Abstract
Staphylococcus aureus is a versatile and dangerous pathogen and one of the major causes of community-acquired and hospital-acquired infections. The rise of multidrug-resistant strains of S. aureus requires the development of new antibiotics with previously unexploited mechanisms of action, such as inhibition of the β-ketoacyl-acyl carrier protein (ACP) synthase III (FabH). This enzyme initiates fatty acid biosynthesis in a bacterial type II fatty acid synthase, catalyzing a decarboxylative condensation between malonyl-ACP and an acyl coenzyme A (CoA) substrate and is essential for viability. We have identified only one fabH in the genome of S. aureus and have shown that it encodes a protein with 57, 40, and 34% amino acid sequence identity with the FabH proteins of Bacillus subtilis (bFabH1), Escherichia coli (ecFabH), and Mycobacterium tuberculosis (mtFabH). Additional genomic sequence analysis revealed that this S. aureus FabH (saFabH) is not mutated in certain methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) strains. saFabH was expressed in E. coli with an N-terminal polyhistidine tag and subsequently purified by metal chelate and size exclusion chromatography. Analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a molecular mass of 37 kDa, while gel filtration demonstrated a mass of 66.7 kDa, suggesting a noncovalent homodimeric structure for saFabH. The apparent Km for malonyl-ACP was 1.76 ± 0.40 μM, and the enzyme was active with acetyl-CoA (kcat, 16.18 min−1; Km, 6.18 ± 0.9 μM), butyryl-CoA (kcat, 42.90 min−1; Km, 2.32 ± 0.12 μM), and isobutyryl-CoA (kcat, 98.0 min−1; Km, 0.32 ± 0.04 μM). saFabH was weakly inhibited by thiolactomycin (50% inhibitory concentration [IC50], >100 μM) yet was efficiently inhibited by two new FabH inhibitors, 5-chloro-4-phenyl-[1,2]-dithiol-3-one (IC50, 1.87 ± 0.10 μM) and 4-phenyl-5-phenylimino-[1,2,4]dithiazolidin-3-one (IC50, 0.775 ± 0.08 μM).
Staphylococcus aureus is a gram-positive coccus that can cause many different types of diseases, ranging from minor skin, soft tissue, and respiratory infections to life-threatening pneumonia, endocarditis, sepsis, and toxic shock syndrome (19, 26). It is also one of the most successful human pathogens due to its efficiency in acquiring antibiotic resistance (19). Shortly after the initial successful use of penicillin for treatment of S. aureus infection, penicillin-resistant strains began to emerge. Methicillin and other semisynthetic penicillins were developed and were effective against penicillin G-resistant S. aureus until the 1980s, when methicillin-resistant S. aureus (MRSA) became endemic in many hospitals (19). Since that time, the development of novel antibiotics has been paralleled by worldwide emergence of multidrug-resistant strains of staphylococci with alarmingly increasing frequency, including isolates that are resistant to methicillin, lincosamides, macrolides, aminoglycosides, and fluoroquinolones or combinations of these antibiotics (19, 21). S. aureus resistant to the glycopeptide vancomycin (VRSA) has also been isolated recently (14, 32, 34). S. aureus has thus acquired resistance to practically all antibiotics and has become one of the major causes of community-acquired and hospital-acquired infections leading to infectious morbidity and mortality (19, 35).
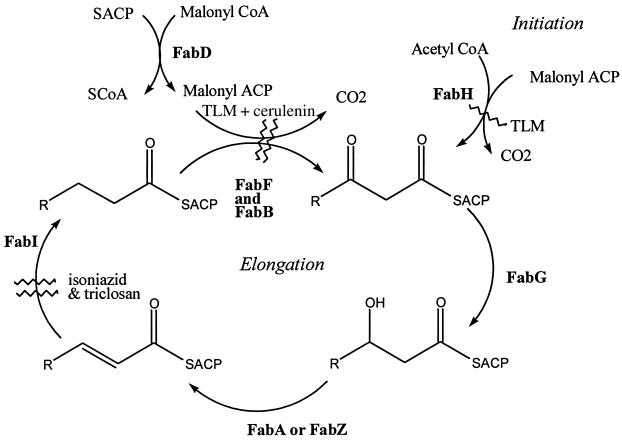
The increasing prevalence of multidrug-resistant S. aureus strains and the appearance of vancomycin resistance has heightened the importance of the development of antibacterials with previously unexploited mechanisms of antibiotic action. Saturated fatty acid biosynthesis has recently emerged as a prime candidate for development of such important and novel antibacterials (25). The ubiquitous type II fatty acid synthase (FAS) in bacteria is not only essential to cell survival but also exhibits structural and organizational differences from that in higher organisms, such as humans. It is generally accepted that highly potent and broad-spectrum antibiotics which selectively target components of this type II FAS can be obtained (17, 27, 28). So far, only the enoyl acyl carrier protein (ACP) reductase is targeted by isoniazid (1, 30) and the multipurpose biocide triclosan (23, 40) (Fig. 1). Compounds developed against other essential components of the type II FAS would potentially be effective against multidrug-resistant bacteria, including MRSA and VRSA.
FIG. 1.
Roles of individual enzymes in a type II fatty acid synthase. Full enzyme names are provided in the text. Enzymes inhibited by triclosan, cerulenin, and TLM are indicated (wavy lines).
In the dissociated type II FAS, each of the reactions is carried out by individual enzymes and an ACP (20), contrasting with the type I FAS of vertebrates, which contains ACP and all of the enzymatic activities encoded on one or two polypeptides (4). The process initiates with a β-ketoacyl-ACP synthase III (KASIII, or FabH)-catalyzed condensation between acyl coenzyme A (CoA) (typically acetyl-CoA) and malonyl-ACP (MACP) to form a 3-ketoacyl-ACP product (Fig. 1) (7, 9, 36). All subsequent extension steps utilize acyl-ACP derivatives and are catalyzed by β-ketoacyl synthase I and II (FabB and FabF) (22, 37). The MACP used in all elongation steps is generated from malonyl-CoA and ACP by the action of FabD (a malonyl-CoA ACP transacylase) (Fig. 1) (20). After each elongation step, the β-ketoacyl-ACP product is reduced to an acyl-ACP by the successive use of a β-ketoacyl-ACP reductase (FabG), β-hydroxyacyl-ACP dehydrase (FabA or FabZ), and NADH-dependent enoyl ACP reductase (FabI). Of all of these enzymes, FabH has attracted considerable interest as a target for drug development, primarily because of its pivotal roles in both initiation and regulation of the fatty acid biosynthesis process (it is inhibited by the ultimate product, palmitoyl-ACP [12, 13, 36, 38]). Until very recently, there were no known effective inhibitors of this enzyme (17). Cerulenin and thiolactomycin (TLM) (Fig. 2), two type II FAS inhibitors which target the ketoacyl-ACP synthases (FabB and FabF), are either inactive (cerulenin) or poor inhibitors (TLM and related compounds) of FabH (15, 17, 37).
FIG. 2.
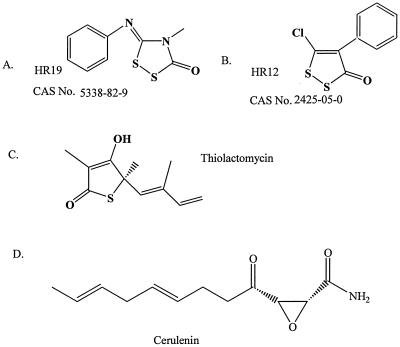
Structures of saFabH inhibitors tested in this study.
In this work, we have identified a single fabH gene in the genome of S. aureus and determined that it is unaltered in both an MRSA (N315) and a VRSA (Mu 50) strain. This S. aureus fabH gene has been cloned, and the corresponding S. aureus FabH (saFabH) recombinant protein has been overexpressed in Escherichia coli and characterized. Two compounds which inhibit this protein as much as 2 orders of magnitude more effectively than TLM have been identified. These results provide the framework for structure-based design of novel inhibitors for saFabH inhibitors and the potential for development of new therapies for treatment of infections with multidrug-resistant strains of S. aureus.
MATERIALS AND METHODS
Materials.
The following reagents were used: E. coli ACP, CoA, malonyl-CoA, imidazole, 2-mercaptoethanol, and protein molecular mass standards (Sigma Chemical Company); [3H]acetyl-CoA (specific activity, 4.6 Ci/mmol), [1-14C]acetyl-CoA (specific activity, 53 Ci/mmol), and [1-14C]butyryl-CoA (specific activity, 53 Ci/mmol) (Moravek Biochemicals); [9,10-3H]myristoyl-CoA (specific activity, 60 Ci/mmol) (American Radiochemical Co.); microbiological media (Difco Laboratories); S. aureus genomic DNA (American Type Culture Collection); restriction enzymes and T4 DNA ligase (New England BioLabs); pET15b vector (Novagen); E. coli XL1-Blue competent cells and the expression strain E. coli BL21-CodonPlus-RP (Stratagene); and Mueller-Hinton broth (Remel). TLM was a gift kindly provided by Pfizer, and the compounds HR12 and HR19 were obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute. The S. aureus RN 450 and MRSA N315 strains were generously provided by G. L. Archer and A. E. Rosato. Radiolabeled isobutyryl-CoA was prepared enzymatically from sodium [1-14C]isobutyric acid (specific activity, 56 Ci/mmol) (Moravek Biochemicals) and CoA as described previously (10). All other chemicals were reagent grade or better and were obtained from VWR Scientific or Fisher Scientific.
Expression and overproduction of saFabH in E. coli.
The putative fabH gene, encoding a 3-ketoacyl-ACP synthase III (FabH) homologue, AP003132, was amplified from S. aureus chromosomal DNA (ATCC 35556). The forward primer 5′-AAGGTGTCTCGAGATGAACGTGGGTA-3′ was designed to introduce an XhoI restriction site (underlined) at the start of the 5′ end of fabH. A BamHI site (underlined) was created downstream of the fabH stop codon in the reverse primer, 5′-ACTCATTCGGGATCCTCCTATTTTCC-3′. PCR was performed using the GeneAmp XL PCR kit (Perkin-Elmer). The resulting PCR product was eluted from agarose gel by using Qiax (Qiagen), digested with XhoI and BamHI, and ligated into the XhoI/BamHI-digested pET15b to construct plasmid pXH29. The resulting plasmid pXH29 was transformed to E. coli XL1-Blue competent cells. Subsequently, pXH29 isolated from E. coli XL1-Blue cells was transformed to the expression strain E. coli BL21-CodonPlus-RP. The inserted coding sequence of PCR-amplified fabH in pXH29 was sequenced and shown to be identical to the genomic fabH of S. aureus. Transformed cells were grown on Luria-Bertani (LB) agar plates supplemented with ampicillin (100 μg/ml). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was used to screen colonies for overexpression of saFabH. One such positive colony was used to inoculate 10 ml of LB medium with 100 μg of ampicillin/ml and grown overnight at 37°C, 1 ml of which was used to inoculate each of the 100-ml cultures of LB medium in 500-ml flasks supplemented with 100 μg of ampicillin/ml. The culture was incubated at 37°C with shaking at 250 rpm and induced with 1 mM isopropyl β-d-thiogalactoside when the optical density reached 0.8 at 600 nm. The culture was then incubated for 12 h at 30°C. Cells were harvested by centrifugation (10,000 × g; 4°C; 30 min) and washed twice in phosphate-buffered saline, and the resultant cell pellet was stored at −80°C.
Purification of saFabH.
All lysis and purification processes were carried out at 4°C. A frozen pellet of E. coli BL21-CodonPlus-RP/pXH29 was rapidly thawed and resuspended in lysis buffer A consisting of 5 mM imidazole, 1 mM 2-mercaptoethanol, 300 mM NaCl, and 50 mM Na2HPO4-NaH2PO4 (pH 8.0). The resulting cell suspension was broken by passage three times through high-pressure homogenization (Avestin) at 12,000 to 15,000 lb/in2 followed by centrifugation (Beckman J2-21) at 16,000 × g for 50 min. The crude cell extract was applied at a rate of 2 ml/min to a 5-ml Hitrap chelating column (Amersham Pharmacia Biotech, Inc.) that had been preequilibrated with buffer A. The column was then washed with 25 ml of buffer A containing 50 mM imidazole followed by a 25-ml linear gradient from 0 to 600 mM imidazole (3 ml/min). The FabH-containing fractions were pooled and concentrated with a Microcon 50-μm concentrator (Amicon). FabH was then applied to a Superdex 200 HR 10/30 size exclusion column (Amersham Pharmacia Biotech, Inc.) equilibrated with buffer B consisting of 10% glycerol, 100 mM Na2HPO4-NaH2PO4 (pH 7.0), and 1 mM 2-mercaptoethanol. The fractions containing the purified FabH pool were pooled and stored in 20% glycerol at −20°C. At each stage of purification, SDS-PAGE analysis with Coomassie blue staining was used to identify FabH-containing fractions. Protein quantitation was carried out using a Dotmetric Protein assay kit (Bioworld).
Native molecular mass determination of FabH.
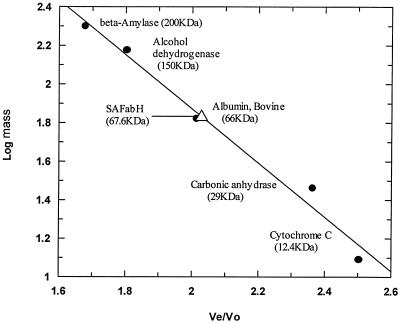
The native molecular mass of FabH was estimated by gel exclusion chromatography using a Superdex 200 HR 10/30 column. The column was preequilibrated and developed with 100 mM sodium phosphate (pH 7.0)-2 mM EDTA at a flow rate of 0.5 ml/min. The column was calibrated using the following molecular mass standards: blue dextran (2,000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). The purified protein sample was loaded onto the column and eluted at a flow rate of 0.5 ml/min. Fractions (0.5 ml) were collected and assayed for FabH activity.
FabH enzyme assays.
FabH activity was determined using standard methodologies to monitor the conversion of radioactive acyl-CoA and MACP substrates to a radiolabeled 3-ketoacyl-ACP (11). The MACP for these assays was generated from malonyl-CoA and the E. coli ACP using FabD and was purified following standard protocols (10, 11). A standard reaction mixture contained the following components in a final volume of 20 μl: 100 mM sodium phosphate buffer (pH 7.2), 15 ng of S. aureus His6-FabH, 1.8 μM MACP, and 6.8 μM [3H]acetyl-CoA (0.625 μCi; specific activity, 4.6 Ci/mmol). The reaction was initiated by adding [3H]acetyl-CoA and incubated at 30°C for 10 min. The reaction was terminated by addition of 10% trichloroacetic acid (TCA) and analyzed in the standard way. A scintillation proximity assay using biotinylated MACP and [9,10-3H]myristoyl-CoA (specific activity, 60 Ci/mmol) was conducted as described previously (11).
Steady-state kinetic determinations.
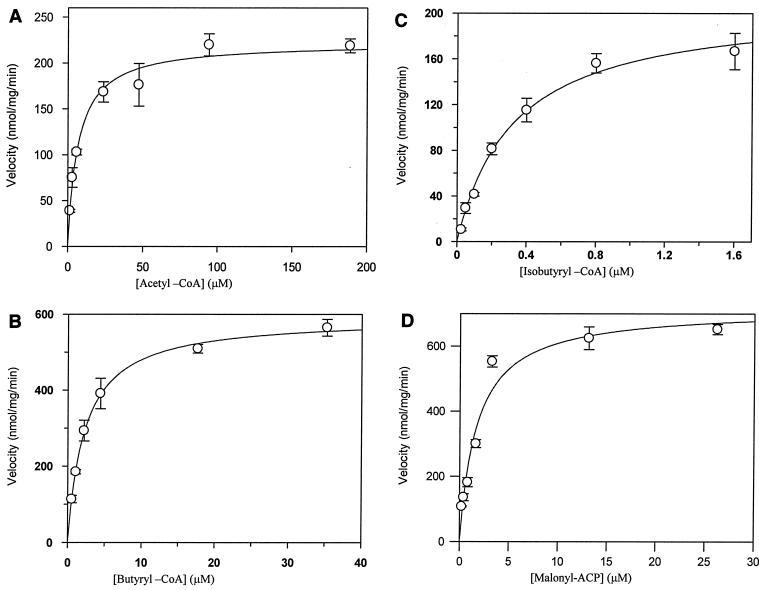
Steady-state kinetic parameters for acetyl-CoA and butyryl-CoA were obtained by determination of FabH activity in the presence of various concentrations of 14C-labeled acetyl-CoA (specific activity, 53 mCi/mmol; 1.48 to 188.6 μM), butyryl-CoA (specific activity, 53 mCi/mmol; 0.27 to 35.3 μM), or isobutyryl-CoA (specific activity, 56 mCi/mmol; 0.025 to 1.6 μM) in the presence of 17.5 μM MACP. Similarly, an apparent Km for MACP was obtained by determining saFabH activity in the presence of 9.43 μM butyryl-CoA and various MACP concentrations. The reactions were terminated at 3 min of incubation during the linear rate of reaction and analyzed using a standard FabH assay (11). Three independent determinations were used to obtain the average and standard deviation for each datum point. Nonlinear regression with Grafit version 4.012 (Erithacus Software, Horley, United Kingdom) was used to determine kcat and Km values (Table 1).
TABLE 1.
Kinetic parameters for saFabH activity
| Substrate and concn (μM) | Km (μM) | Vmax (U/mg of protein)a | kcat (min−1) | kcat/Km (μM·min−1) |
|---|---|---|---|---|
| Acetyl-CoA (1.48-188.6) | 6.18 ± 0.96 | 0.22 | 16.18 | 2.62 |
| Butyryl-CoA (0.27-35.3) | 2.32 ± 0.12 | 0.59 | 42.90 | 18.49 |
| Isobutyryl-CoA (0.025-1.6) | 0.32 ± 0.04 | 0.43 | 31.58 | 98 |
| Malonyl-ACP (0.205-26.25) | 1.76 ± 0.40 |
One unit is defined as the formation of 1 pmol of product 3-ketoacyl-ACP per min.
Inhibition studies.
In vitro inhibition of saFabH activity was evaluated using TLM (0.38 to 238 μM), cerulenin (0.063 to 196 μM), HR12 (0.118 to 236 μM), and HR19 (0.0078 to 6.98 μM). In all inhibitor analyses, the inhibitors were first dissolved in dimethyl sulfoxide (DMSO) and then diluted to the required concentration, followed by incubation with FabH for 15 min at room temperature (23°C). All assays, including controls, were carried out in triplicate in 1% DMSO. The reaction was initiated by the addition of the substrates MACP and [3H]acetyl-CoA under the standard assay conditions described above. The concentrations of MACP (1.8 μM) and [3H]acetyl-CoA (6.2 μM) used in these assays were comparable to the corresponding Km values for these substrates (Table 1). A control experiment in which FabH was preincubated with 1% DMSO for 15 min in the absence of inhibitors was used in all of the experiments. Fifty percent inhibitory concentrations (IC50s) were calculated using Grafit version 4.012.
MIC determination.
Susceptibility testing was carried out by the standard twofold serial broth dilution method with an inoculum of 5 × 105 cells/ml. The data were reported as MICs, the lowest concentration of antibiotic inhibiting visible growth after 24 h of incubation at 37°C.
RESULTS
Identification of fabH from the S. aureus genome.
The genome sequences of S. aureus from an MRSA strain (N315) and a VRSA strain (Mu 50) were analyzed and shown to contain one identical gene encoding a FabH (AP003132 and AP003360) homologue (18). An alignment of the N315 and Mu 50 fabH genes using Clustal W(version 1.4) in MacVector 7.0 (Oxford Molecular) demonstrated that the corresponding proteins were 100% identical to those of the fabH sequence from S. aureus 476 (a hypervirulent United Kingdom methicillin-susceptible S. aureus strain available through the Sanger center) and the fabH PCR amplified in this study using S. aureus chromosomal DNA (American Type Culture Collection). Analysis of genome sequences of MRSA strains 252 (a recent United Kingdom hospital-acquired epidemic MRSA [http://www.sanger.ac.uk]) and NCTC 8325 (http://www.genome.ou.edu/staph.html) revealed FabH proteins whose predicted amino acid sequences contain only two conservative mutations. The calculated molecular mass of the ATCC 35556 S. aureus FabH is 33,877 Da, with a calculated pI of 4.67, which is comparable to those of other FabH proteins (10, 17, 31).
Comparison of FabH sequences from other organisms.
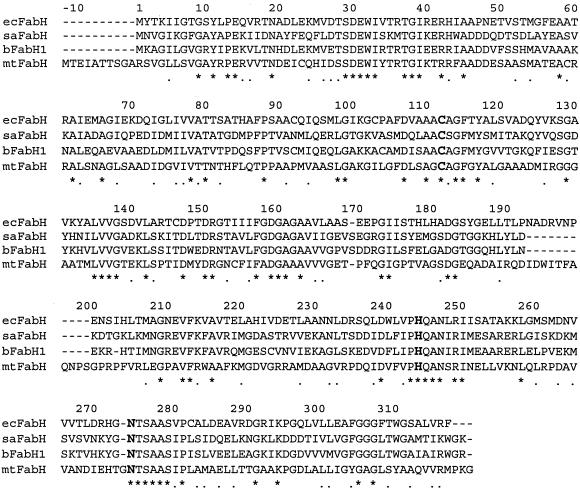
A multiple sequence alignment of the saFabH protein was constructed using Clustal W (version 1.4) in the MacVector 7.0 software package (Fig. 3). The S. aureus FabH protein exhibited 57, 40, and 34% identity with the FabH proteins of Bacillus subtilis (bFabH1) (5), E. coli (ecFabH) (36), and Mycobacterium tuberculosis (mtFabH) (31), respectively. This analysis revealed the 100% conservation of the ecFabH residues Cys112, His244, Asn274, and Gly306 (Fig. 3). Crystallographic studies of ecFabH have recently demonstrated the role of Cys112 as the nucleophilic residue responsible for forming an acyl enzyme intermediate. The backbone nitrogen of the highly conserved Gly306 and Cys112 is proposed to form an oxyanion hole which stabilizes the tetrahedral intermediate that is formed in the initial transacylation and subsequent condensation steps, while the conserved His244 and Asp274 are essential residues presumed to be involved in both binding and promoting decarboxylation of MACP (8, 27, 28).
FIG. 3.
Multiple sequence alignment of saFabH and other bacterial homologues. The amino acid sequence of saFabH is compared with those of bFabH1, ecFabH, and mtFabH. Positions are numbered based on the sequence of ecFabH. The conserved triad (Cys 112, His 244, and Asn 274) at the enzyme active site is in bold face. The asterisks denote sequence identities, and the dots denote sequence similarities.
The sequence alignment also revealed that saFabH has the conserved hydrophobic residues Leu142, Phe157, Leu189, Leu205, and Phe87, proposed to contribute to the formation of a specific binding pocket for acetyl- or propionyl-CoA at the dimer interface of ecFabH (27, 28). This analysis suggested that saFabH might also possess a similar acyl-CoA substrate specificity. In contrast, a recent analysis of the mtFabH crystal structure has revealed that a much larger acyl group can be accommodated at the active site, consistent with the observation that this unusual enzyme prefers lauroyl-CoA and myristoyl-CoA as substrates. This larger acyl group binding channel is created by several mutations, including replacing the ecFabH Phe87 with threonine (31).
Residues which are proposed to interact with the adenosine ring and phosphate groups of CoA, like Trp32, Arg151, and Thr28 (28), are conserved in saFabH. saFabH also contains a highly conserved Arg42, which plays an important role in maintaining the structural integrity of the ligand binding domain (27, 28). A conserved Arg249, shown recently to be important in allowing E. coli ACP to dock to ecFabH (41), is also present in saFabH.
Purification and characterization of saFabH.
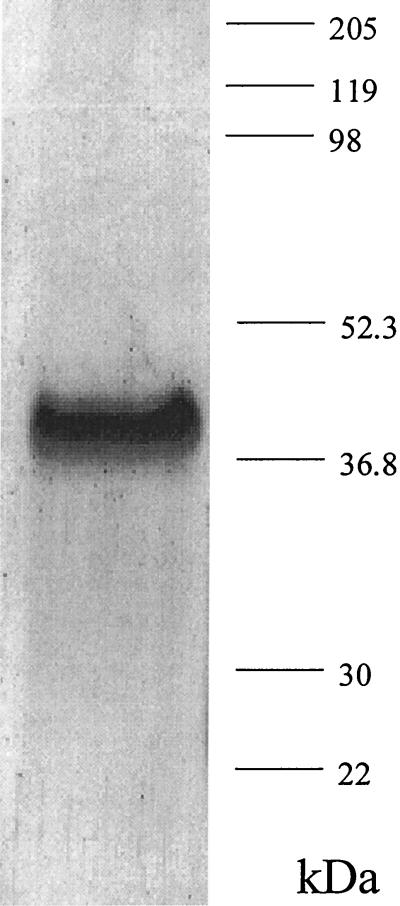
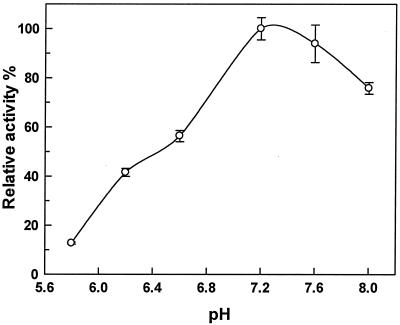
A PCR-based approach was used to generate an E. coli vector for expression of an N-terminal His6-tagged saFabH. This saFabH was purified by metal chelate affinity chromatography and size exclusion chromatography as described in Materials and Methods. The overall yield of the purified saFabH was approximately 3 mg/liter of culture. The purified saFabH appeared as a homogeneous band with a mass of 37 kDa, as judged by SDS-PAGE (Fig. 4), in agreement with the predicted mass of the protein based on the FabH sequence plus the polyhistidine tag. Most condensing enzymes are known to be homodimeric, and in the case of mtFabH and ecFabH, crystallographic studies have shown that the dimer interface is responsible for forming a component of the active site (27, 28, 31). Gel filtration chromatography with saFabH on Superdex 200 demonstrated a mass of 67.6 kDa (Fig. 5), which in conjunction with the SDS-PAGE analysis suggests a noncovalent homodimeric structure for saFabH. The pH optimum for FabH was measured and ranges from 5.8 to 8.0 under the standard assay conditions with acetyl-CoA and MACP as substrates. saFabH showed a significant pH dependence, with maximum activity at pH 7.2 (Fig. 6).
FIG. 4.
SDS-PAGE analysis of purified saFabH.
FIG. 5.
Molecular mass determination of native-form FabH using size exclusion chromatography. Log molecular mass versus elution volume/void volume ratio (Ve/Vo) is shown. The molecular mass standards used were β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). Vo was determined using blue dextran (2,000 kDa). The saFabH mass was measured at approximately 67 kDa.
FIG. 6.
A pH profile for β-ketoacyl-ACP synthase activity of saFabH. Conditions were as described in Materials and Methods. Each datum point is the average of two determinations.
Enzyme kinetics and substrate specificity.
Evidence of a homodimeric structure and the presence of conserved residues such as Phe87, which form the hydrophobic pocket for an acetyl- and propionyl-CoA group at the dimer interface of ecFabH, suggested a similar substrate specificity for saFabH. Consistent with this observation, saFabH was active with acetyl-CoA but not myristoyl-CoA, contrasting with mtFabH, which can use myristoyl-CoA, but not acetyl-CoA or butyryl-CoA (5, 31). Using a direct TCA assay with the E. coli MACP, the purified saFabH demonstrated a high affinity for acetyl-CoA (Km, 6.18 ± 0.9 μM) (Table 1 and Fig. 7), comparable to that observed with Streptomyces glaucescens FabH (2.4 μM) and higher than that reported for either ecFabH (40 μM) (12) or Streptococcus pneumoniae FabH (spFabH) (40 μM) (17). saFabH also demonstrated a high affinity for butyryl-CoA (2.32 μM) and isobutyryl-CoA (0.32 μM) and generated the corresponding 3-ketoacyl-ACP products at rates 2.65- and 1.95-fold faster, respectively, than those observed using acetyl-CoA.
FIG. 7.
Kinetic analysis of saFabH for acetyl-CoA, butyryl-CoA, and MACP. The initial velocities of product formation were measured with purified saFabH and MACP in the presence of increasing concentrations of acetyl-CoA (A), butyryl-CoA (B), and isobutyryl-CoA (C). The initial velocities of product formation were measured with purified saFabH and butyryl-CoA in the presence of increasing concentrations of MACP (D). Each datum point is the average of three determinations.
Inhibition studies.
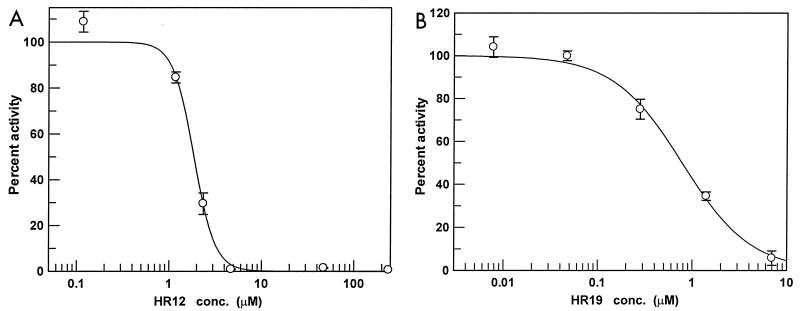
In E. coli, it has been demonstrated that among the condensing enzymes, FabH is inhibited the least by TLM (37). We have previously observed a similar poor inhibition of ecFabH by TLM (124 to 160 μM), although others have reported slightly better inhibition (32 ± 12 μM). The in vitro effectiveness of the TLM of FabH is known to be highly dependent upon both assay conditions (11) and the source of the FabH (17). We used the same set of assay conditions to analyze saFabH as we had previously for ecFabH and observed similarly poor inhibition (IC50, >100 μM). Cerulenin, a known inhibitor of condensing enzymes in both type I and type II FAS systems, is well established as being a poor inhibitor of FabH (15, 17, 25) and was similarly ineffective against saFabH (IC50, >100 μM). The resistance of saFabH to the antibiotics TLM and cerulenin led us to search for new and more effective inhibitors. Numerous compounds bearing some structural similarities to TLM were identified from the National Cancer Institute database and subsequently tested. HR12, 5-chloro-4-phenyl-[1,2]-dithiol-3-one, and HR19, 4-phenyl-5-phenylimino-[1,2,4]dithiazolidin-3-one, both efficiently inhibited saFabH with IC50s of 1.87 ± 0.10 and 0.78 ± 0.08 μM, respectively (Fig. 2 and 8). Thus, under the same assay conditions these two compounds are as much as 2 orders of magnitude more effective inhibitors of saFabH than TLM.
FIG. 8.
Inhibition of saFabH by compounds HR12 (A) and HR19 (B). Inhibitors were tested using a TCA precipitation assay as described in Materials and Methods (n = 3). Inhibitor structures are shown in Fig. 2. conc., concentration.
Antibacterial activity of saFabH inhibitors.
The MICs of HR12, HR19, and TLM against S. aureus and MRSA (Table 2) were determined. With S. aureus RN 450 and MRSA N315, HR12 exhibited significant antibacterial activity at 8 μg/ml (34.9 μM) compared to TLM (>300 μg/ml; 1,429 μM). HR19 was similarly more effective against both strains than TLM although less potent than HR12. Neither compound was as effective as vancomycin or oxacillin, which under comparable conditions with strains N315 (vancomycin) and RN 450 (oxacillin) exhibited MICs of 1 μg/ml (0.67 μM) and 0.2 μg/ml (0.45 μM), respectively.
TABLE 2.
MICs of FabH inhibitors against various S. aureus strains
| Bacterial strain | MIC (μg/ml)
|
||
|---|---|---|---|
| HR12 | HR19 | TLM | |
| S. aureus RN 450 | 8.7 | 35.0 | 300 |
| MRSA N315 | 8.7 | 17.5 | >300 |
DISCUSSION
FabH exists ubiquitously in bacteria (27), is also present in parasites such as Plasmodium falciparum (39), and has a pivotal role in both initiating and regulating fatty acid biosynthesis in type II dissociated fatty acid synthases (24). No successful fabH deletion strains have been reported to date, despite significant efforts by ourselves and others. In Streptomyces coelicolor, it has been observed that disruption of the chromosomal fabH can only be accomplished if a second copy of fabH is first introduced at a separate site on the chromosome (29). The ubiquitous nature of FabH in bacteria and the mounting evidence that it is essential to bacterial viability (27, 29) have made it an attractive potential target for development of new broad-spectrum antibiotics (27, 28). In S. aureus, only one FabH homologue has been observed (B. subtilis, by contrast, contains two fabH genes [5]), and the corresponding FabH is not significantly altered in MRSA and VRSA strains. An inhibitor of saFabH, therefore, may represent an effective treatment of infections with these strains. As a preliminary step in this direction, we expressed and characterized saFabH and discovered two novel inhibitors.
saFabH exhibited properties typical for this class of enzyme. It was able to utilize a variety of short-chain acyl-CoA substrates but exhibited a clear preference for isobutyryl-CoA. The S. glaucescens and B. subtilis FabH proteins have similar relaxed substrate specificities and have also been shown to utilize a wide range of two- to six-carbon straight-chain and branched-chain acyl-CoA substrates (5, 10). This relaxed specificity is thought to be one of the factors contributing to the ability of these organisms to make both straight-chain and branched-chain fatty acids (5). In contrast, the E. coli and S. pneumoniae FabH proteins react preferentially with acetyl-CoA and propionyl-CoA (12, 17), consistent with the observation that these organisms generate straight-chain fatty acids almost exclusively (33). The observation of a relaxed substrate specificity for saFabH is consistent with the ability of this organism to generate both straight-chain and branched-chain fatty acids (16). From the data available to date, it would appear that the qualitative substrate specificity of a FabH protein can be predicted from the type of fatty acids made by an organism. In contrast, the substrate specificity of a FabH protein cannot yet be accurately predicted from analysis of its primary amino acid sequence. The residues in ecFabH which define the acyl group binding pocket are highly conserved in saFabH, yet the enzymes exhibit significantly different substrate specificities. In contrast, these residues are not reported to be rigorously conserved in spFabH, despite both enzymes exhibiting clear substrate preference for acetyl-CoA (17). In particular, the Phe87′ residue, which plays a role in primer recognition in ecFabH, is present in some of the enzymes with more relaxed substrate specificities (bFabH1, bFabH2, and saFabH) but not in spFabH.
TLM was shown in this study to be a poor inhibitor of saFabH. In E. coli, it has been shown that TLM is significantly more effective against the condensing enzyme FabB than against FabH (25). Crystallographic studies of an E. coli FabB-cerulenin and E. coli-TLM complex alongside mutational analysis demonstrated that the two His residues at the active site and the larger hydrophobic binding pocket of FabB are important contributors to effective inhibition by cerulenin and TLM (25). ecFabH, by contrast, has a smaller hydrophobic binding pocket and a histidine-asparagine active-site architecture, which is proposed to contribute to the decreased effectiveness of these antibiotics (25). This theory, however, does not explain why TLM is more effective against spFabH and Haemophilus influenzae FabH, which have similar properties (17), and not effective against mtFabH (25), which has a larger hydrophobic binding pocket (6, 31). The reasons why TLM is a poor inhibitor of ecFabH, saFabH, and other FabH enzymes remains undetermined. In this study we rationalized that an analysis of compounds bearing structural similarity to TLM might provide more potent FabH inhibitors. Such an analysis provided two compounds, HR12 and HR19, which demonstrated similarly dramatic improvements over the inhibition seen with TLM. A similar improvement over inhibition with TLM was recently reported for spFabH using SB418011, a 2,3,6-substituted indole (17) which bears no clear structural resemblance to either TLM or cerulenin. The mechanisms and sites of inhibition of these various FabH inhibitors remain to be determined. Nonetheless, a number of structurally diverse compounds which inhibit FabH from pathogens such as S. pneumoniae and S. aureus have now been discovered. Crystallographic and mechanistic efforts ongoing in our laboratory are aimed at addressing these questions.
5-Chloro-4-phenyl-(1,2)-dithiol-3-one (HR12) was recently identified as RWJ-3981, one of three new compounds which inhibit the bacterial enzyme MurA (2). This enzyme, UDP-N-acetylglucosamine enolpyruvyl transferase, catalyzes the first step in peptidoglycan biosynthesis and is essential in that its deletion is lethal in both E. coli and S. pneumoniae. MurA and FabH catalyze different reactions in different biological processes. Despite these differences, both enzymes form covalent adducts between one of their substrates and an active-site cysteine (2, 3) and are efficiently inhibited by the same cyclic disulfide. Ultrafiltration and mass spectrometry experiments coupled with molecular modeling have been used to suggest that this inhibitor binds noncovalently with the centroid of the ring approximately 7 Å from the active-site cysteine residue of MurA (2). The exact mode of binding to either MurA or FabH, however, remains to be determined.
The two saFabH inhibitors HR12 and HR19 were shown to have significantly higher antibacterial activity against S. aureus and MRSA than TLM (Table 2). A similar activity was observed for HR12 (RWJ-3981) against S. aureus strains in the recent studies of a novel MurA inhibitor (2). In the latter case, addition of RJW-3981 at the MIC was observed to inhibit DNA, RNA, protein, and cell wall synthesis from 78 to 99%. Thus, it seems unlikely that the antibacterial activity of this compound is due specifically to inhibition of either MurA (2) or FabH. Other studies with RWJ-3981 have suggested that it does not affect the bacterial cell wall. The biochemical basis for the antibacterial activity thus has yet to be determined. The compound does not cause overt toxicity at doses of 80 mg/kg of body weight but does not protect mice from death in an S. aureus lethal-infection model (2).
In conclusion, we have cloned and characterized the saFabH homologue and identified several potent inhibitors. With the availability of an appropriate FabH scintillation proximity assay (11) and significant progress in crystallographic studies of this class of proteins (8, 27, 28, 31), it is reasonable to suggest that additional FabH inhibitors will be obtained through screening and structure-based drug design approaches. Such compounds might eventually lead to a new and effective treatment for infections with MRSA and VRSA.
Acknowledgments
We are grateful to G. L. Archer and A. E. Rosato (Virginia Commonwealth University) for providing S. aureus RN 450 and MRSA N315 strains for MIC determination and for their helpful suggestions. We thank the Central Research Division, Pfizer Inc., for providing TLM. We also thank the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, for providing the chemical samples for the biological evaluations in this study.
This work was supported by NIAID Grant AI44772 from the National Institutes of Health.
REFERENCES
- 1.Baldock, C., J. B. Raffert, S. E. Sedelnikova, P. J. Baker, A. R. Stuitje, A. R. Slabas, T. R. Hawkes, and D. W. Rice. 1996. A mechanism of drug action revealed by structural studies of enoyl reductase. Science 274:2107-2110. [DOI] [PubMed] [Google Scholar]
- 2.Baum, E. Z., D. A. Montenegro, L. Licata, I. Turchi, G. C. Webb, B. D. Foleno, and K. Bush. 2001. Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme. Antimicrob. Agents Chemother. 45:3182-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, E. D., J. L. Marquardt, J. P. Lee, C. T. Walsh, and K. S. Anderson. 1994. Detection and characterization of a phospholactoyl-enzyme adduct in the reaction catalyzed by UDP-N-acetylglucosamine enolpyruvoyl transferase, MurZ. Biochemistry 33:10638-10645. [DOI] [PubMed] [Google Scholar]
- 4.Chirala, S. S., W. Y. Huang, A. Jayakumar, K. Sakai, and S. J. Wakil. 1997. Animal fatty acid synthase: functional mapping and cloning and expression of the domain I constituent activities. Proc. Natl. Acad. Sci. USA 94:5588-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, K. H., R. J. Heath, and C. O. Rock. 2000. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K. H., L. Kremer, G. S. Besra, and C. O. Rock. 2000. Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J. Biol. Chem. 275:28201-28207. [DOI] [PubMed] [Google Scholar]
- 7.Clough, R. C., M. Matthis, S. R. Barnum, and J. G. Jaworski. 1992. Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach; a condensing enzyme utilizing aceyl coenzyme A to initiate fatty acid synthesis. J. Biol. Chem. 267:20992-20998. [PubMed] [Google Scholar]
- 8.Davies, C., R. J. Heath, S. W. White, and C. O. Rock. 2000. The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure Fold. Des. 8:185-195. [DOI] [PubMed] [Google Scholar]
- 9.Gulliver, B. S., and A. R. Slabas. 1994. Acetoacetyl-acyl carrier protein synthase from avocado: its purification, characterisation and clear resolution from acetyl CoA:ACP transacylase. Plant Mol. Biol. 25:179-191. [DOI] [PubMed] [Google Scholar]
- 10.Han, L., S. Lobo, and K. A. Reynolds. 1998. Characterization of 3-ketoacyl acyl carrier protein synthase III from Streptomyces glaucescens: its role in the initiation of fatty acid biosynthesis. J. Bacteriol. 180:4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, X., J. P. Mueller, and K. A. Reynolds. 2000. Development of a scintillation proximity assay for beta-ketoacyl-acyl carrier protein synthase III. Anal. Biochem. 282:107-114. [DOI] [PubMed] [Google Scholar]
- 12.Heath, R. J., and C. O. Rock. 1996. Inhibition of β-ketoacyl-acyl carrier protein synthase-III (Fab H) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:10996-11000. [DOI] [PubMed] [Google Scholar]
- 13.Heath, R. J., and C. O. Rock. 1996. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271:1833-1836. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 15.Jackowski, S., C. M. Murphy, J. J. E. Cronan, and C. O. Rock. 1989. Acetoacetyl-acyl carrier protein synthase. A target for the antibiotic thiolactomycin. J. Biol. Chem. 264:7624-7629. [PubMed] [Google Scholar]
- 16.Kaneda, T., E. J. Smith, and D. N. Naik. 1983. Fatty acid composition and primer specificity of de novo fatty acid synthetase activity in Bacillus globisporus, Bacillus insolitus, and Bacillus psychrophilus. Can. J. Microbiol. 29:1634-1641. [DOI] [PubMed] [Google Scholar]
- 17.Khandekar, S. S., D. R. Gentry, G. S. Van Aller, P. Warren, H. Xiang, C. Silverman, M. L. Doyle, A. K. Konstantinidis, M. Brandt, R. A. Daines, and J. T. Lonsdale. 2001. Identification, substrate specificity, and inhibition of the Streptococcus pneumoniae β-ketoacyl-acyl carrier protein synthase III (FabH). J. Biol. Chem. 274:30024-30030. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 20.Magnusson, K., S. Jackowski, C. O. Rock, and J. J. E. Cronan. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57:522-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moellering, R. C. 1998. Problems with antimicrobial resistance in gram-positive cocci. Clin. Infect. Dis. 26:1177-1178. [DOI] [PubMed] [Google Scholar]
- 22.Nishida, I., A. Kawaguchi, and M. Yamada. 1986. Effect of thiolactomycin on the individual enzymes of the fatty acid synthase system in Escherichia coli. J. Biochem. 99:1447-1454. [DOI] [PubMed] [Google Scholar]
- 23.Parikh, S. L., G. Xiao, and P. J. Tonge. 2000. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39:7645-7650. [DOI] [PubMed] [Google Scholar]
- 24.Payne, D. J., P. V. Warren, D. J. Holmes, Y. Ji, and J. T. Lonsdale. 2001. Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov. Today 6:537-544. [DOI] [PubMed] [Google Scholar]
- 25.Price, A. C., K. H. Choi, R. J. Heath, Z. Li, S. W. White, and C. O. Rock. 2000. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin: structure and mechanism. J. Biol. Chem. 276:6551-6559. [DOI] [PubMed] [Google Scholar]
- 26.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human diseases. Churchill Livingstone, New York, N.Y.
- 27.Qiu, X., C. A. Janson, A. K. Konstantinidis, S. Nwagwu, C. Silverman, W. W. Smith, S. Khandekar, J. Lonsdale, and S. S. Abdel-Meguid. 1999. Crystal structure of beta-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis. J. Biol. Chem. 274:36465-36471. [DOI] [PubMed] [Google Scholar]
- 28.Qiu, X., C. A. Janson, W. W. Smith, M. Head, J. Lonsdale, and A. K. Konstantinidis. 2001. Refined structures of beta-ketoacyl-acyl carrier protein synthase III. J. Mol. Biol. 307:341-356. [DOI] [PubMed] [Google Scholar]
- 29.Revill, W. P., M. J. Bibb, A. K. Scheu, H. J. Kieser, and D. A. Hopwood. 2001. β-Ketoacyl acyl carrier protein synthase III (FabH) is essential for fatty acid biosynthesis in Streptomyces coelicolor A3(2). J. Bacteriol. 183:3526-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozwarski, D. A., G. A. Grant, D. H. Barton, W. R. Jacobs, Jr., and J. C. Sacchettini. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98-102. [DOI] [PubMed] [Google Scholar]
- 31.Scarsdale, J. N., G. Kazanina, X. He, K. A. Reynolds, and H. T. Wright. 2001. Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III. J. Biol. Chem. 276:20516-20522. [DOI] [PubMed] [Google Scholar]
- 32.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 33.Smirnova, N., and K. A. Reynolds. 2001. Branched-chain fatty acid biosynthesis in Escherichia coli. J. Ind. Microbiol. Biotechnol. 26:246-251. [DOI] [PubMed] [Google Scholar]
- 34.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, W. R. Jarvis, et al. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 35.Steinberg, J. P., C. C. Clark, and B. O. Hackman. 1996. Nosocomial and community-acquired Staphylococcus aureus bacteremias from 1980 to 1993: impact of intravascular devices and methicillin resistance. Clin. Infect. Dis. 23:255-259. [DOI] [PubMed] [Google Scholar]
- 36.Tsay, J. T., W. Oh, T. J. Larson, S. Jackowski, and C. O. Rock. 1992. Isolation and characterization of the β-keto-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J. Biol. Chem. 267:6807-6814. [PubMed] [Google Scholar]
- 37.Tsay, J. T., C. O. Rock, and S. Jackowski. 1992. Overproduction of β-keto-acyl carrier protein synthase I imparts thiolactomycin resistance to Escherichia coli K-12. J. Bacteriol. 174:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verwoert, G. S., II, K. H. van der Linden, M. C. Walsh, H. J. J. Nijkamp, and A. R. Stuitje. 1995. Modification of Brassica napus seed oil by expression of the Escherichia coli fabH gene, encoding 3-ketoacyl-acyl carrier protein synthase III. Plant Mol. Biol. 27:875-886. [DOI] [PubMed] [Google Scholar]
- 39.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, W. H., G. A. Holdgate, S. Rowsell, E. G. McLean, R. A. Pauptit, E. Clayton, W. W. Nichols, J. G. Colls, C. A. Minshull, D. A. Jude, A. Mistry, D. Timms, R. Camble, N. J. Hales, C. J. Britton, and I. W. Taylor. 1999. Kinetic and structural characteristics of the inhibition of enoyl (acyl carrier protein) reductase by triclosan. Biochemistry 38:12514-12525. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y. M., M. S. Rao, R. J. Heath, A. C. Price, A. J. Olson, C. O. Rock, and S. W. White. 2001. Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III. J. Biol. Chem. 276:8231-8238. [DOI] [PubMed] [Google Scholar]