Abstract
A new oxytetracycline (OTC) resistance (Otcr) determinant, Tet 34, was cloned from chromosomal DNA of Vibrio sp. no. 6 isolated from intestinal contents of cultured yellowtail (Seriola quinqueradiata). The transformant, containing cloned Tet 34, could grow in broth containing 25 μg of drug per ml with 10 mM MgCl2. Tet 34 encoded an open reading frame (ORF) 154 amino acids long. The amino acid sequence of the ORF was homologous to sequences of several bacterial xanthine-guanine phosphoribosyltransferases (XPRTs), which act in purine nucleotide salvage synthesis. Mg2+ binding site residues and the active site were highly conserved in XPRT and the ORF of Tet 34. The results suggest that Tet 34 encodes a new Mg2+-dependent Otcr mechanism.
Oxytetracycline (OTC) has widely been used in aquaculture, and there are many reports on the occurrence of OTC-resistant (Otcr) fish-pathogenic bacteria (3, 5, 9, 16, 17). The Otcr bacteria were found not only among the pathogenic bacteria, but also in aquacultural environments (7, 13, 16). In our previous study, a high prevalence of Otcr bacteria appeared in intestinal contents and rearing water of yellowtail (Seriola quinqueradiata) (14). The frequency of Otcr reached 56 to 91% of the total viable bacteria. Several Tet determinants (Tet A to G) were reported from fish-pathogenic bacteria or the marine environment (1, 2, 5, 16). As a first step toward a better understanding of the dynamics of the Otcr determinant in aquatic environments, we cloned the Otcr determinant from Vibrio sp. isolated from intestinal contents of cultured yellowtail.
Otcr strains of Vibrio spp. were isolated from cultured yellowtail obtained during 1999 in Japan (14). We isolated 288 Otcr isolates from intestinal contents and seawater at that time. We checked the Otcr strains used for cloning. These strains were not PCR positive for one of the known tet genes, tet(G) (3). Only a small number of tet genes among marine bacteria have been reported, which reminds us to be able to find unknown factors. We had performed cloning with 10 strains of them and got a new determinant successfully from Vibrio sp. no. 6. The MIC of OTC for this strain was 500 μg/ml.
Since it is known that the efflux system of tetracycline resistance is sometimes dependent on various divalent cations, we assessed whether Vibrio sp. no. 6 requires MgCl2 for the Otcr. Cell growth was determined in broth containing 100 μg of OTC per ml with or without MgCl2 at concentrations of 10, 1, and 0.1 mM at 25°C. The broth used contained 5 g of polypeptone (Wako, Osaka, Japan); 1 g each of yeast extract (Difco, Detroit, Mich.), proteose peptone (Difco), beef extract (Wako); and 20 g of NaCl per liter of distilled water. Growth of Vibrio sp. no. 6 was sensitive to OTC when MgCl2 was absent, whereas addition of MgCl2 allowed growth in a dose-dependent manner in the presence of OTC (data not shown). In the presence of 10 mM MgCl2, the growth of Vibrio sp. no. 6 was almost the same as the growth in broth without OTC. This indicates that the Otcr of Vibrio sp. no. 6 is dependent on MgCl2.
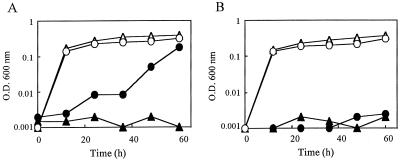
The Otcr determinant was cloned from chromosomal DNA, because Vibrio sp. no. 6 seemed not to possess a plasmid (data not shown). However, it is possible that there exists a low-copy-number plasmid that could not be detected by agarose gel electrophoresis and/or a megaplasmid, which might be removed with the chromosome during extraction. The chromosomal DNA was extracted from the bacteria as follows. The bacterial pellet was suspended in extraction solution (0.15 M NaCl, 0.1 M EDTA, 0.5 mg of RNase A per ml, and 0.5% sodium dodecyl sulfate [SDS]) and incubated at 65°C for 5 min. This solution was mixed with phenol saturated with 10 mM Tris-HCl (pH 8.0) plus 1 mM EDTA (TE) and centrifuged (17,000 × g) for 3 min. The DNA in the supernatant was extracted two times with TE-saturated phenol-chloroform-iso-amyl alcohol (25:24:1 [vol/vol/vol]) and with chloroform. Precipitated DNA with ethanol was resuspended in 50 μl of TE and stored at −20°C. After the chromosomal DNA and vector pUC119 (TaKaRa, Kyoto, Japan) were digested by restriction endonucleases PstI (TaKaRa) and EcoRI (TaKaRa), they were ligated with a DNA ligation kit, version 2 (TaKaRa). Escherichia coli JM109 was transformed and spread on Luria-Bertani (LB) medium containing 50 μg of ampicillin per ml, 10 μg of OTC per ml, 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml, and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After three passages of candidate colonies, we obtained a viable transformant. This transformant possessed a 919-bp inserted fragment, and the recombinant plasmid was designated pOV. Growth of transformants (pOV/JM109) with the cloned fragment was examined with or without MgCl2. The results are shown in Fig. 1. pOV/JM109 grew in the LB broth with 25 μg of OTC per ml (Fig. 1A), but not in the broth without MgCl2 (Fig. 1B). The same growth profiles were observed in the case of cultures containing 12.5 μg of tetracycline per ml (data not shown). This suggests that pOV/JM109 is also a tetracycline resistance determinant acting in the same manner as OTC and that the resistance ability of the transformed E. coli is dependent on MgCl2 as well as Vibrio sp. no. 6. Furthermore, we have retransformed pOV to different host E. coli DH5α cells. The transformants (pOV/DH5α) showed resistance in the presence of 10 mM MgCl2, the same as in JM109 (data not shown). On the other hand, the transformant pUC119/DH5α could not grow on the plate with OTC, regardless of the presence and absence of MgCl2. This suggests that Tet 34 can be expressed in both strains JM109 and DH5α.
FIG. 1.
Growth of E. coli JM109 transformed with pOV (○, •) or pUC119 (▵, ▴). LB broth contained 25 μg of OTC and 50 μg of ampicillin per ml. Panel A shows results with 10 mM MgCl2, and panel B shows results without MgCl2. Open symbols, without OTC; solid symbols, with OTC. O.D., optical density.
To confirm that the cloned fragment originated from the chromosomal DNA of Vibrio sp. no. 6, Southern hybridization was performed. The cloned fragment was labeled with a DIG-Chem-Link labeling and detection set (Roche Diagnostics, Mannheim, Germany) in accordance with the manufacturer's directions. Ten micrograms of DNA was digested with PstI and EcoRI and employed for Southern transfer. After denaturation and neutralization were performed, DNA was fixed with UV light (254 nm) by irradiation for 1 min. Fifty nanograms of digoxigenin (DIG)-labeled DNA probe was added to the blotted filter and hybridized at 65°C overnight. The membrane was washed three times with wash solution (40 mM NaH2PO4, 0.1% SDS [pH 7.2]). The DIG-labeled DNA was reacted with nitroblue tetrazolium chloride-9-bromo-4-chloro-3-indoyl phosphate toluidine salt (NBT/BCIP), with the enzymatic reaction followed by an anti-DIG antibody-conjugated alkaline phosphatase (Roche Diagnostics). Tet 34 probes hybridized to Vibrio sp. no. 6, but not to E. coli JM109 chromosome (data not shown). This result confirms that Tet 34 is coded on the chromosome DNA of Vibrio sp. no. 6.
Tet 34 was sequenced by using an ABI PRISM BigDye Terminator cycle sequencing FS Ready Reaction kit (PE Biosystems, Tokyo, Japan) with an ABI PRISM 310 DNA sequencer (PE Biosystems, Tokyo, Japan) in the Center for Gene Research, Ehime University. An open reading frame (ORF) of 465 bp (154 amino acids) was found on this fragment, which existed between 306 and 770 bp. Sequence analysis was performed with the homology search program FASTA. However, the DNA sequence of the determinant did not show homology with any Tet determinants, as described above. This determinant was named Tet 34 according to the recommendation proposed in reference 11. The deduced amino acid sequence of the ORF showed 79.8% homology with xanthine-guanine phosphoribosyltransferase (XPRT) of Vibrio cholerae (8) and 68 and 66% homology with XPRTs of E. coli (15) and Salmonella enterica serovar Typhimurium (12), respectively. The alignment with the known XPRTs is shown in Fig. 2.
FIG. 2.
Alignment of the amino acid sequence of the ORF on Tet 34 with five XPRTs obtained from the database. The PRib-PP motif is boxed, and the conserved amino acid residues related to activity of XRPT are in boldface. Asterisks denote the consensus amino acid residues. See references 6 (noted by no. 1) and 19 (noted by no. 2) for review.
From our studies, we propose another mechanism to explain Otcr by Tet 34. XPRT is an enzyme that acts in purine nucleotide salvage synthesis. This enzyme catalyzes the transfer of the phosphoribosyl moiety from 5-phospho-α-d-ribosyl-1-pyrophosphate (PRib-PP) to the 6-oxo-guanine and -xanthine (20). GMP and XMP are formed by this reaction, and then these nucleotides are converted to triphosphate forms. The fact that Tet 34 has homology to XPRT suggests it has a similar function. It is known that the Mg2+ ion interacts with the Asp-89 in the PRib-PP binding site motif of XRPT (20). Asp-89 was found conserved in the ORF on Tet 34 (Fig. 2). The fact that Vibrio sp. no. 6 requires MgCl2 for Otcr might relate to the Mg2+-binding property of the PRib-PP-like protein coded in Tet 34. The PRib-PP binding site motif in Tet 34 consists of the residues 85-IVEDLVDSG-96 (Fig. 2). In Tet 34, the region of the motif revealed high similarity to the PRib-PP binding site of the previously reported phosphoribosyltransferase (PRT) (20). XRPT comprises three subunits (A, B, and C). The active residues Pro-25, Ser-26, Trp-29, and Arg-53 are present in subunit A; Arg-37 and Asp-89 are present in subunit B; and Arg-152 is present in subunit C (20). The Pro-25, Trp-29, Arg-37, Arg-53, and Asp-89 residues were highly conserved in the sequence of the Tet 34 determinant. The PRib-PP binding site sequence forms part of the active site, and its relative position is completely conserved among several PRT structures by structural analysis (20). These findings suggest that the Tet 34 determinant possesses a similar function to PRTs, including XRPT.
Tetracycline inhibits bacterial growth by inhibiting protein synthesis in ribosomes (4, 18). The inhibition of protein synthesis is caused by disruption of codon-anticodon interactions. Elongation factor Tu (EF-Tu) (10) is a protein that plays a crucial role in the polypeptide elongation step of protein biosynthesis. This factor is required for the correct binding of the aminoacyl-tRNA to the ribosomal acceptor site and is a key factor in maintaining translational fidelity. During protein synthesis, the EF-Tu·GTP complex binds strongly to aminoacyl-tRNA, thereby preventing spontaneous hydrolysis of the aminoacyl-tRNA ester bond. When the EF-Tu · GTP · tRNA complex interacts with the ribosome, the GTP is hydrolyzed, and then EF-Tu·GDP is released from the ribosome (10). The GDP is thereafter recycled to the GTP (10). The similarity of protein sequence between Tet 34 and XPRT, which catalyzes formation of GMP, XMP, and IMP from guanine, xanthine, and hypoxanthine, respectively, suggests that the Tet 34 product supplies the purine nucleotides needed for the translational step described above. Consequently, an excess supply of GTP may have some effect on accelerating the binding of aminoacyl-tRNA and EF-Tu·GTP, which attenuates inhibition by OTC.
In conclusion, we cloned a new Tet determinant and determined that the possible function of the determinant is activation of Mg2+-dependent purine nucleotide synthesis, which finally protects protein synthesis.
Nucleotide sequence accession number.
The sequence of the Tet 34 determinant has been submitted to the DDBJ database under accession no. AB061440.
Acknowledgments
We thank S. Nakano for useful suggestions and D. E. Taylor, University of Alberta, for critical review of the manuscript.
This work was partly supported by a grant-in-aid from MEXT. Genetic experiments were performed in the Center for Gene Research, Ehime University.
REFERENCES
- 1.Adams, C. A., B. Austin, P. G. Meaden, and D. McIntosh. 1998. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl. Environ. Microbiol. 64:4194-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, S. R., and R.-A. Sandaa. 1994. Distribution of tetracycline resistance determinants among gram-positive bacteria isolated from polluted and unpolluted marine sediments. Appl. Environ. Microbiol. 60:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, T., T. Satoh, and T. Kitao. 1987. New tetracycline resistance determinant on R plasmid from Vibrio anguillarum. Antimicrob. Agents Chemother. 31:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopra, I., P. M. Hawkey, and M. Hinton. 1992. Tetracyclines, molecular and clinical aspects. J. Antimicrob. Chemother. 29:245-277. [DOI] [PubMed] [Google Scholar]
- 5.DePaola, A., P. A. Flynn, R. M. McPhearson, and S. B. Levy. 1988. Phenotypic and genotypic characterization of tetracycline- and oxytetracycline-resistant Aeromonas hydrophila from cultured channel catfish (Ictalurus punctatus) and their environments. Appl. Environ. Microbiol. 54:1861-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. G. Sutton, W. FitzHugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 7.Guardabassi, L., L. Dijkshoorn, J.-M. Collard, J. E. Olsen, and A. Dalsgaard. 2000. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49:929-936. [DOI] [PubMed] [Google Scholar]
- 8.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, D. H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, S.-P., T.-Y. Hsu, M.-H. Chen, and W.-S. Wang. 2000. Antibacterial effect of chloramphenicol, thiamphenicol and florfenicol against aquatic animal bacteria. J. Vet. Med. Sci. 62:479-485. [DOI] [PubMed] [Google Scholar]
- 10.Kjeldgaad, M., and J. Nyborg. 1992. Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol. 223:721-742. [DOI] [PubMed] [Google Scholar]
- 11.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui, M., T. Sofumi, and T. Nhomi. 1996. Regionally-targeted mutagenesis by metabolically-activated steviol: DNA sequence analysis of steviol-induced mutants of guanine phosphoribosyltransferase (gpt) gene of Salmonella typhimurium. Mutagenesis 11:565-572. [DOI] [PubMed] [Google Scholar]
- 13.McPhearson, R. M., A. DePaola, S. R. Zywno, Z. M. Motes, Jr., and A. M. Guarino. 1991. Antibiotic resistance in Gram-negative bacteria from cultured catfish and aquaculture ponds. Aquaculture 99:203-211. [Google Scholar]
- 14.Nonaka, L., T. Isshiki, and S. Suzuki. 2000. The occurrence of the oxytetracycline resistant bacteria in the fish intestine and seawater environment. Microb. Environ. 15:223-228. [Google Scholar]
- 15.Pratt, D., and S. Subramani. 1983. Nucleotide sequence of the Escherichia coli xanthine-guanine phosphoribosyl transferase gene. Nucleic Acids Res. 11:8817-8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes, G., G. Huys, J. Swings, P. McGann, M. Hiney, P. Smith, and R. W. Pickup. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn 1721 in dissemination of the tetracycline resistance determinant Tet A. Appl. Environ. Microbiol. 66:3883-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, A. S., M. S. Bruun, I. Dalsgaard, K. Pedersen, and J. L. Larsen. 2000. Occurrence of antimicrobial resistance in fish-pathogenic and environmental bacteria associated with four Danish rainbow trout farms. Appl. Environ. Microbiol. 66:4908-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnappinger, D., and W. Hillen. 1996. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 165:359-369. [DOI] [PubMed] [Google Scholar]
- 19.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 20.Vos, S., J. Jersey, and J. L. Martin. 1997. Crystal structure of Escherichia coli xanthine phosphoribosyltransferase. Biochemistry 36:4125-4134. [DOI] [PubMed] [Google Scholar]