Abstract
The entire simocyclinone biosynthetic cluster (sim gene cluster) from the producer Streptomyces antibioticus Tü6040 was identified on six overlapping cosmids (1N1, 5J10, 2L16, 2P6, 4G22, and 1K3). In total, 80.7 kb of DNA from these cosmids was sequenced, and the analysis revealed 49 complete open reading frames (ORFs). These ORFs include genes responsible for the formation and attachment of four different moieties originating from at least three different pools of primary metabolites. Also in the sim gene cluster, four ORFs were detected that resemble putative regulatory and export functions. Based on the putative function of the gene products, a model for simocyclinone D8 biosynthesis was proposed. Biosynthetic mutants were generated by insertional gene inactivation experiments, and culture extracts of these mutants were analyzed by high-performance liquid chromatography. Production of simocyclinone D8 was clearly detectable in the wild-type strain but was not detectable in the mutant strains. This indicated that indeed the sim gene cluster had been cloned.
Simocyclinone D8 (Fig. 1) is produced by Streptomyces antibioticus Tü60 40. It is active against gram-positive bacteria and also shows distinct cytostatic activities against human tumor cell lines (39, 40, 46). Simocyclinone D8 consists of four different moieties, an angucyclic polyketide core, a deoxyhexose (d-olivose), a tetraene side chain, and a halogenated amino-coumarin. The aromatic polyketide moiety is characterized by a large number of unusually placed hydroxyl groups and an oxiran bridge at positions C-12a and C-6a. It contains a C-glycosidically linked d-olivose at position C-9. Attached to the 4-OH group of d-olivose is an acetyl group, and attached to the 3-OH group is a tetraene side chain. Both are linked to the deoxysugar by ester bonds. The final amino-coumarin moiety is linked to the tetraene chain by an amide bond, resulting in an unusual polyene-amide structure. Features that distinguish simocyclinone from other angucycline antibiotics are the enormous size of the molecule and the fact that it originates from at least three different pools of primary metabolites. S. antibioticus Tü6040 also produces other simocyclinones, which can be seen as intermediates of simocyclinone D8. These compounds include simocyclinones of the A-series, the B-series, and the C-series, consisting either of the polyketide moiety (series A), the polyketide moiety plus d-olivose (series B), and the polyketide moiety plus d-olivose plus the tetraene side chain (series C) (40). Genetic engineering and combinatorial biosynthesis in bacteria provide an important new tool for drug discovery and drug design (16, 19, 24). Knowledge of the sequence and function of genes involved in the biosynthesis of natural products is prerequisite for this new approach. In the present study we describe the isolation of the simocyclinone biosynthetic gene cluster. Sequencing of the entire gene cluster revealed the presence of 49 open reading frames (ORFs) probably involved in simocyclinone biosynthesis. Insertional inactivation of genes by homologous recombination effected simocyclinone production, confirming that the cloned genes belong to the simocyclinone biosynthetic gene cluster.
FIG. 1.
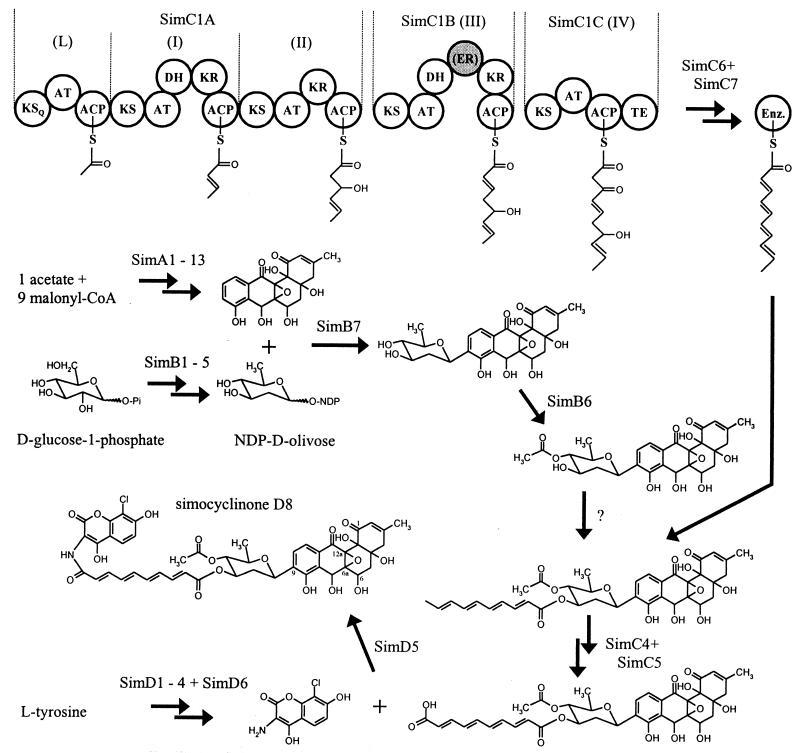
Biosynthetic scheme for simocyclinone D8. Domain organization and biosynthetic intermediates for SimC1A, SimC1B, and SimC1C are shown on top. Domain designations: KS, ketosynthase; AT, acyltransferase; ACP, acyl carrier protein; DH, dehydratase; KR, ketoreductase; ER, enoyl reductase; TE, thioesterase. The ER domain, shown in grey, is believed to be inactive. Single enzymatic steps are indicated by a single arrow, and multiple reactions steps are indicated by two arrows.
MATERIALS AND METHODS
Strains, growth conditions, media, and vectors.
For standard purposes, S. antibioticus Tü6040 and mutant strains Sim-C2 and Sim-PKSII were grown on 2% mannitol and 2% soybean meal, pH 7.5, prepared as solid or liquid medium at 28°C. For maintenance of mutants, erythromycin was added to a final concentration of 50 μg/ml. For simocyclinone production, liquid medium (20 ml in a single-baffled 100-ml Erlenmeyer flask) was used (39, 40, 46). DNA manipulation was carried out in Escherichia coli XL-1 Blue MRF′ (Stratagene). Before transforming S. antibioticus Tü6040, plasmids were propagated in E. coli ET12567 (dam deficient, dcm deficient, hsdS, Cmr) (12) to obtain unmethylated DNA. E. coli strains were grown on Luria-Bertani agar or liquid medium containing the appropriate antibiotic for selection. The vectors pBluescript SK(−) (pBSK−) and pBCSK− were from Stratagene, and pSP1, carrying the erythromycin resistance gene, was used for gene disruption (34). Cosmid pKC505 (36) was obtained from Christiane Bormann (Tübingen, Germany).
General genetic manipulation and PCR.
Standard molecular biology procedures were performed as described previously (37). Isolation of plasmid DNA from E. coli and DNA restriction-ligation were performed following the protocols of the manufacturers of the kits, enzymes, and reagents (Amersham Pharmacia [Freiburg, Germany], Macherey & Nagel [Dueren, Germany], New England Biolabs [Frankfurt, Germany], and Promega [Mannheim, Germany]). A cosmid library was prepared using cosmid pKC505. For preparation of DNA, mycelia were embedded in agarose. Isolation, partial digestion, and separation were performed as described elsewhere (35). PCRs were performed on a Perkin-Elmer GeneAmp 2400 thermal cycler. PCR conditions were similar to those described previously (9). Primers were purchased from Amersham Pharmacia. For the amplification of nucleoside diphosphate (NDP)-4-keto-6-deoxyhexose 2,3-dehydratase genes, primers PCR-D1a (5′ CAGGCSACSWSSAACTACAC 3′) and PCR-D1b (5′ SWRGAASCGSCCSCCCTCCT 3′) were used. For the amplification of coumarate ligase genes, the primers were PCR-D2a (5′ TACACSWSSGGSACSACSGG 3′) and PCR-D2b (5′ GTCSCCSGTGTGSTSCCASCCGTC 3′). (R, AG; W, AT; S, CG.) Primers SFA (5′ GTTCCTCACCTGATCCAGGAGATCC 3′) and SFB (5′ CCGAACTCGCCAGGA CGTACG 3′) were used to amplify probe F. Primers Int1 (5′ AAGCATTGGTAACTGTCAGTCCAAG 3′) and Int2 (5′ AGGAAAGAACATGTGAGCAAAAGGC 3′) were used to verify the integration of pSP1-C2 and pSP1-PKSII. Screening of the cosmid library was performed on robotically produced high-density colony arrays (Hybond N+; Amersham Pharmacia). Prehybridization and hybridization were performed following standard procedures (Church buffer, 65°C). DNA probes were labeled with digoxigenin. The detection was performed by inoculation with anti-digoxigenin antibodies conjugated with alkaline phosphatase and by incubation with attophos, following the protocols given by the manufacturer (Boehringer, Mannheim, Germany).
DNA sequencing and computer-assisted sequence analysis.
Nucleotide sequences were determined on an ABI sequencer at the 4-Base Lab GmbH (Reutlingen, Germany) and at Invitek GmbH (Berlin, Germany) by using either standard primers (M13 universal and reverse, T3, and T7) or customized, internal primers. Computer-assisted analysis was done with the DNASIS software (version 2.1.; Hitachi Software Engineering) and the frameplot software available at the website http://www.nih.go.jp/jun/cgi-bin/frameplot.pl (18). Database comparison was performed with the BLAST search tools on the server of the National Center for Biotechnology Information, Bethesda, Md. (1).
Generation of chromosomal mutants of S. antibioticus Tü6040.
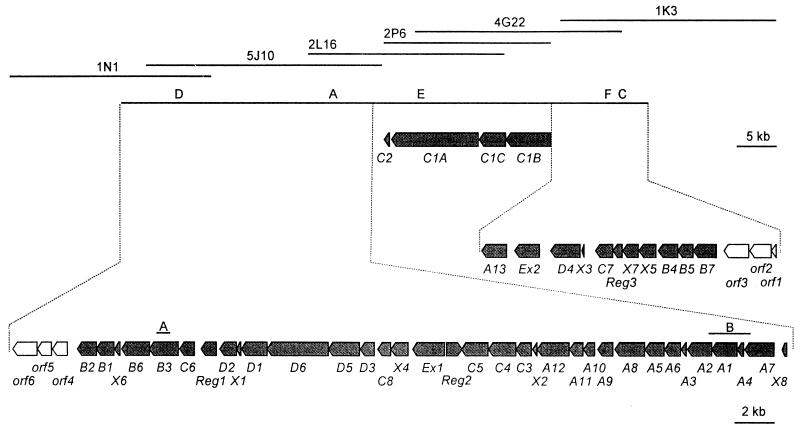
For generation of chromosomal mutants of S. antibioticus Tü6040 by homologous recombination, two gene disruption plasmids, pSP1-C2 and pSP1-PKSII, were constructed. pSP1-C2 was generated by ligating a 0.8-kb PCR fragment (Fig. 2, fragment A) containing internal DNA of simB3 into the singular XbaI site of pSP1. To generate pSP1-PKSII, a 2.1-kb BamHI fragment from cosmid 5J10 (Fig. 2, fragment B) containing simA1, simA4, and parts of simA2 and simA7 was first cloned into the BamHI site of pBSK− and then transferred to pSP1 using the EcoRI/XbaI sites. DNA of pSP1-C2 and pSP1-PKSII was isolated from the methylase-negative E. coli strain ET12567 and used to transform protoplasts of S. antibioticus Tü6040. S. antibioticus Tü6040 protoplast preparation, transformation, and protoplast regeneration were performed as described previously (25) using polyethylene glycol 2000 (Merck, Darmstadt, Germany) instead of polyethylene glycol 1000 (Merck). After 16 h of incubation at 30°C, cells with integrated plasmids were selected by covering the transformation plates with 2.5 ml of soft agar containing 500 μg of erythromycin. DNA denaturation by alkaline treatment (32) led to a significant increase in transformation efficiency. The integration of pSP1-C2 and pSP1-PKSII into the chromosome was shown by PCR analysis.
FIG. 2.
Organization of the simocyclinone biosynthetic gene cluster from S. antibioticus Tü6040. Cosmid clones isolated are shown above the map. The solid line indicates the region sequenced during this study, and letters above the line give the position of probes used for hybridization. ORFs are shown as arrows indicating the size and direction of transcription. Fragments A and B were used for insertional inactivation.
Analysis of simocyclinone D8 and its intermediates.
Conditions for production, extraction of simocyclinone, and analysis by high-pressure liquid chromatography with UV and visible spectrum (HPLC-UV/Vis) were as described elsewhere (39, 40).
Nucleotide sequence accession number.
The sequence of the simocyclinone biosynthesis gene cluster reported here has been deposited in the GenBank database under the accession number AF32483.
RESULTS AND DISCUSSION
Cloning of the simocyclinone cluster.
Due to the chemical structure of simocyclinone, it was likely that a polyketide synthase (PKS) type II, an NDP-glucose-4,6-dehydratase, and a glycosyltransferase are involved in the biosynthesis of simocyclinone. Therefore, approximately 3,000 colonies containing recombinant cosmids were probed by colony hybridization using as probes (i) a 3.9-kb BglII DNA fragment of the landomycin cluster containing the PKSII genes lanC, lanD, and parts of lanB and lanP (54) (putatively involved in landomycinone formation) (probe A); (ii) an internal fragment of a NDP-glucose-4,6-dehydratase gene obtained by PCR amplification using chromosomal DNA of S. antibioticus Tü6040 (8) (probe B); and (iii) urdGT2 (a glycosyltransferase gene of the urdamycin cluster) (11) (probe C). Fifteen cosmids were hybridized to probe A (inter alia cosmid 5J10), 6 hybridized to probe B, and 8 hybridized to probe C. Two cosmids (cosmid 3N5 and 3M22) hybridized to probes B and C, but random sequencing of fragments subcloned from these cosmids did not reveal any significant homology to secondary metabolite genes (data not shown). Restriction analysis of cosmids hybridizing to probe A revealed that most of them contained overlapping DNA.
It was considered likely that an NDP-4-keto-6-deoxyhexose 2,3-dehydratase and a coumarate ligase are involved in the biosynthesis of simocyclinone D8. The comparison of the sequence of known NDP-4-keto-6-deoxyhexose 2,3-dehydratases from Streptomyces fradiae Tü2717 (UrdS) (accession number AAF67505), Streptomyces cyanogenus S136 (LanS) (accession number AAD13549), Streptomyces violaceoruber Tü22 (Gra-ORF27) (accession number CAA09648), and Streptomyces peucetius (DnmT) (accession number AAD94713) revealed two regions of high similarity (QATRSNYT and EEGGRFL/Y). Based on the sequences of these regions, primers were prepared that took into account the codon usage of actinomycetes (57) (primers PCR-D1a and PCR-D1b). For the amplification of a putative coumarate ligase gene, we compared the sequence of a novobiocic acid synthetase from Streptomyces spheroides NCIMB11891 (NovL) (accession number AAF67505) and other adenylate forming enzymes (e.g., a coumarate-coenzyme A [CoA] ligase from Lolium perenne (accession number AAF37734), a coumarate-CoA ligase from Solanum tuberosum (accession number AAA33842), an acyl-CoA ligase from Mycobacterium tuberculosis (accession number CAB08962), an acyl-CoA ligase from Streptomyces coelicolor A3(2) (accession number CAB55668), the adenylation domain RapA from Streptomyces hygroscopicus (accession number AAC38061), and the adenylation domain RifA from Amycolatopsis mediterranei S699 (accession number AAC01710). Two consensus sequences (YTSGTTG and DGWVLHTGD) were detected which were then used for the design of oligonucleotide primers (primers PCR-D2a and PCR-D2b). With cosmid 5J10 as a template, PCR fragments were obtained using PCR-D1a and PCR-D1b and PCR-D2a and PCR-D2b. Sequencing analysis of these fragments indicated that an NDP-4-keto-6-deoxy-d-glucose 2,3-dehydratase gene and a gene similar to coumarate ligase genes are located on cosmid 5J10.
The fragment obtained with primers PCR-D1a and PCR-D1b was used as the probe (probe D) in hybridization experiments to screen for further cosmids putatively involved in simocyclinone biosynthesis. Restriction analysis revealed that cosmid 5J10 (see above), cosmid 1N1 (hybridizing to probe D), and cosmid 2L16 (hybridizing to probe A) contained overlapping DNA encompassing approximately 75 kb of S. antibioticus Tü6040 DNA. A 2-kb PstI fragment (2L16P15) subcloned from cosmid 2L16 was used as a probe (probe E) to screen for further overlapping cosmids, resulting in the isolation of cosmids 2P6 and 4G22. Finally, based on sequence data of DNA from cosmid 4G22, primers were designed to amplify a fragment using cosmid 4G22 as template (probe F). This fragment was used to identify cosmid 1K3, which also hybridized to probe C. Together, cosmids 1N1, 5J10, 2L16, 2P6, 4G22, and 1K3 contain a region of approximately 120 kb from the S. antibioticus Tü6040 chromosome that includes the entire simocyclinone biosynthetic gene cluster (Fig. 2). In total, 80.7 kb of DNA located on cosmids 1N1, 5J10, 2L16, 2P6, 4G2, and 1K3 were sequenced. Sequenced genes and the actual or putative function of their gene products are listed in Table 1. Figure 2 shows the genetic organization of the simocyclinone biosynthetic gene cluster (sim gene cluster). Although the boundaries of the sim gene cluster have not been determined experimentally, it is reasonable to propose that the cluster is flanked by simB7 and simB2.
TABLE 1.
Deduced function of ORFs shown in Fig. 2
| Polypeptide or ORF | Sequence similarity | No. of identical amino acids (%) | Accession no. | Refer- ence |
|---|---|---|---|---|
| ORF1 | ACP from T. gondii | 34 | AAC63953 | 51 |
| ORF2 | 3-oxo-ACP synthase II from Mesorhizobium loti | 33 | AP003012 | 23 |
| ORF3 | 3-oxo-ACP synthase II from Caulobacter crescentus | 42 | AE000752 | 31 |
| SimB7 | d-Olivosyltransferase (UrdGT2) from S. fradiae Tü2717 | 55 | AAF00209.1 | 11 |
| SimB5 | Putative 4-KR (LanR) from S. cyanogenus S136 | 58 | AAD13548 | 54 |
| SimB4 | Putative 3-KR (LanT) from S. cyanogenus S136 | 60 | AAD13550 | 54 |
| SimX5 | 3-oxo-ACP synthase III (FabH) from A. aeolicus | 30 | AAC07144 | 10 |
| SimX7 | —a | — | — | |
| SimReg3 | Hemolysin activator (SlyA) from Salmonella enterica serovar Typhimurium | 29 | P40676 | 27 |
| SimC7 | Putative hydroxylase/dehydratase (SnoAW) from S. nogalater | 50 | AAF01810 | 47 |
| SimX3 | — | — | — | |
| SimD4 | Halogenase (PCZA361.26) from Amycolatopsis orientalis | 38 | CAA11780 | 48 |
| SimEx2 | Putative Na+/H+ antiporter (CZA382.28) from Amycolatopsis orientalis | 53 | CAB45049 | 48 |
| SimA13 | Cytochrome P450 oxygenase (EryF) from Saccharopolyspora erythraea | 28 | AAA26496.1 | 53 |
| SimC1B, SimC1C, SimC1A | SimC1B, SimC1C, and SimC1A resemble several PKS type enzymes.b | |||
| SimC2 | Putative TE (PimI) from Streptomyces natalensis | 50 | CAC20922 | 2 |
| SimX8 | — | — | — | |
| SimA7 | Putative oxygenase (UrdE) from S. fradiae Tü2717 | 70 | CAA60567 | 8 |
| SimA4 | Putative cyclase (UrdF) from S. fradiae Tü2717 | 79 | CAA60568 | 8 |
| SimA1 | Putative KS (UrdA) from S. fradiae Tü2717 | 79 | CAA60569 | 8 |
| SimA2 | Putative KS (CLF) (Jad-CLF) from Streptomyces venezuelae ISP5230 | 72 | AAB36563 | 13 |
| SimA3 | Putative ACP (Jad-ACP) from S. venezuelae ISP5230 | 70 | AAB36564 | 13 |
| SimA6 | Putative KR (Jad-KR) from S. venezuelae ISP5230 | 80 | AAB36565 | 13 |
| SimA5 | Putative cyclase (UrdL) from S. fradiae Tü2717 | 67 | AAF00205 | 8 |
| SimA8 | Putative oxygenase (LanM) from S. cyanogenus S136 | 56 | AAD13541 | 54 |
| SimA9 | Putative KR (LanV) from S. cyanogenus S136 | 62 | AAD13552 | 54 |
| SimA10 | Putative KR (UrdO) from S. fradiae Tü2717 | 61 | AAF00220 | 8 |
| SimA11 | Putative phosphopantetheinyl transferase (JadM) from S. venezuelae ISP5230 | 47 | AAF34678 | 13 |
| SimA12 | Putative decarboxylase (LanP) from S. cyanogenus S136 | 78 | AAD13544 | 54 |
| SimX2 | Protein of unknown function (Sc1c2.1) from S. coelicolor A3(2) | 56 | CAA19984 | 35 |
| SimC3 | Putative TE (Ty1ORF5) from S. fradiae | 38 | AAA21345 | 28 |
| SimC4 | Putative dioxygenase (ScF12.12c) from S. coelicolor A3(2) | 49 | CAB56238 | 35 |
| SimC5 | Putative aldehyde dehydrogenase (ALDH3) from Homo sapiens | 47 | AAH04370 | 45 |
| SimReg2 | Putative regulatory protein (repressor) (Sc2H2.22c) from S. coelicolor A3(2) | 41 | CAC16726 | 35 |
| SimEx1 | Putative proton dependent transporter (LanJ) from S. cyanogenus S136 | 45 | AAD13557 | 54 |
| SimX4 | Protein of unknown function (Sc5a7.22) from S. coelicolor A3(2) | 61 | CAA19951 | 35 |
| SimC8 | Putative phosphopantetheinyl transferase (NysF) from S. noursei | 50 | AAF71762 | 38 |
| SimD3 | Putative oxidoreductase (NovK) from S. spheroides NCIMB11891 | 42 | AAF67504 | 44 |
| SimD5 | Novobiocic acid synthetase (NovL) from S. spheroides NCIMB11891 | 41 | AAF67505 | 44 |
| SimD6 | l-Tyroxyl-AMP-forming enzyme (NovH) S. spheroides NCIMB11891 | 54 | AAF67501 | 44 |
| SimD1 | Cytochrome P450-type monooxygenase (NovI) S. spheroides NCIMB11891 | 61 | AAF67502 | 44 |
| SimX1 | Protein of unknown function (MbtH) from M. tuberculosis | 61 | CAB08480 | 7 |
| SimD2 | Putative 3-ketoacyl-(ACP)-reductase (NovJ) from S. spheroides NCIMB11891 | 59 | AAF67503 | 44 |
| SimReg1 | Transcriptional activator of the response regulator type (JadR1) from S. venezuelae ISP5230 | 44 | AAB36584 | 60 |
| SimC6 | Putative reductase (DR1943) from Deinococcus radiodurans R1 | 48 | AAF11496 | 55 |
| SimB3 | Putative dNDP-4-keto-6-deoxy-glucose 2,3-dehydratase (SpnO) from Saccharopolyspora spinosa | 54 | AAG23276 | 50 |
| SimB6 | Benzyl alcohol AT (Beat1) from Clarkia species | 28 | AAF04782 | 29 |
| SimX6 | — | — | — | — |
| SimB1 | Putative glucose-1-phosphate dNDP-transferase (DesIII) from S. venezuelae ISP5230 | 54 | AAC68682 | 59 |
| SimB2 | Putative dNDP-glucose-4,6-dehydratase (MtmE) from Streptomyces argillaceus | 56 | CAA71847 | 26 |
| ORF4 | Protein of unknown function (Sch17.03c) from S. coelicolor A3(2) | 93 | CAB44549 | 35 |
| ORF5 | Protein of unknown function (Sch17.02c) from S. coelicolor A3(2) | 81 | CAB44549 | 35 |
| ORF6 | Putative serine protease (Sch17.01c) from S. coelicolor A3(2) | 83 | CAB44547 | 35 |
—, no similar protein was detected in the database.
For more detailed information on SimC1B, SimC1C, and SimC1A, see the text.
Genes putatively involved in the formation of the angucycline moiety.
The deduced amino acid sequences encoded by simA1, simA2, simA3, simA4, simA5, and simA6 strongly resemble gene products of type II iterative PKSs. The closest resemblance was found for gene products of the jadomycin (13), urdamycin (8), and landomycin (54) biosynthetic gene clusters. simA1, simA2, and simA3 encode the minimal PKS, and simA4 and simA5 (encoding cyclases) and simA6 (encoding a ketoreductase [KR]) are involved in modification of the nascent polyketide chain. The products of two genes (simA7 and simA8) were very similar to oxygenases also found in the urdamycin and landomycin clusters, indicating that they are involved in oxygenation of the angucycline polyketide moiety. simA13 encodes a third putative P450 hydroxylase. SimA13 does not resemble proteins known to be involved in the biosynthesis of other angucycline antibiotics, but it is similar to EryF, a 6-deoxyerythronolide β-hydroxylase from Saccharopolyspora erythraea (53). We assume that SimA13 is involved in oxygenation reactions on the angucycline moiety rather than in oxygenation of other parts of the molecule. Further genes involved in modifying steps are simA9 and simA10 (putatively involved in reduction steps) and simA12, encoding a decarboxylating enzyme. A putative phosphopantetheinyl transferase is encoded by simA11. As SimA11 resembles the gene product of jadM found in the jadomycin biosynthetic gene cluster, we can speculate that both are involved in modifying the acyl carrier protein (ACP) of the minimal PKS needed for biosynthesis of the angucycline polyketide moiety in either compound. Genes simA1 through simA12, which are involved in the formation of the angucycline moiety, seem to be organized in one single transcription unit, as indicated by short intergenic regions. Upstream to simA7, a putative regulatory region is located, as this region (bp 41470 to 41800) has a low G-C content of 49.8% compared to an average of 70.1% for the whole sequenced region.
Genes putatively involved in the formation of dTDP-d-olivose.
Computer-based sequence analysis revealed that simB1, simB2, simB3, simB4, and simB5 are involved in the biosynthesis of dTDP-d-olivose. In the case of simB4, simB5, and simB7, again, genes involved in the biosynthesis of urdamycin A (15) and landomycin A (54) are most similar. In contrast, gene products of simB1 (59), simB2 (26), and simB3 (50) showed high sequence similarities to deoxysugar biosynthetic genes from other clusters. Based on the sequencing data, the biosynthetic pathways from glucose-1-phosphate to NDP-d-olivose should comprise five steps. This would include an NDP-d-glucose synthase (SimB1), a 4,6-dehydratase (SimB2), a 2,3-dehydratase (SimB3), and a 3-ketoreductase (SimB4) to yield NDP-4-keto-2,6-dideoxy-d-glucose. The remaining step would be catalyzed by a 4-ketoreductase (SimB5) to prepare NDP-d-olivose. The deduced amino acid sequence of simB7 is very similar to that of UrdGT2 (11) and LanGT2 (54), both identified as glycosyltransferases, and SimB6 resembles acetyltransferases from different organisms (22, 29), indicating that it catalyzes the acetylation of the d-olivose residue at position C-4. Interestingly, simB1, simB2, simB3, and simB6 are located on one end, and simB4, simB5, and simB7 are on the other end of the cluster. The average G-C content of simB1, simB2, simB3, and simB6 is 67%, whereas the average G-C content of simB4, simB5, and simB7 is 72%. This difference might be explained by different evolutionary origins of these two parts of the d-olivose pathway.
Genes putatively involved in the formation of the tetraene side chain.
The synthesis of type I polyketides is a processive process in which each individual module in PKS is responsible for adding a specific extender unit to the growing polyketide chain. In the last 10 years a high number of type I PKSs have been cloned from different strains. As three ORFs of the sim biosynthetic gene cluster encode large, multifunctional type I PKSs, they seem to be involved in the formation of the tetraene moiety. simC1A encodes an enzyme with three modules (one loading module and two extension modules), while simC1B and simC1C encode PKSs, both with just one extension module (Fig. 1). The first module (module L) of SimC1A contains a β-ketoacyl synthase (KS) domain, an acyltransferase (AT) domain, and an ACP domain, indicating that this module performs the loading of the PKS. Module I consists of a KS domain, an AT domain, a dehydratase (DH), a KR domain, and an ACP domain and might catalyze the first extension reaction. Module II of SimC1A contains KS, AT, and ACP domains and an additional KR domain. SimC1B consists of one large module (module III) with KS, AT, DH, enoyl reductase (ER), KR, and ACP domains. Finally, SimC1C includes a thioesterase (TE) domain at the end of the module (module IV) which includes a KS, an AT, and an ACP domain.
In all KS domains of the extension molecules, the signature active site sequence (DTACSS) (2) with an invariant cysteine residue can be found, and these domains also contain two histidine residues located 135 and 175 amino acids C-terminal of the active-site cysteine. The KS domain of the putative loading module of SimC1A has a glutamine in place of the cysteine residue, and the second histidine residue is located 173 amino acids C-terminal of the glutamine. A similar type of KS is found in the loading domain of other biosynthetic gene clusters, and it has been shown that this KS domain is able to perform decarboxylation of a malonyl starter unit to prime polyketide synthesis (2, 4). All modules contain one ACP domain, which includes the 4′-phosphopantetheine binding site L(M)GxxS (49). Three of the five AT domains (AT of the loading domain, AT of module II, and AT of module III) contain amino acid sequence motifs that can be found in ATs that incorporate malonyl-CoA extender units. The other two ATs (AT of module I and AT of module IV) contain motifs that are considered to control the incorporation of methylmalonyl-CoA or other extender units in other organisms (14). All three KR domains contain the putative NAD(P)H binding site motif GxGxxGxxxA located near the N terminus and a characteristic Lx(S,G)Rx(G,T,A) motif with an invariant arginine (20, 41). The DH domains in SimC1A and SimC1B were identified by their conserved active motifs LxxHxxxG/DxxxxP (2), and the ER domain in SimC1B contains a GGVGxAAxQxA motif, which can be found in the ER of other PKSs (21). The sequence of the TE domain of SimC1C includes the invariant GxSxG motif and the PGxH motif, both of which seem to be essential for TE activity (56). The domains found in SimC1A, SimC1B, and SimC1C do not all correspond to the pattern expected for the proposed structure of the PKS product. KR and DH activity in modules I, II, III, and IV are predicted to be required for the biosynthesis of the tetraene side chain, but there is obviously no KR domain in module IV and no DH domain in module II and IV. The ER domain of module IV is not needed for the formation of the polyketide side chain, and the specificity of the AT domains in module I and IV for methylmalonyl-CoA substrates is also not consistent with biosynthetic expectations. However, similar observations have been made during the analysis of the biosynthetic gene clusters for other antibiotics (3, 42, 50, 58).
There are several genes in the cluster that might encode enzymes involved in tailoring reactions on the assembled polyketide chain. SimC6 is similar to several putative KRs (55), and SimC7 is most similar to SnoAW, a putative hydroxylase or DH from Streptomyces nogalater (47). SimC6 and SimC7 might catalyze the KR and DH reactions needed to complete the biosynthesis of the tetraene moiety. Similar enzymes acting on the assembled polyketide have been described for the biosynthesis of avermectin (17). SimC4 is similar to a putative dioxygenase from S. coelicolor A3(2) (35) and a neoxanthine-cleaving enzyme from Arabidopsis thaliana (30), and the deduced amino acid sequence of simC5 is similar to aldehyde dehydrogenases from different sources (45). Both SimC4 and SimC5 might be involved in the formation of the second carboxyl group of the tetraene moiety, which seems to be generated after the formation of the side chain and the attachment of the side chain to d-olivose (Fig. 1). The deduced amino acid sequences of simC2 (2) and simC3 (28) are similar to putative TEs, providing yet further examples of TE genes located in an antibiotic biosynthetic gene cluster. The deduced amino acid sequence of simC8 is similar to that of NysF, a putative phosphopantetheinyl transferase from Streptomyces noursei (5), and it is also similar to Svp, a phosphopantetheinyl transferase from Streptomyces verticillus (38). SimC8 might posttranslationally modify the ACP domains of SimC1A, SimC1B, and SimC1C. As Svp was characterized as a flexible phosphopantetheinyl transferase which is able to modify different types of ACPs and peptidyl carrier proteins (PCPs), we propose that SimC8 might also be involved in modifying the PCP domain of SimD6 (38). Further experiments have to be performed to study the specificities of SimA11 and SimC8 and their exact functions during simocyclinone biosynthesis.
Genes putatively involved in the formation of the amino-coumarin moiety.
There are six genes in the cluster that are believed to be involved in the biosynthesis and attachment of the amino-coumarin moiety of simocyclinone. The deduced amino acid sequences of these genes mostly resemble proteins known to be involved in the biosynthesis of coumermycin in Streptomyces rishiriensis DSM40489 (52) and novobiocin in Streptomyces spheroides NCIMB11891 (44). SimD6 resembles CumC and NovH, which were shown to be involved in activating tyrosine (6). However, SimD6 is a larger protein than NovH and CumC, with an extension of 400 amino acids at the N terminus. This 400-amino-acid region also shows similarity to peptide synthases from different sources. SimD1, which is similar to NovI and CumD, is most likely responsible for the conversion of tyrosine to a β-OH-tyrosine intermediate which is covalently tethered to SimD6. SimD2 resembles NovJ, CumE, and other putative 3-ketoacyl-(ACP) reductases and, therefore, it is probably involved in oxidation of the β-OH-tyrosine to a β-keto intermediate. SimD3, which is closely related to NovK and CumF, might catalyze the formation of an oxidative cyclization reaction to yield the amino-coumarin moiety. Walsh and Chen (6) postulated that NovJ and NovK together are involved in the oxidation of the β-OH-tyrosine and that NovC, a putative flavine-dependent monooxygenase, would hydroxylate the ortho position of the phenyl ring of β-keto-tyrosine to facilitate the cyclization. Anyway, we could not detect a novC-like gene in the simocyclinone biosynthetic gene cluster. The attachment of the amino-coumarin to the tetraene moiety is catalyzed by SimD5, which is similar to CumG and NovL, which have been shown to catalyze the formation of an amide bond between dimethylallyl-4-hydroxybenzoic acid and 3-amino-4,7-dihydroxy-8-methyl coumarin (43). Finally, the gene product of simD4 is similar to halogenases from other organisms (48) and most likely performs the formation of the chlorinated antibiotic.
Genes putatively involved in regulation and resistance.
There are two putative transporter genes located in the cluster. The deduced amino acid sequence of simEx2 shows homology to putative integral membrane ion transporters (48), and SimEx1 resembles transporters known as proton-dependent transporters of different drugs (54). Three putative regulatory genes were found in the cluster. The deduced amino acid sequence of simReg1 is similar to that of transcriptional activators of the response regulator type (60). simReg2 encodes a repressor protein belonging to the TetR family of bacterial repressors (35), and SimReg3 is similar to different activator proteins (27). simReg2 is the only gene in the cluster that is transcribed in the opposite direction. The fact that simReg2 is transcribed divergently from simEx1 provides the possibility that SimEx1 could be regulated by SimReg2 at the level of transcription (33).
Genes of unknown function.
SimX5 resembles 3-oxo-ACP synthase type III enzymes from different sources. It is most similar to a protein from Aquifex aeolicus (10). SimX1 shows significant homology to MbTH, a protein with unknown function from M. tuberculosis (7). Homologues of this gene are present in various biosynthetic gene clusters from actinomycetes, suggesting a conserved function of these genes in secondary metabolite formation or regulation. Directly downstream and translationally coupled to simA12, the gene simX2 was identified. The gene product of simX2 showed significant homology to Sc1c2.1, a protein of unknown function from S. coelicolor A3(2) (35). simX4 is located directly upstream from simC8. Similar genes (sc5A7.22 and sc5A7.23) also located in close proximity were found in the S. coelicolor A3(2) genome, in M. tuberculosis (rv2795v and rv2796c) (7), and in S. verticillus (orf1 and svp) (38). simX3, simX6, simX7, and simX8 all encode short ORFs which do not resemble any protein in the databases.
Genes putatively located outside the cluster.
Three putative ORFs (orf1 to orf3) were found to reside close to the margin on one side of the cluster, and three other putative ORFs (orf4 to orf6) are near the opposite margin. They probably do not belong to the simocyclinone biosynthetic gene cluster. ORF1 is similar to an ACP from Toxoplasma gondii (51), and ORF2 and ORF3 resemble 3-oxo-ACP synthase type II enzymes from different sources (23, 31). The deduced amino acid sequences of ORF4 and ORF5 resemble proteins of unknown function found in the genome of S. coelicolor A3(2), and ORF6 might be an alkaline serine protease very similar to a protease from S. coelicolor A3(2) (35).
Insertional inactivation experiments.
The similarities between the gene products of the cluster identified in this study and the gene products of other previously identified gene clusters made it very likely that we had indeed cloned the simocyclinone biosynthetic gene cluster. After establishing a gene transfer system, functional proof for this hypothesis was provided by insertional gene inactivation experiments. simB3, very likely involved in the biosynthesis of dTDP-d-olivose, was chosen for this experiment. Plasmid pSP1-C2 was constructed, allowing the inactivation of simB3 by integration. The plasmid was introduced into S. antibioticus Tü6040 by protoplast transformation, and recombinant strains were detected after selection for erythromycin resistance. PCR analysis confirmed the integration of pSP1-C2 into the chromosome. Mutant Sim-C2 was chosen for further experiments.
Plasmid pSP1-PKSII was constructed, allowing the interruption of the transcriptional unit within the minimal PKS genes. We expected that an insertion of this plasmid would strongly depress simocyclinone production. Plasmid pSP1-PKSII was introduced into S. antibioticus Tü6040 and erythromycin-resistant clones were obtained. Again, PCR analysis confirmed the integration of pSP1-PKSII into the chromosome. Mutant Sim-PKSII was chosen for further experiments.
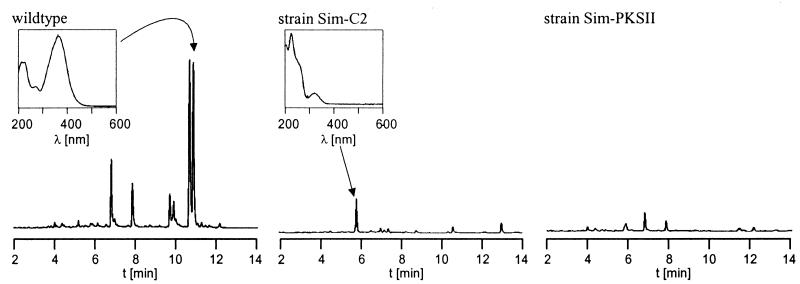
Culture extracts from the wild-type strain and from mutant strains Sim-C2 and Sim-PKSII were analyzed by HPLC-UV/Vis. Simocyclinone D8 production was clearly detected in the wild-type strain but was not detectable in mutant Sim-C2 and mutant Sim-PKSII. HPLC-UV/Vis analysis showed the accumulation of simocyclinone A1 in mutant Sim-C2, indicating that the biosynthesis of dTDP-d-olivose was completely abolished and, as expected, the biosynthesis of the angucycline moiety was retained (Fig. 3). All of these data clearly show that we indeed cloned the simocyclinone biosynthetic gene cluster.
FIG. 3.
HPLC traces of simocyclinones produced by S. antibioticus Tü6040 (wild type), mutant Sim-C2, and mutant Sim-PKSII. HPLC conditions were as indicated in the text. The retention times were as follows: simocyclinone D8, 10.8 min; simocyclinone A1, 5.8 min. UV/Vis spectra of simocyclinone D8 and simocyclinone A1 are shown above the HPLC chromatograms.
The sim biosynthetic gene cluster represents the most versatile gene cluster so far investigated. It contains genes responsible for the formation and attachment of four different moieties originating from at least three different pools of primary metabolites. The hypothetical biosynthetic scheme for simocyclinone D8 is given in Fig. 1. We hope that we will learn to use these genes to increase the number of unnatural natural products made by combinatorial biosynthesis in the future.
Acknowledgments
This work was supported by a grant from the European Union (QLRT-1999-00095) and a grant from the Bundesministerium für Bildung und Forschung, both to A.B.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, J. F., R. Fouces, M. V. Mendes, N. Olivera, and J. F. Martin. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7:895-905. [DOI] [PubMed] [Google Scholar]
- 3.August, P. R., L. Tang, Y. J. Yoon, S. Ning, R. Müller, T. W. Yu, M. Taylor, D. Hoffmann, C. G. Kim, X. Zhang, C. R. Hutchinson, and H. G. Floss. 1998. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 5:69-79. [DOI] [PubMed] [Google Scholar]
- 4.Bisang, C., P. F. Long, J. Cortes, J. Westcott, J. Crosby, A. L. Matharu, R. J. Cox, T. J. Simpson, J. Staunton, and P. F. Leadlay. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502-505. [DOI] [PubMed] [Google Scholar]
- 5.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. Stroem, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., and C. T. Walsh. 2000. Coumarin formation in novobiocin biosynthesis: beta-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem. Biol. 8:301-312. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, S. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, S. Skelton, S. Squares, R. Sqares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Decker, H., and S. Haag. 1995. Cloning and characterization of a polyketide synthase gene from Streptomyces fradiae Tü2717, which carries the genes for biosynthesis of the angucycline antibiotic urdamycin A and a gene probably involved in its oxygenation. J. Bacteriol. 177:6126-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker, H., S. Gaisser, S. Pelzer, P. Schneider, L. Westrich, W. Wohlleben, and A. Bechthold. 1996. A general approach for cloning and characterizing dNDP-glucose dehydratase genes from actinomycetes. FEMS Microbiol. Lett. 141:195-201. [DOI] [PubMed] [Google Scholar]
- 10.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 11.Faust, B., D. Hoffmeister, G. Weitnauer, L. Westrich, S. Haag, P. Schneider, H. Decker, E. Künzel, J. Rohr, and A. Bechthold. 2000. Two new tailoring enzymes, a glycosyltransferase and an oxygenase, involved in biosynthesis of the angucycline antibiotic urdamycin A in Streptomyces fradiae Tü2717. Microbiology 146:147-154. [DOI] [PubMed] [Google Scholar]
- 12.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 13.Han, L., K. Yang, E. Ramalingam, R. H. Mosher, and L. C. Vining. 1994. Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiology 140:3379-3389. [DOI] [PubMed] [Google Scholar]
- 14.Haydock, S. F., J. F. Aparacio, I. Molnar, T. Schwecke, L. E. Khaw, A. Konig, A. F. Marsden, I. S. Galloway, J. Staunton, and P. F. Leadlay. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl) malonyl-CoA: acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374:246-248. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmeister, D., K. Ichinose, S. Domann, B. Faust, A. Trefzer, G. Draeger, A. Kirsching, E. Kuenzel, J. Rohr, and A. Bechthold. 2000. The NDP-sugar cosubstrate concentration and the enzyme expression level influence the substrate specificity of glycosyltransferases: cloning and characterization of deoxysugar biosynthetic genes of the urdamycin biosynthetic gene cluster. Chem. Biol. 7:821-831. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson, C. R. 1998. Combinatorial biosynthesis for new drug discovery. Curr. Opin. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 17:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 19.Jacobsen, J. R., C. R. Hutchinson, D. E. Cane, and C. Khosla. 1997. Precursor-directed biosynthesis of erythromycin analogs by an engineered polyketide synthase. Science 277:367-369. [DOI] [PubMed] [Google Scholar]
- 20.Julien, B., S. Shah, R. Ziermann, R. Goldman, L. Katz, and C. Khosla. 2000. Isolation and characterization of the epothilone biosynthetic gene cluster from Sorangium cellulosum. Gene 249:153-160. [DOI] [PubMed] [Google Scholar]
- 21.Kakavas, S. J., L. Katz, and D. Stassi. 1997. Identification and characterization of the niddamycin polyketide synthase genes from Streptomyces caelestis. J. Bacteriol. 179:7515-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, T., T. Katoh, S. Sato, Y. Nakamura, E. Asamizu, H. Kotani, N. Miyajima, and S. Tabata. 1999. Structural analysis of Arabidopsis thaliana chromosome 5. IX. Sequence features of the regions of 1,011,550 bp covered by seventeen P1 and TAC clones. DNA Res. 6:183-195. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 31:331-338. [DOI] [PubMed] [Google Scholar]
- 24.Katz, L., and R. McDaniel. 1999. Novel macrolides through genetic engineering. Med. Res. Rev. 19:543-558. [DOI] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2001. Practical Streptomyces genetics. John Innes Centre, Norwich, United Kingdom.
- 26.Lombo, F., K. Siems, A. F. Brana, C. Mendez, K. Bindseil, and J. A. Salas. 1997. Cloning and insertional inactivation of Streptomyces argillaceus genes involved in the earliest steps of biosynthesis of the sugar moieties of the antitumor polyketide mithramycin. J. Bacteriol. 179:3354-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig, A., C. Tengel, S. Bauer, A. Bubert, R. Benz, H. J. Mollenkopf, and W. Goebel. 1995. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474-486. [DOI] [PubMed] [Google Scholar]
- 28.Merson-Davies, L. A., and E. Cundliffe. 1994. Analysis of five tylosin biosynthetic genes from the tylIBA region of the Streptomyces fradiae genome. Mol. Microbiol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 29.Nam, K. H., N. Dudareva, and E. Pichersky. 1999. Characterization of benzylalcohol acetyltransferases in scented and non-scented Clarkia species. Plant Cell Physiol. 40:916-923. [DOI] [PubMed] [Google Scholar]
- 30.Neill, S. J., E. C. Burnett, R. Desikan, and J. T. Hancock. 1998. Cloning of a wilt-responsive cDNA from an Arabidopsis thaliana suspension culture cDNA library that encodes a putative 9-cis-epoxy-carotenoid dioxygenase. J. Exp. Bot. 49:1893-1894. [Google Scholar]
- 31.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 27:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh, S. H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otten, S. L., J. Ferguson, and C. R. Hutchinson. 1995. Regulation of daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J. Bacteriol. 177:1216-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelzer, S., W. Reichert, M. Huppert, D. Heckmann, and W. Wohlleben. 1997. Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system. J. Biotechnol. 56:115-128. [DOI] [PubMed] [Google Scholar]
- 35.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, M. A., S. Kuhstoss, P. Solenberg, N. A. Schaus, and R. N. Rao. 1987. A new shuttle cosmid vector, pKC505, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene 61:231-241. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sanchez, C., L. Du, D. J. Edwards, M. D. Toney, and B. Shen. 2001. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC 15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 8:725-738. [DOI] [PubMed] [Google Scholar]
- 39.Schimana, J., H.-P. Fiedler, I. Groth, R. Süssmuth, W. Beil, M. Walker, and A. Zeeck. 2000. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü6040. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 53:779-787. [DOI] [PubMed] [Google Scholar]
- 40.Schimana, J., M. Walker, A. Zeeck, and H.-P. Fiedler. 2001. Simocyclinones: diversity of metabolites is dependent on fermentation conditions. J. Ind. Microbiol. Biotechnol. 27:144-148. [DOI] [PubMed] [Google Scholar]
- 41.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38-43. [DOI] [PubMed] [Google Scholar]
- 42.Silakowski, B., G. Nordsiek, B. Kunze, H. Blöcker, and R. Müller. 2001. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem. Biol. 8:59-69. [DOI] [PubMed] [Google Scholar]
- 43.Steffensky, M., S. M. Li, and L. Heide. 2000. Cloning, overexpression, and purification of novobiocic acid synthetase from Streptomyces spheroides NCIMB11891. J. Biol. Chem. 275:21754-21760. [DOI] [PubMed] [Google Scholar]
- 44.Steffensky, M., A. Mühlenweg, Z. X. Wang, S. M. Li, and L. Heide. 2000. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIMB11891. Antimicrob. Agents Chemother. 44:1214-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinmetz, C. G., P. Xie, H. Weiner, and T. D. Hurley. 1997. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 15:701-711. [DOI] [PubMed] [Google Scholar]
- 46.Theobald, U., J. Schimana, and H.-P. Fiedler. 2000. Microbial growth and production kinetics of Streptomyces antibioticus Tü6040. Antonie Leeuwenhoek 78:307-313. [DOI] [PubMed] [Google Scholar]
- 47.Torkkell, S., L. Ylihonko, J. Hakala, M. Skurnik, and P. Mantsala. 1997. Characterization of Streptomyces nogalater genes encoding enzymes involved in glycosylation steps in nogalamycin biosynthesis. Mol. Gen. Genet. 256:203-209. [DOI] [PubMed] [Google Scholar]
- 48.van Wageningen, A. M., P. N. Kirkpatrick, D. H. Williams, B. R. Harris, J. K. Kershaw, N. J. Lennard, M. Jones, S. J. Jones, and P. J. Solenberg. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5:155-162. [DOI] [PubMed] [Google Scholar]
- 49.Wakil, S. J. 1989. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 11:4523-4530. [DOI] [PubMed] [Google Scholar]
- 50.Waldron, C., P. Matsushima, P. R. Rosteck, M. C. Broughton, J. Turner, K. Madduri, K. P. Crawford, D. J. Merlo, and R. H. Baltz. 2001. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 8:487-499. [DOI] [PubMed] [Google Scholar]
- 51.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Z. X., S. M. Li, and L. Heide. 2000. Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM40489. Antimicrob. Agents Chemother. 44:3040-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber, J. M., J. O. Leung, S. J. Swanson, K. B. Idler, and J. B. McAlpine. 1991. An erythromycin derivative produced by targeted gene disruption in Saccharopolyspora erythraea. Science 252:114-117. [DOI] [PubMed] [Google Scholar]
- 54.Westrich, L., S. Domann, B. Faust, D. Bedford, D. A. Hopwood, and A. Bechthold. 1999. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol. Lett. 170:381-387. [DOI] [PubMed] [Google Scholar]
- 55.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witkowski, A., J. Naggert, B. Wessa, and S. Smith. 1991. A catalytic role for histidine 237 in rat mammary gland thioesterase II. J. Biol. Chem. 5:18514-18519. [PubMed] [Google Scholar]
- 57.Wright, F., and M. J. Bibb. 1992. Codon usage in the G + C-rich Streptomyces genome. Gene 1:55-65. [DOI] [PubMed] [Google Scholar]
- 58.Wu, K., L. Chung, W. P. Revill, L. Katz, and C. D. Reeves. 2000. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251:81-90. [DOI] [PubMed] [Google Scholar]
- 59.Xue, Y., L. Zhao, H. W. Liu, and D. H. Sherman. 1998. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 95:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, K., L. Han, and L. C. Vining. 1995. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J. Bacteriol. 177:6111-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]