreviations: S, STR; R, RIF; H, INH; E, ethambutol, Z, pyrazinamide.
DISCUSSION
This study was undertaken to gain further insight into the molecular basis of the INH resistance of M. tuberculosis clinical strains circulating in northwestern Russia. The majority of INHr strains collected over the entire study period (1996 to 2001) from both patients with previously diagnosed cases of TB (89.3%) and patients with new cases of TB (71.7%) showed the triple-drug-resistance profile (resistance to STR, INH, and RIF) (Table 1). Nearly the same percentages were observed for the 3-year period from 1999 to 2001: 92.4 and 69.2% for patients with previously diagnosed and new TB cases, respectively (Table 2). In general, the resistance profiles corroborated the expected pattern of acquisition of resistance to particular drugs, i.e., INH resistance preceding resistance to RIF. The high prevalence of STR resistance (200 of 204 INHr strains; Table 1) is a feature characteristic of the current TB epidemic in Russia and may be explained by overuse of the drug for treatment of other nontuberculous diseases and poor adherence to treatment protocols. Furthermore, STR was invariably used for TB treatment in prisons, and many prisoners were released back into society in the 1990s.
Our results on the variation at katG codon 315 (Table 2) are in concordance with data published by other researchers. The prevalence of the katG S315T substitution in M. tuberculosis strains from around the world varies, especially with regard to the prevalence of TB. In the regions where the prevalence of TB is intermediate and low, this mutation has been reported relatively infrequently: in 26 to 30% of isolates in Singapore (11) and Madrid (19) and rarely in isolates from Scotland (6) and Finland (13). In contrast, the S315T mutation accounted for INH resistance in 52 to 64% of strains in Africa (4, 7, 37), 79% of strains in Peru (5), 91% of strains in Russia (24 strains studied in 1993 to 1994 [12]), and 58% of strains in New York City (19). The present study demonstrated the high prevalence of the 315ACC mutant allele among M. tuberculosis isolates in an area of northwestern Russia: in 93 to 100% of INHr isolates, depending on the patient group and genotype, versus an average of 60 to 65% isolates in other countries with a high prevalence of TB. It may be explained by noncompliance in some patient groups (e.g., homeless persons, refugees, and drug abusers) and a lack of resources for TB control programs in different regions of Russia (18).
We also investigated the distribution of the katG S315T mutation among different strains of M. tuberculosis by molecular typing. Two different typing methods were used: IS6110 RFLP analysis and direct repeat (DR)-based spoligotyping. It has been demonstrated that the housekeeping genes of M. tuberculosis exhibit a high degree of conservation and that genotypic discrimination of individual strains is possible by evaluation of insertion and repetitive elements. In particular, the DR locus is characteristic of the M. tuberculosis complex and consists of multiple tandem 36-bp repeats interspersed with variable spacers of about the same size (8, 33). The DR and the adjacent variable sequence form a direct variant repeat (DVR). Polymorphism of the DR locus (the absence or presence of single DVRs) has been exploited widely for the differentiation of strains of the M. tuberculosis complex by the spoligotyping method on the basis of 43 distinct DVRs (8, 26). Twenty-five additional DR spacers have recently been described (33), but use of these sequences has provided only slight improvement in the ability to discriminate among strains of M. tuberculosis (24, 33). Spoligotyping is less discriminatory than IS6110-based fingerprinting, especially for strains with high IS6110 copy numbers (1, 8, 26, 30), and is most suitable for the preliminary discrimination of strains. Subsequently, the strains with identical spoligoprofiles may be subtyped by IS6110 RFLP analysis, which is widely used as a standardized epidemiological typing technique (30, 32).
The Beijing family of M. tuberculosis is recognized by specific IS6110 RFLP profiles: the DR locus, which consists of 15 units in virtually all strains; and the typical spoligoprofile, which consists of nine signals (1, 24, 33, 35). This genetic family, initially found to be endemic in the countries of East Asia (35), is marked by high transmissibility and is distributed worldwide (26). Previously, we showed its predominance in the northwestern region of Russia (16, 17); the Beijing family strains were identified in more than 50% of patients (about 450 strains examined from 1996 to 2001 [O. Narvskaya and I. Mokrousov, unpublished data]) by using IS6110 RFLP typing and spoligotyping. In the present study we compared the distribution of the katG S315T mutation in the Beijing family of strains versus that in strains of other genotypes. In our sample, the proportion of Beijing strains was 60% (123 of 204 INHr strains; Tables 2 and 3). This frequency was slightly higher than expected, but this may be explained by biased evaluation of INHr strains. Of note, the Beijing strains studied, even those differentiated by IS6110 RFLP analysis, was a closely related group (Fig. 3). The S315T substitution in katG was observed in 97.8% of INHr Beijing strains and in 84.6% of INHr non-Beijing strains isolated from 1999 to 2001 and in 96.7 and 89.0% of strains, respectively, isolated over the entire study period, 1996 to 2001 (odds ratio, 3.6; 95% confidence interval, 1.0 to 14.6; Table 2). We also compared the distributions of the katG S315T mutation among Beijing and non-Beijing INHr strains isolated from the subgroup of patients with new cases of TB (Table 2; Fig. 3). The katG 315ACC mutant allele was present in 100.0% (55 of 55) and 86.5% (32 of 37) of Beijing and non-Beijing INHr strains, respectively, from patients with new cases of TB (Table 2). Our data obtained by IS6110 RFLP typing demonstrate that a majority of drug-resistant strains from patients with newly diagnosed cases belonged to the Beijing family and that all of the INHr strains had the katG S315T mutation (Fig. 3). These results confirm our previous observation that the epidemic spread of MDR TB in northwestern Russia is due to a greater extent to the clonal dissemination of MDR strains of the Beijing genotype than to the dissemination of strains of other genotypes (17). We assume that ongoing transmission of these strains could be the driving force of such a high prevalence of the katG S315T mutation.
Generally, the Beijing family strains do not appear to be inherently MDR (1, 21, 30). In the present study, 10 of 39 (25.6%) pansusceptible strains, 4 of 10 (40%) STR-monoresistant strains, and 1 of 4 INH-monoresistant strains belonged to the Beijing type. However, the majority of these strains currently circulating in Russia are apparently highly transmissible and MDR. Our results suggest that strains of the Beijing genotype more readily acquire the S315T mutation in katG, but further studies are required to confirm this assumption.
The evaluation of a limited number of gene codons in the genome of M. tuberculosis reliably predicts resistance to major drugs in the majority of M. tuberculosis strains, especially in areas of the world with a high prevalence of TB (34). For RIF, however, a minimum of three codons should be surveyed, and neither wild-type nor mutant alleles can be determined with any restriction endonuclease (22, 28). In contrast, analysis of a single codon (katG codon 315) will identify a majority of INHr isolates in countries with a high prevalence of MDR TB. Methods described so far for the detection of changes in katG codon 315 include DNA sequencing (5, 6, 9, 12, 13), single-strand conformation polymorphism analysis (20, 29), and cleavase fragment length polymorphism analysis (2) assays, which permit surveillance for all katG mutations, and dot blot hybridization (34, 36) and HapII (or its isoschizomers like MspI) digestion (4, 7, 31), which identify specific katG mutations. It should be noted that the use of single-strand conformation polymorphism analysis for the screening of katG for mutations in one case failed to reveal the S315T mutation due to the inappropriate positioning of the PCR primers, which resulted in an underestimation of the prevalence of this mutation (20, 37). Other primer pairs have been used to amplify a larger portion of katG and to detect simultaneously mutations in katG codons 315 and 463 by PCR-RFLP analysis (7, 31). However, it is accepted that variation in codon 463 presents a natural polymorphism unrelated to INH resistance (22). Therefore, we did not study this site in katG.
In conclusion, the katG S315T mutation can serve as a reliable marker for the detection of INH resistance in M. tuberculosis isolates in northwestern Russia. The PCR-RFLP assay that we have described is rapid, easy to perform, and easy to interpret. Furthermore, the procedure is inexpensive and requires standard PCR and electrophoresis equipment and can therefore be implemented in many of the clinical microbiology laboratories in northwestern Russia and other regions with a high incidence of TB where this mutation is predominant. Detection of INH resistance by this rapid genetic approach should facilitate the appropriate and timely delivery of antituberculous therapy.
Acknowledgments
We thank Anna Vyazovaya for technical assistance. We thank Christophe Sola for use of his spoligotype database for comparison. We are grateful to Alessandra Riva for critical reading of the manuscript and English-language corrections. We also acknowledge two anonymous reviewers for their valuable comments and suggestions.
This study was partly supported by the “Reseau International des Instituts Pasteur et Instituts Associes,” Institut Pasteur, Paris, France, and by the International Atomic Energy Agency (research contract no. 9924).
REFERENCES
- 1.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brow, M. A., M. C. Oldenburg, V. Lyamichev, L. M. Heisler, N. Lyamicheva, J. G. Hall, N. J. Eagan, D. M. Olive, L. M. Smith, L. Fors, and J. E. Dahlberg. 1996. Differentiation of bacterial 16S rRNA genes and intergenic regions and Mycobacterium tuberculosis katG genes by structure-specific endonuclease cleavage J. Clin. Microbiol. 34:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cockerill, F. R., III. 1999. Genetic methods for assessing antimicrobial resistance. Antimicrob. Agents Chemother. 43:199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobner, P., S. Rusch-Gerdes, G. Bretzel, K. Feldmann, M. Rifai, T. Loscher, and H. Rinder. 1997. Usefulness of Mycobacterium tuberculosis genomic mutations in the genes katG and inhA for the prediction of isoniazid resistance. Int. J. Tuberc. Lung Dis. 1:365-369. [PubMed] [Google Scholar]
- 5.Escalante, P., S. Ramaswamy, H. Sanabria, H. Soini, X. Pan, O. Valiente-Castillo, and J. M. Musser. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuberc. Lung Dis. 79:111-118. [DOI] [PubMed] [Google Scholar]
- 6.Fang, Z., C. Doig, A. Rayner, D. T. Kenna, B. Watt, and K. J. Forbes. 1999. Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Schotland (1990-1997). J. Clin. Microbiol. 37:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, R. B. Fourie, G. Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiepiela, P., K. S. Bishop, A. N. Smith, L. Roux, and D. F. York. 2000. Genomic mutations in the katG, inhA, and ahpC genes are useful for the prediction of isoniazid resistance in Mycobacterium tuberculosis isolates from Kwazulu Natal, South Africa. Tuberc. Lung Dis. 80:47-56. [DOI] [PubMed] [Google Scholar]
- 10.Kelley, C. L., D. A. Rouse, and S. L. Morris. 1997. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:2057-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, A. S. G., I. H. K. Lim, L. L. H. Tang, A. Telenti, and S. Y. Wong. 1999. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob. Agents Chemother. 43:2087-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marttila, H. J., H. Soini, P. Huovinen, and M. K. Viljanen. 1996. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob. Agents Chemother. 40:2187-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazars, E., S. Lesjean, A.-L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution mini-satellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mdluli, K., D. R. Sherman, M. J. Hickey, B. N. Kreiswirth, S. Morris, C. K. Stover, and C. E. Barry III. 1996. Biochemical and genetic data suggest that InhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J. Infect. Dis. 174:1085-1090. [DOI] [PubMed] [Google Scholar]
- 16.Narvskaya, O., I. Mokrousov, E. Limeschenko, T. Otten, L. Steklova, O. Graschenkova, and B. Vyshnevskiy. 2000. Molecular characterization of Mycobacterium tuberculosis strains from northwestern region of Russia. EpiNorth 1:22-24. [Google Scholar]
- 17.Narvskaya, O., I. Mokrousov, T. F. Otten, and B. I. Vyshnevskiy. 1999. Genetic marking of polyresistant Mycobacterium tuberculosis strains isolated in the north-west of Russia. Probl. Tuberk. N3:39-41. (In Russian.) [PubMed]
- 18.Perelman, M. I. 2000. Tuberculosis in Russia. Int. J. Tuberc. Lung Dis. 4:1097-1103. [PubMed] [Google Scholar]
- 19.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implication for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pretorius, G. S., P. D. van Helden, F. Sirgel, K. D. Eisenach, and T. C. Victor. 1995. Mutations in katG gene sequences in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis are rare. Antimicrob. Agents Chemother. 39:2276-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prodinger, W. M., P. Bunyaratvej, R. Prachaktam, and M. Pavlic. 2001. Mycobacterium tuberculosis isolates of Beijing genotype in Thailand. Emerg. Infect. Dis. 7:483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramaswami, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis. Update. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 23.Rozwarski, D. A., G. A. Grant, D. H. R. Barton, W. R. Jakobs Jr., and J. C. Sacchettini. 1998. Isoniazid modifies the NADH of its target enzyme (InhA) from Mycobacterium tuberculosis. Science 279:98-102. [DOI] [PubMed] [Google Scholar]
- 24.Sebban, M., I. Mokrousov, N. Rastogi, and C. Sola. 2002. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics 18:235-243. [DOI] [PubMed] [Google Scholar]
- 25.Slayden, R. A., and C. E. Barry III. 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659-669. [DOI] [PubMed] [Google Scholar]
- 26.Sola, C., I. Filliol, M. C. Guttieres, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreevatsan, S., X. Pan, Y. Zhang, V. Deretic, and J. M. Musser. 1997. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob. Agents Chemother. 41:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 29.Temesgen, Z., K. Satoh, J. R. Uhl, B. C. Kline, and F. R. Cockerill III. 1997. Use of polymerase chain reaction single strand conformation polymorphism (PCR-SSCP) analysis to detect a point mutation in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis. Mol. Cell. Probes 11:59-63. [DOI] [PubMed] [Google Scholar]
- 30.Tuyen, L. T. K., B. K. Hoa, H. M. Ly, L. N. Van, N. T. N. Lan, D. Chevrier, and J.-L. Guesdon. 2000. Molecular fingerprinting of Mycobacterium tuberculosis strains isolated in Vietnam using IS6110 as probe. Tuberc. Lung Dis. 80:75-83. [DOI] [PubMed] [Google Scholar]
- 31.Uhl, J. R., G. S. Sandhu, B. C. Kline, and F. R. Cockerill III. 1996. PCR-RFLP detection of point mutations in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis associated with isoniazid resistance, p. 144-149. In D. Persing (ed.), PCR protocols for emerging infectious disease. ASM Press, Washington, D.C.
- 32.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Embden, J. D. A., T. Van Gorkom, K. Kremer, T. Jansen, B. A. M. van der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Quing, D. Enkhasaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Victor, T., A. M. Jordaan, A. van Rie, G. D. van der Spuy, M. Richardson, P.D. van Helden, and R. Warren. 1999. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuberc. Lung Dis. 79:343-348. [DOI] [PubMed] [Google Scholar]
- 37.Victor, T. C., G. S. Pretorius, J. V. Felix, A. M. Jordaan, P. D. van Helden, and K. D. Eisenach. 1996. katG mutations in isoniazid-resistant strains of Mycobacterium tuberculosis are not infrequent. Antimicrob. Agents Chemother. 40:1572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viljanen, M. K., B. I. Vyshnevskiy, T. F. Otten, E. Vyshnevskaya, M. Marijamaki, H. Soini, P. J. Laippala, and A. V. Vasilyef. 1998. Survey of drug-resistant tuberculosis in northwestern Russia from 1984 through 1994. Eur. J. Clin. Microbiol. Infect. Dis. 17:177-183. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organization. 1998. Laboratory services in tuberculosis control. Part III. Culture, p. 77. World Health Organization, Geneva, Switzerland.
- 40.World Health Organization. 1993. Treatment of tuberculosis. Guidelines for national programs. World Health Organization, Geneva, Switzerland.