Abstract
The mechanisms of resistance to macrolides in seven group A streptococcal (Streptococcus pyogenes) isolates that were the cause of pharyngitis in children who were unsuccessfully treated with azithromycin (10 mg/kg of body weight/day for 3 days) were evaluated. All posttreatment strains were found to be genetically related to the pretreatment isolates by random amplified polymorphism DNA analysis and pulsed-field gel electrophoresis. Two isolates had acquired either a mef(A) or an erm(B) gene, responsible for macrolide efflux and ribosomal modification, respectively. Three isolates displayed mutations in the gene encoding the L4 ribosomal protein that is part of the exit tunnel within the 50S subunit of the bacterial ribosome. In the two remaining posttreatment strains, the mechanisms of macrolide resistance could not be elucidated.
The recommended treatment for group A Streptococcus (GAS) pharyngitis, a 10-day course of penicillin, was established 50 years ago and remains the “gold standard.”
However, for patients hypersensitive to β-lactam antibiotics and in whom therapy with these drugs fails, macrolides are often the recommended substitute. Azithromycin is an azalide antibiotic chemically related to erythromycin and has a long half-life and excellent tissue penetration. Short-course azithromycin therapy has been reported to be effective for the eradication of oropharyngeal GAS (14).
In a prospective, comparative, randomized, multicenter trial that was conducted between November 1997 and July 1998 and that involved 350 children with GAS pharyngitis, we observed better eradication rates on days 14 and 30 after a 3-day course of therapy with azithromycin at 20 mg/kg of body weight/day than after a 3-day course of therapy with azithromycin at 10 mg/kg/day. In the per protocol analysis, the failure rates on day 14 were 57 of 135 (42.2%) in the 10-mg/kg treatment arm and 8 of 139 (5.8%) in the 20-mg/kg treatment arm (8). Furthermore, analysis of bacterial isolates by random amplified polymorphic DNA (RAPD) analysis revealed seven cases of bacteriological treatment failure: azithromycin MICs for genetically related pre- and posttreatment GAS strains increased after treatment with this antimicrobial at 10 mg/kg/day for 3 days. In contrast, azithromycin MICs were not increased for any of the strains from any of the patients with treatment failure in the group treated with azithromycin at 20 mg/kg/day (8). The known mechanisms of macrolide resistance in streptococci are modification of the ribosomal target by a methylase encoded by erm genes (13, 20) and a macrolide-specific efflux mechanism encoded by the mef(A) gene (7). Mutation of the ribosomal target of macrolides is a rare resistance mechanism in streptococci and has been reported in only a few clinical pneumococcal isolates (21).
In the study described here we investigated the mechanisms of macrolide resistance among the GAS strains that were the cause of bacteriological treatment failures in the group treated with azithromycin at 10 mg/kg/day.
MATERIALS AND METHODS
Bacterial isolates.
GAS isolates were obtained by swabbing of the throat on day 0 (pretreatment isolate [V1]) and day 14 or day 30 (posttreatment isolates [V2]) after the onset of low-dose azithromycin treatment in children. All GAS isolates were associated with bacteriological treatment failure (persistence or relapse) (6, 10), and the azithromycin MICs for the posttreatment isolates had increased. The isolates were identified as GAS by colony morphology, beta-hemolysis on blood agar, and a commercial agglutination technique (Murex Diagnostics, Dartford, United Kingdom).
MIC determination.
The MICs of erythromycin, azithromycin, josamycin, and clindamycin were determined by the agar dilution method with Mueller-Hinton medium supplemented with 5% defibrinated sheep blood (15). The plates were incubated overnight at 35°C in ambient air.
Molecular analysis.
RAPD analysis and pulsed-field gel electrophoresis (PFGE) were used to compare pre- and posttreatment GAS isolates in order to distinguish between persistence-relapse and the acquisition of new strains (6, 10). RAPD analysis and PFGE were based on previously described methods (2, 10). For PFGE, SmaI and SfiI chromosomal digests were separated by using a CHEF-MAPPER apparatus (Bio-Rad, Marnes la Coquette, France) with a switch time of 0.85 to 35.38 s for 22 h and 35 min at a 120° angle with a voltage gradient of 6 V/cm at 14°C. The DNA size standard was a bacteriophage lambda DNA ladder (Bio-Rad). The PFGE banding patterns were compared visually. Strains were considered genetically distinguishable if their restriction patterns differed by three or more bands (23).
Detection of erythromycin resistance genes.
All erythromycin-resistant isolates were screened for erythromycin resistance genes. The erm(B), erm(A) subclass erm(TR), and mef(A) genes were detected by PCR amplification as described previously (3). Streptococcus pyogenes 02C1061, S. pyogenes 02C1110, and S. pyogenes 02C1064 were used as positive PCR controls for the erm(B), erm(A) subclass erm(TR), and mef(A) genes, respectively (3).
DNA from PFGE was transferred to nitrocellulose membranes (Hybond-N+; Amersham, Little Chalfont, England) by vacuum blotting. DNA was hybridized with a digoxigenin-labeled probe of 616 bp obtained by PCR amplification of the erm(B) gene from reference strain S. pyogenes 02C1061 containing the erm(B) gene (3). Hybridization and colorimetric detection were performed as recommended by the manufacturers (Roche Molecular Biochemicals, Meylan, France).
Detection of mutations in the ribosomal target of macrolides.
The nucleotide sequences of the 23S rRNA and L4 and L22 ribosomal proteins in Escherichia coli were obtained from The Institute for Genomic Research website (http://www.tigr.org.), and homologs in GAS were detected by using BLAST software (9). Specific oligonucleotide primers were then designed. We amplified a portion of the rrl gene for domain II from nucleotides (nt) 580 to 852 (E. coli numbering) with primers 5′-CGGCGATTACGATATGATGC-3′ and 5′-CTCTAATGTCGACGCTAGCC-3′ and two fragments of domain V of 23S rRNA (nt 1990 to 2134 and nt 2331 to 2769) with two pairs of primers (primers 5′-CTGTCTCAACGAGAGACTC-3′ and 5′-CTTAGACTCCTACCTATCC-3′ and primers 5′-GTATAAGGGAGCTTGACTG-3′ and 5′-GGGTTTCACACTTAGATG-3′). The entire L22 (rplV) and L4 (rplD) genes were amplified with pairs of primers (primers 5′-GCTGACGACAAGAAAACACG-3′ and 5′-GCCGACACGCATACCAATTG-3′ and primers 5′-CAAGTCAGGAGTTAAAGCTGC-3′ and 5′-CAACTTCGAAAGTGTATTTGCC-3′, respectively). The three amplified rrl fragments (two for domain V and one for domain II) included bases critical for erythromycin resistance (G2057, A2058, A2059, A2062, G2505, C2611, A754, and A752). PCR products were sequenced by the rhodamine dye terminator method with an ABI Prism 377 sequencer (Perkin-Elmer Corp., Norwalk, Conn.). For the two pairs of pre- and posttreatment isolates of strains 133 and 233, the sequences of the rrl genes were obtained as follows. Ten overlapping fragments were amplified with the primers shown in Table 1 and sequenced. Amplicons were also analyzed by single-strand conformation polymorphism analysis as described previously (5). Briefly, aliquots of amplified fragments were heat denaturated, and the single-strand PCR products were then separated by nondenaturant polyacrylamide gel electrophoresis. Fragments bearing mutations could be distinguished from wild-type fragments on the basis of different electrophoretic mobilities.
TABLE 1.
Oligonucleotides used for amplification of rrl gene (23S rRNA)
| Primer designation | Primer sequence (5′ to 3′)a | Positionb | Product size (bp) |
|---|---|---|---|
| 23SCPU1 | + GTTAATAAGGGCGCACGG | 7-24 | 247 |
| 23SCPL1 | − GCTCGCCGCTACTAAGG | 237-253 | |
| 23SCPU2 | + TTAGTAGCCGCAGGAAGAG | 192-210 | 336 |
| 23SCPL2 | − GTAGGCACACGGTTTCAGG | 509-527 | |
| 23SCPU3 | + GAACCAGTACCGTGAGGG | 452-469 | 384 |
| 23SCPL3 | − TAGCCCTAAAGCTATTTCGG | 816-835 | |
| 23SCPU4 | + GATGACTTGTGGGTAGCGG | 760-778 | 381 |
| 23SCPL4 | − GCCCCGGTACATTTTCGG | 1123-1140 | |
| 23SCPU5 | + GGTTGCCCAGACAACTAGG | 1038-1056 | 367 |
| 23SCPL5 | − GTACAGGAATATCAACCTG | 1386-1404 | |
| 23SCPU6 | + GCTCGTCCGCCCTGGG | 1324-1339 | 377 |
| 23SCPL6 | − TCTCCCGAAGTTACGGGG | 1683-1700 | |
| 23SCPU7 | + GTACCGCAAACCGACACAG | 1601-1619 | 406 |
| 23SCPL7 | − GTCTCTCGTTGAGACAGTG | 1988-2006 | |
| 23SCPU8 | + GACCCGCACGAAAGGCG | 1959-1975 | 401 |
| 23SCPL8B | − GTAGCTCTCGCAGTCAAG | 2342-2359 | |
| 23SCPU9 | + GTTCCCTCAGATTGGTTGG | 2290-2308 | 386 |
| 23SCPL9 | − GCGTGCCGCTTTAATGGG | 2558-2575 | |
| 23SCPU10 | + GTAGTCGGTCCCAAGGG | 2529-2545 | 372 |
| 23SCPL10 | − ATAAGTCCTCGAGCGATTAG | 2880-2900 |
+, sense primer; −, antisense primer.
E. coli numbering.
RESULTS
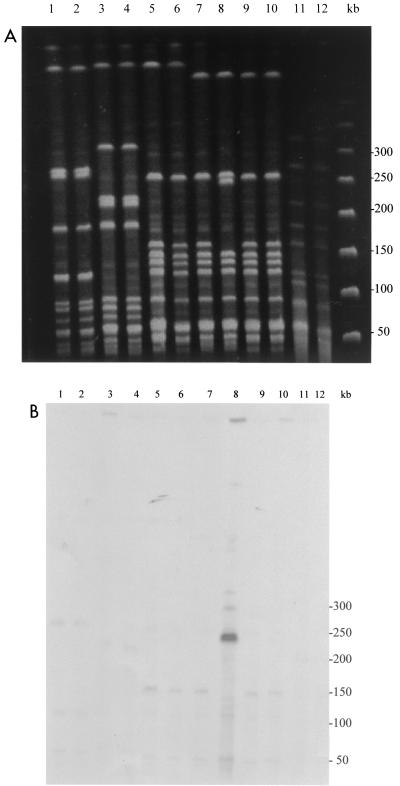
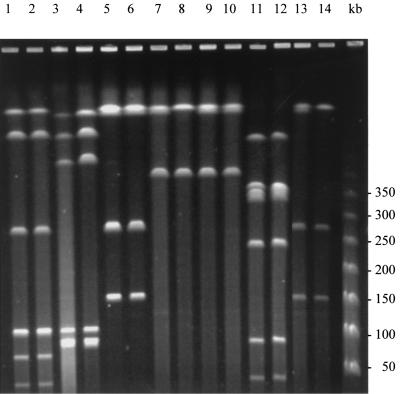
Using RAPD analysis and PFGE, we found that the seven paired pre- and posttreatment GAS isolates associated with persistence or relapse were genetically related (Fig. 1 and 2). Two of the seven pairs of strains were of the same PFGE type, although the patients were epidemiologically unrelated. For patient 508, the DNA of the posttreatment strain could not be analyzed by PFGE with SmaI, despite repeated attempts. Macrolide MICs for the paired GAS strains are shown in Table 2. All pretreatment isolates were sensitive to 14-, 15-, and 16-membered-ring macrolides and clindamycin, while the posttreatment isolates showed different levels of resistance according to the antibiotic tested. The azithromycin MIC for the posttreatment isolate of strain 286 was >128 μg/ml, whereas the azithromycin MIC for the isolate was 0.12 μg/ml before treatment. PCR and Southern hybridization showed that the azithromycin resistance in this strain was due to acquisition of an erm(B) gene. The erm(B) gene was located on a chromosomal fragment of 250 kb which resulted from the insertion of a DNA sequence of ∼90 kb in a ∼160-kb SmaI fragment of the pretreatment isolate's DNA (Fig. 1B). For the posttreatment isolate of the pair of isolates from patient 508, acquisition of a mef(A) gene, as identified by PCR, resulted in resistance to azithromycin (MIC, 8 μg/ml). For pairs of isolates of three strains (strains 11, 124, and 390), azithromycin MICs increased from 0.125 or 0.25 μg/ml to 1 or 2 μg/ml. No erm or mef genes were detected in the three posttreatment isolates of these strains. However, sequence analysis of structures belonging to the macrolide binding site revealed mutations in a highly conserved region of ribosomal protein L4 (61KPWRQKGTGRAR72). The mutations consisted of a WR deletion at positions 64 and 65, an RA insertion after position 72, and a TG deletion at positions 69 to 70 (S. pyogenes numbering) in posttreatment isolates of strains 11, 124, and 390, respectively. Since no mutation was found in posttreatment isolates of strains 133 and 323, the entire rrl genes from these strains and their parents were sequenced. The DNA sequences were identical to those of the respective parent strains. Since S. pyogenes harbors six copies of genes for rRNAs, we could not exclude the possibility that only a minority of copies were mutated and therefore not detected by sequencing of genomic DNA as a bulk. To explore this possibility, we used PCR-single-strand conformation polymorphism analysis. In Streptococcus pneumoniae, which contains four copies of the rrl genes, this technique allowed investigators to distinguish a single wild-type copy from three mutated copies on the basis of heterogeneous electrophoretic profiles (5). The migration profiles of the PCR products were homogeneous and identical for the parent and the resistant mutated strains. Therefore, no explanation for the azithromycin resistance of the posttreatment isolates of strains 133 and 323 was found.
FIG. 1.
(A) PFGE patterns of the pre- and posttreatment GAS isolates digested with SmaI. Lane 1, isolate 11 V1; lane 2, isolate 11 V2; lane 3, isolate 124 V1; lane 4, isolate 124 V2; lane 5, isolate 390 V1; lane 6, isolate 390 V2; lane 7, isolate 286 V1; lane 8, isolate 286 V2; lane 9, isolate 133 V1; lane 10, isolate 133 V2; lane 11, isolate 323 V1; lane 12, isolate 323 V2. (B) Southern blot of the gel with an erm(B) probe. Lane 8, isolate 286 V2 shows a signal with the probe. The molecular sizes of the standards (in kilobases) are shown to the right of the gels.
FIG. 2.
PFGE patterns of the pre- and posttreatment GAS isolates digested with SfiI. Lane 1, isolate 11 V1; lane 2, isolate 11 V2; lane 3, isolate 124 V1; lane 4, isolate 124 V2; lane 5, isolate 390 V1; lane 6, isolate 390 V2; lane 7, isolate 286 V1; lane 8, isolate 286 V2; lane 9, isolate 133 V1; lane 10, isolate 133 V2; lane 11, isolate 323 V1; lane 12, isolate 323 V2; lane 13, isolate 508 V1; lane 14, isolate 508 V2.
TABLE 2.
MICs of macrolides and clindamycin for the seven pre- and posttreatment GAS isolates by mechanism of resistance
| Patient no. | GAS isolatea | Isolation date (mo/day/yr) | MIC (μg/ml)
|
Mechanism of resistance | |||
|---|---|---|---|---|---|---|---|
| Erythromycin | Azithromycin | Josamycin | Clindamycin | ||||
| 11 | 11 V1 | 11/19/97 | 0.064 | 0.125 | 0.5 | 0.125 | |
| 11 | 11 V2 | 12/03/97 | 0.5 | 2 | 1 | 0.125 | L4 mutation |
| 124 | 124 V1 | 01/12/98 | 0.064 | 0.25 | 0.5 | 0.064 | |
| 124 | 124 V2 | 01/23/98 | 0.5 | 1 | 1 | 0.064 | L4 mutation |
| 390 | 390 V1 | 05/05/98 | 0.064 | 0.25 | 0.5 | 0.125 | |
| 390 | 390V2 | 05/23/98 | 1 | 2 | 2 | 0.125 | L4 mutation |
| 286 | 286 V1 | 03/23/98 | 0.064 | 0.125 | 0.5 | 0.125 | |
| 286 | 286 V2 | 04/06/98 | >128 | >128 | >128 | >128 | erm(B) |
| 133 | 133 V1 | 01/15/98 | 0.064 | 0.125 | 0.5 | 0.125 | |
| 133 | 133 V2 | 01/27/98 | 8 | 8 | 2 | 0.125 | Unknown |
| 323 | 323 V1 | 04/08/98 | 0.064 | 0.125 | 0.5 | 0.125 | |
| 323 | 323 V2 | 05/11/98 | 32 | >128 | 128 | 1 | Unknown |
| 508 | 508 V1 | 06/05/98 | 0.064 | 0.25 | 0.5 | 0.125 | |
| 508 | 508 V2 | 06/16/98 | 8 | 8 | 0.5 | 0.125 | mef(A) |
V1, isolate obtained before treatment; V2, isolate obtained after treatment.
DISCUSSION
Resistance to macrolides has been widely reported in GAS, although its incidence varies considerably among countries. A previous study from Finland demonstrated that a decrease in the incidence of macrolide resistance in S. pyogenes could be managed by limiting erythromycin use through a national program (19). However, the rate of use of newer macrolides with fewer daily dosages increased during the course of the study, which suggests that the overall numbers of patients treated with macrolides did not decrease (19).
Resistance to macrolides was reported to be associated with dissemination of clonal strains (24) but rarely with the in vivo acquisition of resistance during therapy. In our work, the number of GAS isolates (7 of 135) that developed resistance to macrolides after low-dose azithromycin treatment was strikingly high (8). This phenomenon was not observed in three recent clinical trials of azythromycin treatment in patients with tonsillopharyngitis (4, 16, 18). In two studies (4, 18) the susceptibilities of the posttreatment isolates were not reported. In the last study (16), no acquisition of resistance was observed. However, the large number of patients included in our study may account for the difference with the latter study.
The efficacy of treatment against streptococcal pharyngitis should be evaluated both clinically and bacteriologically (6). If microbiological treatment failure occurs, persistence or relapse due to the original GAS strain should be distinguished from recurrence due to acquisition of a new strain (6, 10). Genomic typing methods such as RAPD analysis and PFGE have been used to discriminate among S. pyogenes isolates (2, 10). We applied these techniques to the analysis of GAS strains isolated during a clinical trial of azithromycin therapy (10 or 20 mg/kg/day) for GAS pharyngitis in children. RAPD analysis and PFGE suggested that seven cases of microbiological treatment failure were due to GAS persistence or relapse, with acquisition of macrolide resistance after treatment with azithromycin at 10 mg/kg/day (8). However, we cannot exclude the possibility of the presence at the onset of the study of a double population of organisms, some of which had already acquired the gene for resistance, leading to the selection of the resistant organism by therapy.
In our study, among the seven cases of emergent azithromycin resistance, one was due to acquisition of a mef gene and another was due to the acquisition of an erm gene. The acquisition of a DNA fragment of about 90 kb bearing an erm gene by posttreatment strain 286 could correspond to acquisition of a mobile element or, rather, to the integration of a plasmid in the chromosome. Previous studies have shown that the commensal flora serves as a potential source of macrolide resistance determinants (1, 17). The location of erm on conjugative transposons and of mef on a transposon (12) might facilitate the circulation of these genes among the oropharyngeal flora under selective antibiotic pressure. Interestingly, we observed three cases of microbiological persistence or relapse with acquisition of L4 ribosomal mutations. So far, the only streptococcal species reported to have acquired resistance to macrolides by a mutation in 23S rRNA or ribosomal proteins is S. pneumoniae, both among clinical isolates (21) and among strains selected by multiple passages in the presence of azithromycin in vitro (22). Mutations in the gene encoding the L4 proteins of GAS strains and S. pneumoniae were clustered in an identical conserved region of the protein. In E. coli, L4 mutations perturb the three-dimensional structure of 23S rRNA at multiple sites and could therefore hypothetically prevent macrolide binding (11).
In the case of pneumococci and macrolides, the mutations detected in laboratory-derived mutants obtained with subinhibitory concentrations of azithromycin could be used to predict the mutations observed in clinical strains (22). The observation that repeated exposure to low concentrations of azithromycin selects resistant pneumococcal strains in vitro might help provide an understanding of the selection of GAS mutants in vivo. In this analysis, we found that three of seven clinical strains contained mutations in the gene encoding ribosomal protein L4, with modest increases in the MICs for strains detected. Similar mutations were not found among strains from patients in the 20-mg/kg azithromycin treatment arm with bacteriological treatment failures; either the L4 mutation and the resulting low MICs do not occur for a total dose of 60 mg, or the sample size for either treatment arm does not permit one to draw definitive conclusions. However, the higher dose of azithromycin gave significantly better bacterial eradication rates, thereby minimizing the chance for the emergence of resistance. Finally, no L22, L4, or 23S rRNA mutations were detected in two posttreatment strains; their mechanisms of resistance are still under investigation. Characterization of the mechanisms of resistance to macrolides, lincosamides, and streptogramin B antibiotics are important for the clinical management of antibiotic therapy and can provide clinicians with alternative treatment choices.
Acknowledgments
We thank Joyce Sutcliffe for providing reference strains S. pyogenes 02C1061, 02C1110, and 02C1064 and Maëlle Coquemont for technical assistance.
REFERENCES
- 1.Aracil, B., M. Minambres, J. Oteo, C. Torres, J. L. Gomez-Garces, and J. I. Alos. 2001. High prevalence of erythromycin-resistant and clindamycin-susceptible (M phenotype) viridans group streptococci from pharyngeal samples: a reservoir of mef genes in commensal bacteria. J. Antimicrob. Chemother. 48:587-595. [DOI] [PubMed] [Google Scholar]
- 2.Bert, F., C. Branger, and N. Lambert-Zechovsky. 1997. Pulsed-field gel electrophoresis is more discriminating than multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for typing phylogenic streptococci. Curr. Microbiol. 34:226-229. [DOI] [PubMed] [Google Scholar]
- 3.Bingen, E., F. Fitoussi, C. Doit, R. Cohen, A. Tanna, R. George, C. Loukil, N. Brahimi, I. Le Thomas, and D. Deforche. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boccazzi, A., P. Tonelli, M. De Angelis, L. Bellussi, D. Passali, and P. Careddu. 2000. Short course therapy with ceftibuten versus azythromycin in pediatric streptococcal pharyngitis. Pediatr. Infect. Dis. J. 19:963-967. [DOI] [PubMed] [Google Scholar]
- 5.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, A. W., C. B. Hall, J. O. Klein, R. B. Kammer, R. D. Meyer, and J. S. Remington. 1992. Evaluation of new anti-infective drugs for treatment of respiratory tract infections. Clin. Infect. Dis. 15(Suppl.):S62-S88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, R., P. Reinert, F. de la Rocque, C. Levy, M. Boucherat, M. Robert, M. Navel, N. Brahimi, D. Deforche, B. Palestro, and E. Bingen. Comparison of two dosages of azithromycin for 3 days versus penicillin V for 10 days in acute group A streptococcal tonsillopharyngitis. Pediatr. Infect. Dis. J., in press. [DOI] [PubMed]
- 9.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, S. Sezate, A. N. Suvorov, C. Primeaux, S. Kenton, H. Lai, S. Lin, Y. Qian, H. Jia, H. Zhu, Q. Ren, F. Z. Najar, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitoussi, F., R. Cohen, G. Brami, C. Doit, N. Brahimi, F. de la Rocque, and E. Bingen. 1997. Molecular DNA analysis for differentiation of persistence or relapse from recurrence in treatment failure of Streptococcus pyogenes pharyngitis. Eur. J. Clin. Microbiol. Infect. Dis. 16:233-237. [DOI] [PubMed] [Google Scholar]
- 11.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 12.Kataja, J., P. Huovinen, M. Skurnik, The Finnish Study Group for Antimicrobial Resistance, and H. Seppala. 1999. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob. Agents Chemother. 43:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita, J. Y., E. Kahn, T. Thompson, L. Laclaire, B. Beall, G. Gherardi, K. L. O'Brien, and B. Schwartz. 2000. Impact of azithromycin on oropharyngeal carriage of group A Streptococcus and nasopharyngeal carriage of macrolide-resistant Streptococcus pneumoniae. Pediatr. Infect. Dis. J. 9:41-46. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 5th ed. M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.Pacifico, L., F. Scopetti, A. Ranucci, M. Pataracchia, F. Savignoni, and C. Chiesa. 1996. Comparative efficacy and safety of a 3-day azithromycin and 10-day penicillin V treatment of group A beta-hemolytic streptococcal pharyngitis in children. Antimicrob. Agents Chemother. 40:1005-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reig, M., J. C. Galan, F. Baquero, and J. C. Perez-Diaz. 2001. Macrolide resistance in Peptostreptococcus spp. mediated by ermTR: possible source of macrolide-lincosamide-streptogramin B resistance in Streptococcus pyogenes. Antimicrob. Agents Chemother. 45:630-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaad, U., G. Heynen, and the Swiss Tonsillopharyngitis Study Group. 1996. Evaluation of the efficacy, safety and toleration of azithromycin vs penicillin V in the treatment of acute streptococcal pharyngitis in children: results of a multicentre open comparative study. Pediatr. Infect. Dis. J. 15:791-795. [DOI] [PubMed] [Google Scholar]
- 19.Seppäla, H., T. Klaukka, J. Vuopio-Varkila, A. Muotiala, H. Helenus, K. Lager, P. Huovinen, and the Finnish Study Group for Antimicrobial Resistance. 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N. Engl. J. Med. 337:441-446. [DOI] [PubMed] [Google Scholar]
- 20.Seppäla, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinene. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York, M. K., L. Gibbs, F. Perdreau-Remington, and G. F. Brooks. 1999. Characterization of antimicrobial resistance in Streptococcus pyogenes isolates from the San Francisco Bay Area of Northern California. J. Clin. Microbiol. 37:1727-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]