Abstract
Tetracycline resistance is common among isolates of the animal commensal and opportunistic pathogen Arcanobacterium pyogenes. The tetracycline resistance determinant cloned from two bovine isolates of A. pyogenes was highly similar at the DNA level (92% identity) to the tet(W) gene, encoding a ribosomal protection tetracycline resistance protein, from the rumen bacterium Butyrivibrio fibrisolvens. The tet(W) gene was found in all 20 tetracycline-resistant isolates tested, indicating that it is a widely distributed determinant of tetracycline resistance in this organism. In 25% of tetracycline-resistant isolates, the tet(W) gene was associated with a mob gene, encoding a functional mobilization protein, and an origin of transfer, suggesting that the determinant may be transferable to other bacteria. In fact, low-frequency transfer of tet(W) was detected from mob+ A. pyogenes isolates to a tetracycline-sensitive A. pyogenes recipient. The mobile nature of this determinant and the presence of A. pyogenes in the gastrointestinal tract of cattle and pigs suggest that A. pyogenes may have inherited this determinant within the gastrointestinal tracts of these animals.
Resistance to tetracycline is the most common bacterial antibiotic resistance found in nature and similarly is the most prominent type of resistance among bacteria isolated from animals (10). Tetracycline binds to the 30S ribosomal subunit and inhibits protein synthesis (32), and resistance to this drug is most commonly mediated either by active efflux of tetracycline from the cell or by ribosomal protection from the action of tetracycline (10). In rare cases, tetracycline resistance is mediated through direct inactivation of the antibiotic (30) or by mutations in the 16S rRNA that prevent binding of tetracycline to the ribosome (26).
Arcanobacterium pyogenes is a common inhabitant of the respiratory and urogenital tracts of domestic animals, including cattle and swine (8). It has also been isolated from the rumen of feedlot cattle (22) and more recently from the gastric mucosa of pigs (B. H. Jost, K. W. Post, J. G. Songer, and S. J. Billington, unpublished data), suggesting that A. pyogenes may also be a common inhabitant of the gastrointestinal tract of these species. Under conditions of physical trauma or previous microbial infection, A. pyogenes can disseminate to cause a variety of suppurative infections, such as liver abscesses and mastitis in cattle (15, 18, 21) and suppurative pneumonia and polyarthritis in pigs (14, 35). Virulence, at least in an intraperitoneal mouse model, appears to be mediated through a pore-forming toxin, pyolysin (6, 16). The development of antibiotic resistance in normal flora secondary pathogens, such as A. pyogenes, is of particular significance, not only because of the ability of resistant A. pyogenes to avoid antibiotic therapy, but also because of the potential for transfer of resistance to other pathogens.
Resistance to the tetracycline antibiotics among A. pyogenes isolates is widespread. A French study indicated that 67% of 103, predominantly bovine, A. pyogenes isolates were resistant to tetracycline, doxycycline, and minocycline (13), while a study in Japan found that 85.7% of porcine and 57.1% of bovine isolates were resistant to oxytetracycline and doxycycline (40). These results were confirmed by a recent study of predominantly North American isolates of A. pyogenes, which indicated that 42% were resistant to tetracycline, chlortetracycline, and oxytetracycline, with more than 70% of porcine isolates being resistant (34).
No information is currently available on the mechanism of tetracycline resistance in A. pyogenes or the determinants involved in this resistance. In this work, we present evidence that tetracycline resistance in A. pyogenes isolates is predominantly conferred by a Tet W determinant previously found only in obligately anaerobic bacteria isolated from the rumen of cattle and the human gut (2, 5, 27, 28) and that this determinant, in at least some strains, is carried on a mobile genetic element capable of transfer between strains of A. pyogenes.
MATERIALS AND METHODS
Bacterial strains, bacteriophage, plasmids, and growth conditions.
Escherichia coli strains and plasmids are described in Table 1. E. coli strains were grown on either Luria-Bertani (LB) agar or in LB broth (Difco) at 37°C. Antibiotics (Sigma) were added as appropriate at the following concentrations: chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 10 μg/ml; tetracycline, 10 μg/ml. λGEM12 derivatives were propagated on E. coli LE392 as previously described (3). A. pyogenes isolates used in this study were obtained from veterinary diagnostic laboratories or personal culture collections in North America and include 20 tetracycline-resistant isolates (7 of bovine origin, 12 of porcine origin, and 1 from a macaw) and 10 tetracycline-susceptible isolates (6 of bovine origin and 4 of porcine origin). Specific A. pyogenes strains used in mating experiments are described in Table 1. A. pyogenes strains were grown on brain heart infusion (BHI) agar (Difco), supplemented with 5% bovine blood, at 38°C and 5% CO2 in a humidified incubator. Liquid cultures of A. pyogenes were grown in BHI broth supplemented with 5% fetal bovine serum (Omega Scientific, Inc.) at 37°C. Antibiotics were added as appropriate at the following concentrations: erythromycin, 15 μg/ml; kanamycin, 30 μg/ml; nalidixic acid, 10 μg/ml; tetracycline, 5 μg/ml.
TABLE 1.
Relevant characteristics of bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Sourcea or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 thi-1 phoA supE44 gyrA96 relA1 λ− | Gibco-BRL |
| LE392 | F− e14−mcrA hsdR514(rK− mK+) supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 | Promega |
| S17-1 | recA pro hsdR RP4-2-Tc::Mu-Kan::Tn7 Tmpr Sptr Strr | 29 |
| A. pyogenes | ||
| BBR1 | Bovine isolate, Tetr | 6 |
| JGS190 | Bovine isolate, Tetr | CSU |
| 3 | Bovine isolate, Tetr | 25 |
| 4 | Bovine isolate, Tetr | 25 |
| 98-1508 | Macaw isolate, Tetr | AZ VDL |
| 3274 | Bovine isolate, Tets | CSU |
| JGS478 | nanP::Kan derivative of strain 3274 | This study |
| JGS603 | Tetr Kanr BBR1 × JGS478 transconjugant | This study |
| JGS605 | Tetr Kanr JGS190 × JGS478 transconjugant | This study |
| JGS610 | nanH::Erm derivative of strain 3274 | This study |
| Plasmids | ||
| pBC KS | Chlr lacZ′, ColE1 origin | Stratagene |
| pCR-Script Cam SK(+) | pBC SK with incorporated SrfI site | Stratagene |
| pWSK129 | KanrlacZ′, p15A origin | 38 |
| pJRD215 | Kanrmob+, RSF1010 origin | 12 |
| pJGS259 | pBC KS BamHI Ω 4-kb partial Sau3AI fragment containing the tet(W) gene of strain 4 | This study |
| pJGS264 | pJGS259 Δ 1.3-kb NotI fragment | This study |
| pJGS279 | pBC KS BamHI Ω 12-kb BamHI fragment from λJGS9 containing the tet(W) gene of BBR1 | This study |
| pJGS324 | pCR-Script Cam SK SfiI Ω 1,710-bp PCR product containing mob and oriT | This study |
| pJGS416 | pBC KS SacI-KpnI Ω 1,524-bp PCR product containing mob | This study |
| pJGS417 | pWSK129 SacI-KpnI Ω 202-bp PCR product containing oriT | This study |
CSU, Colorado State University; AZ VDL, Arizona Veterinary Diagnostic Laboratory.
Determination of MICs of tetracycline antibiotics.
A broth microdilution technique was used to determine the MICs of tetracycline antibiotics for A. pyogenes isolates. The inoculum was prepared by resuspending several isolated colonies in sterile Mueller-Hinton Broth (Difco) containing 2% lysed, defibrinated horse blood (BBL Microbiology Systems) and adjusting the suspension to an optical density at 600 nm (OD600) of 0.001 (approximately 106 CFU/ml). The antimicrobial agents tetracycline, chlortetracycline, and oxytetracycline (Sigma) were prepared according to the guidelines of the National Committee for Clinical Laboratory Standards (23). The antibiotics were diluted in a doubling dilution pattern over the range 0.06 to 64 μg/ml in the wells of sterile, 96-well, round-bottom microtiter plates (Falcon) in 50-μl volumes. Fifty microliters of inoculum was dispensed into each well, and the plates were sealed and incubated at 38°C and 5% CO2 for 24 h. The MIC was read visually as the lowest concentration of the antibiotic to prevent growth (turbidity) compared with that of the control (no antibiotic added). All MICs were tested in duplicate on at least two independent cultures. MICs for A. pyogenes isolates were also determined after growth as described above on BHI-5% bovine blood agar, supplemented with 1 μg of tetracycline per ml, to determine the inducibility of the determinant. MICs for E. coli strains were performed essentially as described above, except that Mueller-Hinton broth was used without supplementation, and plates were incubated at 37°C for 18 to 24 h.
DNA techniques.
Procedures for E. coli transformation and plasmid extraction, bacteriophage DNA extraction, DNA restriction, ligation, agarose gel electrophoresis, and DNA dot blotting were performed essentially as described previously (3). Genomic DNA was prepared from A. pyogenes strains by the method of Pospiech and Neumann (24). Preparation of DNA probes, DNA hybridization, and probe detection were performed with the digoxigenin (DIG) DNA labeling and detection kit (Roche Molecular Biochemicals). For dot blot analysis against genomic DNA from A. pyogenes, a tet(W)-specific gene probe spanning bases 117 to 1362 of tet(W) was generated by PCR with primers (Sigma-Genosys), 5′-AAGCGGGAGCGGCGTAACAGAC-3′ and 5′-GACAACGAGAACGGACACTATG-3′, and a mob-specific gene probe spanning bases 450 to 1177 of mob was generated by PCR with primers 5′-CTACCACCTCCATGTGGTCTAT-3′ and 5′-GCGCAGACCTCGTAAATCCTGG-3′. PCR DNA amplification was performed with Taq DNA polymerase (Promega) for 35 cycles consisting of 1 min at 94°C (DNA denaturation), 1 min at 55°C (primer annealing), and 1 min/kb at 72°C (DNA synthesis).
Cloning of the A. pyogenes tetracycline resistance genes.
A library of partially Sau3AI-digested genomic DNA from A. pyogenes strain 4 was prepared in the plasmid vector pBC KS (Stratagene). The ligation mixture was introduced into E. coli strain DH5αMCR by electroporation, and tetracycline-resistant transformants were selected on LB agar containing 10 μg of tetracycline per ml. One tetracycline-resistant transformant contained the plasmid pJGS259 with a 4-kb insert.
A DIG-labeled probe was prepared from pJGS259 and used to screen a λGEM12 (Promega) library of partial Sau3AI fragments from our standard laboratory A. pyogenes strain, BBR1. A 12-kb BamHI fragment from one probe-positive λGEM12 recombinant, λJGS9, was cloned into the BamHI site of pBC KS to create pJGS279.
Nucleotide sequence determination.
Nucleotide sequencing of tetracycline resistance determinants was performed on a 377A DNA sequencer (Applied Biosystems, Inc.) at the Automated DNA Sequencing Facility at the University of Arizona. Sequence was determined from both strands, crossing all restriction sites. Sequence data were determined from primary clones and appropriate subclones by using vector- and insert-specific primers. To determine the sequence of the strong rho-independent terminator of tet(W), it was necessary to design primers to the sequence immediately upstream and downstream of the stem-loop. These primers were used in two-step (96°C, 60°C) cycle sequencing reactions performed in the presence of 5% dimethyl sulfoxide.
Computer analysis.
Nucleotide sequence data were compiled by using the Sequencher 3.1 program (GeneCodes, Ann Arbor, Mich.). Database searches were performed with the BlastN, BlastX, and BlastP algorithms (1). Sequence analysis was performed with the suite of programs available through the Genetics Computer Group, Inc. (University of Wisconsin). Similarity was determined from optimized sequence alignments by using the CLUSTAL W program (33).
Construction of plasmids for mobilization experiments.
The mob gene and oriT of pJGS279 were amplified by PCR with the primers 5′-CTGGGGGAGGCAACCGCACACC-3′, which binds 150 bp upstream of oriT, and 5′-GTCCACGATTTCCGCCGCACAC-3′, which binds 265 bp downstream of mob. The 1,710-bp PCR product was polished with the PCR-Script Cam cloning kit (Stratagene) and inserted into the SfiI site of pCR-Script Cam SK(+) to create pJGS324 (Table 1). The mob gene was amplified from pJGS324 with the M13 reverse primer (5′-AGCGGATAACAATTTCACACAGGA-3′) and the primer 5′-TCCCGCAGGAGGTACCTCCCCTGAAC-3′, containing a KpnI site. The 1,524-bp PCR product, containing only the mob gene, was digested with KpnI and SacI and inserted into similarly digested pBC KS to create pJGS416 (Table 1). oriT was amplified from pJGS324 by using the M13 universal primer (5′-ACGTTGTAAAACGACGGCCAGT-3′) and the primer 5′-TCGAGGACGGGAGCTCAGGGGAGGC-3′, containing a SacI site. The 202-bp PCR product, containing only oriT, was digested with KpnI and SacI and inserted into similarly digested pWSK129 (Table 1) to create pJGS417.
E. coli mobilization experiments.
Recombinant plasmids to be assessed for mobilization were introduced into E. coli S17-1 (Table 1) by transformation. S17-1 supplies conjugative functions in trans from a chromosomal copy of RP4. These strains were then used as donors in filter matings with the nalidixic acid-resistant recipient strain DH5αMCR. Donor and recipient strains were grown overnight under appropriate selection, diluted to an OD600 of 0.05 in the same medium, and grown to an OD600 of 1.0. The two cultures (0.5 ml of each) were mixed and filtered through a 0.45-μm-pore-size filter. The filter was incubated for 2 h on an LB agar plate at 37°C, and the cells were resuspended in LB broth. Serial dilutions were plated onto LB agar containing the appropriate antibiotics to select for transconjugants. Bacterial viable counts were obtained from identically treated filters containing either the donor or recipient strain alone. Mobilization frequencies were expressed as transconjugants per donor cell and were determined as an average of at least three independent experiments.
A. pyogenes filter matings.
Tetracycline-resistant A. pyogenes isolates were used in filter matings with the kanamycin-resistant A. pyogenes recipient strain JGS478. JGS478 is a derivative of the tetracycline-susceptible isolate 3274, which contains a kanamycin resistance cassette inserted into the neuraminidase gene, nanP (Table 1). Donor and recipient strains were grown overnight under appropriate selection, diluted to an OD600 of 0.1 in the same medium, and grown to an OD600 of 1.0. The two cultures (0.5 ml of each) were mixed and filtered through a 0.45-μm-pore-size filter. Following overnight incubation on a BHI-5% blood agar plate at 37°C under 5% CO2, the cells were resuspended from the filter in BHI broth. Serial dilutions were plated onto BHI-5% blood agar supplemented with 5 μg of tetracycline and 30 μg of kanamycin per ml to select for transconjugants. Bacterial viable counts were obtained from identically treated filters containing either the donor or recipient strain alone. Conjugation frequencies were expressed as transconjugants per donor cell and were determined as an average of at least three independent experiments.
Selected transconjugants from the conjugations described above were used as donors in separate conjugation experiments with JGS610 (Table 1). JGS610 is an erythromycin-resistant derivative of strain 3274, which contains an erythromycin resistance cassette inserted into the nanH gene, constructed as previously described (17).
Nucleotide sequence accession number.
The nucleotide sequences of the A. pyogenes BBR1 mob and tet(W) genes have been deposited in the GenBank nucleotide sequence database under accession no. AY049983.
RESULTS
Cloning of an A. pyogenes tetracycline resistance determinant.
The tetracycline resistance determinant from A. pyogenes strain 4 was cloned, by direct selection, on a 4-kb partial Sau3AI fragment, in the vector pBC KS, to generate pJGS259 (Fig. 1). The MICs of tetracycline, chlortetracycline, and oxytetracycline for E. coli DH5αMCR carrying pJGS259 were significantly higher than that for a control strain carrying the vector pBC KS (Table 2). A NotI deletion derivative, pJGS264 (Fig. 1), did not confer tetracycline resistance upon DH5αMCR (Table 2), suggesting this deletion removed all or part of the tetracycline resistance determinant. Nucleotide sequence data from this region of pJGS259 allowed the identification of a 1,920-bp open reading frame (ORF) that spanned the NotI site of pJGS259 and showed 92% DNA sequence identity to the tet(W) gene of the rumen bacterium Butyrivibrio fibrisolvens (5). The putative product of this ORF had 91% identity and 96% similarity to the B. fibrisolvens TetW protein. Since current convention insists that 80% amino acid identity is the cutoff between tetracycline resistance determinants (19), the A. pyogenes determinant was also designated Tet W. Like the TetW protein of B. fibrisolvens and other ribosomal protection tetracycline resistance proteins, the A. pyogenes TetW protein shared similarity with the elongation factors, particularly in N-terminal regions associated with GTP binding (32).
FIG. 1.
Restriction maps of plasmids containing the tet(W) genes of strains 4 (pJGS259) and BBR1 (pJGS279). pJGS264, the NotI deletion derivative of pJGS259 that does not confer tetracycline resistance in E. coli, is also shown. Only the A. pyogenes-derived insert is shown for each plasmid. The large arrows indicate the positions of ORFs, which are labeled. The oriT upstream of mob is indicated by the solid rectangle. Restriction enzyme sites are as follows: B, BglII; Bm, BamHI; C, ClaI; E, EagI; N, NotI; Nd, NdeI; P, PstI; Sa, Sau3AI; S, SacI; Sc, ScaI; V, EcoRV; and X, XhoI. Coordinates (in kilobases) are given below the pJGS279 map.
TABLE 2.
MICs of tetracycline, chlortetracycline, and oxytetracycline for E. coli clones and A. pyogenes strain BBR1
| Strain | MIC (μg/ml) of:
|
||
|---|---|---|---|
| Tetracycline | Chlortetracycline | Oxytetracycline | |
| DH5αMCR(pBC KS) | 0.25 | 0.12 | 1 |
| DH5αMCR(pJGS259) | 16 | 8 | 32 |
| DH5αMCR(pJGS264) | 0.25 | 0.12 | 0.5 |
| DH5αMCR(pJGS279) | 32 | 32 | 64 |
| BBR1 | 8 | 8 | 8 |
| BBR1 + tetracyclinea | 8 | 8 | 8 |
BBR1 was grown on BHI-blood agar supplemented with 1 μg of tetracycline per ml prior to determination of the MIC.
Since the tet(W) gene in pJGS259 abuts one ends of the clone, a tet(W)-specific probe was used to identify homologous sequences in a λGEM12 library of our standard laboratory strain, BBR1, which is also tetracycline resistant. Homologous sequences were subcloned into pBC KS to create pJGS279 (Fig. 1), which overlaps considerably in restriction sites with pJGS259 and also confers tetracycline, chlortetracycline, and oxytetracycline resistance to DH5αMCR (Table 2). Nucleotide sequencing of the BBR1 tet(W) gene indicated that it was identical to that of strain 4. However, pJGS279 contains at least 6 kb upstream of the tet(W) gene (Fig. 1).
The percent G+C content of the tet(W) genes of strain 4 and BBR1 was 52.2, which is considerably lower than that of the A. pyogenes housekeeping genes we have sequenced (average G+C = 62.5%) (S. J. Billington, S. T. Gilbert, and B. H. Jost, unpublished data). Downstream of tet(W) was a strong rho-independent terminator (ΔG = −35.7 kcal/mol) composed of a 14-bp stem of completely base-paired G and C residues. Upstream of tet(W) were sequences similar to those that control the regulation of the inducible tet(M) gene of Tn916 (31), including the presence of a significant secondary structure (ΔG= −26.3 kcal/mol) 36 bp upstream of tet(W), which could act as a rho-independent terminator. However, the MIC of tetracycline for BBR1 was not increased following incubation on plates containing tetracycline (Table 2), suggesting that this tet(W) gene is not induced by tetracycline.
Distribution of the tet(W) gene among A. pyogenes isolates.
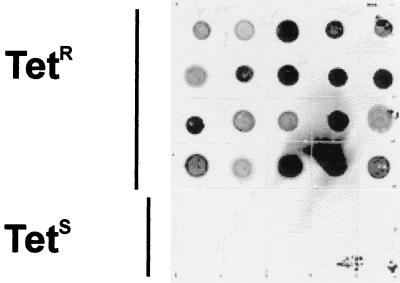
Genomic DNA from 20 tetracycline-resistant and 10 tetracycline-susceptible isolates were subjected to dot blot analysis with a tet(W)-specific gene probe (Fig. 2). All 20 tetracycline-resistant isolates, but none of the tetracycline-susceptible isolates, hybridized to the tet(W)-specific probe. To confirm these results, oligonucleotide primers internal to the tet(W) gene were used to screen all 30 isolates by PCR for the presence of the gene. Again, a direct correlation was obtained between resistance of the isolate to tetracycline and the amplification of a 694-bp DNA fragment internal to tet(W) (data not shown). These results indicate that tet(W) is widely distributed among tetracycline-resistant A. pyogenes strains.
FIG. 2.
Dot blot hybridization of tetracycline-resistant (Tetr) and tetracycline-susceptible (Tets) isolates with a tet(W)-specific probe. Approximately 500 ng of genomic DNA from 20 tetracycline-resistant and 10 tetracycline-sensitive isolates was spotted onto a nylon membrane in the arrangement indicated and hybridized with the 1,246-bp tet(W)-specific probe.
Identification of a tet(W)-associated mobilization gene.
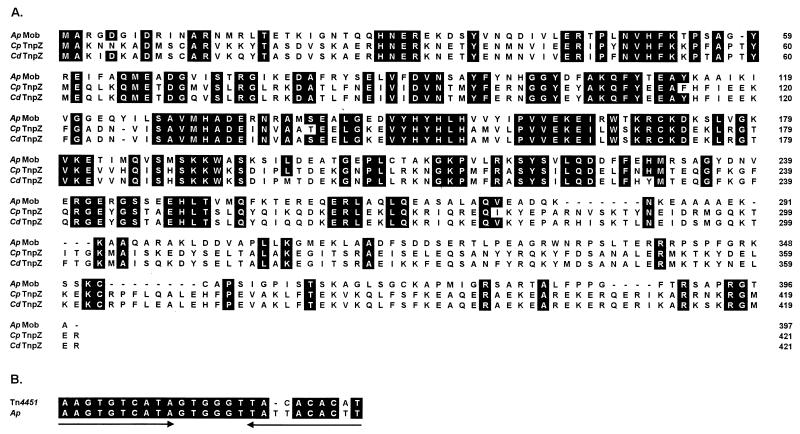
A 1,194-bp ORF, designated mob, was identified 657 bp upstream of the BBR1 tet(W) gene (Fig. 1). The product of the mob gene had significant similarity to members of the Pre/Mob family of gram-positive mobilization proteins (4). Pre/Mob proteins can effect mobilization of DNA elements, such as plasmids and transposons, in the presence of conjugative machinery by creating a nick at an origin of transfer, oriT (11). The BBR1 Mob protein had most similarity to the TnpZ proteins of the clostridial mobilizable transposons Tn4451 (35.4% identity, 65.8% similarity) (11) and Tn4453a (35.6% identity, 67.5% similarity) (20) (Fig. 3A). TnpZ nicks the circular form of these transposons at an upstream oriT, to create the leading strand for mobilization. Immediately upstream of the BBR1 mob gene is a sequence with identity of 22 of 24 bp to the oriT of Tn4451 and Tn4453a (11, 20) (Fig. 3B).
FIG. 3.
(A) Amino acid sequence alignment of the A. pyogenes Mob protein (Ap Mob) with the clostridial TnpZ proteins from Tn4451 of Clostridium perfringens (Cp TnpZ) (11) and Tn4453a of C. difficile (Cd TnpZ) (20). Amino acids identical to those in Mob are boxed. Amino acid numbers are shown on the right. (B) Nucleotide sequence comparison of the oriT upstream of mob with the oriT of Tn4451. The oriT of Tn4451 and that of Tn4453a are identical (11, 20). Identical nucleotides are boxed, and the region of dyad symmetry is indicated by the divergent arrows.
The tet(W)-associated mob gene and oriT can function in mobilization.
E. coli plasmids carrying the BBR1 mob gene and oriT were assessed for their ability to be mobilized from E. coli strain S17-1 to DH5αMCR (Table 3). pJGS279 could be mobilized at a frequency >4 logs more efficiently than the negative control plasmid pBC KS, with which no transconjugants were obtained. pJGS324, containing only the mob gene and oriT, could be transferred at a frequency similar to that of pJGS279, suggesting that the mob gene and oriT can function in DNA mobilization. When the mob gene (pJGS416) or oriT (pJGS417) was cloned independently, neither of these plasmids could be mobilized. However, if these plasmids were present in the same cell, pJGS417, but not pJGS416, could be mobilized to the recipient strain. Thus, the product of the mob gene expressed from pJGS416 is able to act in trans on oriT to effect mobilization of pJGS417.
TABLE 3.
Mobilization frequencies of mob and oriT plasmids
| Donor | Mobilization frequencya |
|---|---|
| S17-1(pJRD215) | 3.5 × 10−2 |
| S17-1(pBC KS) | <7.5 × 10−8 |
| S17-1(pWSK129) | <7.4 × 10−8 |
| S17-1(pJGS279) | 1.0 × 10−3 |
| S17-1(pJGS324) | 3.2 × 10−4 |
| S17-1(pJGS416) | <6.6 × 10−8 |
| S17-1(pJGS417) | <2.3 × 10−8 |
| S17-1(pJGS416 | <2.9 × 10−7 |
| + pJGS417) | 1.4 × 10−4 |
Number of transconjugants per donor cell.
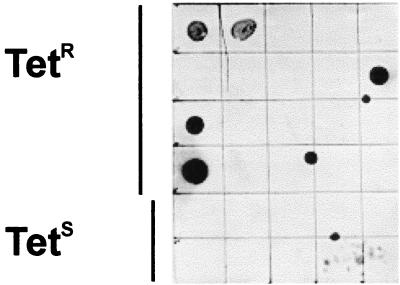
The mob gene is only present in 25% of tetracycline-resistant A. pyogenes strains.
A mob-specific gene probe was used to screen genomic DNA from 20 tetracycline-resistant and 10 tetracycline-susceptible A. pyogenes isolates (Fig. 4). Despite the fact that each of the 20 tetracycline-resistant isolates carries tet(W), only 5 (25%) of these isolates hybridized to the mob-specific probe. The five mob+ strains represent four of the seven tetracycline-resistant bovine isolates examined as well as the single avian isolate, 98-1508. Thus, none of the 12 tetracycline-resistant porcine isolates examined carried the mob gene. The tetracycline-susceptible isolates did not hybridize to the mob probe (Fig. 4).
FIG. 4.
Dot blot hybridization of tetracycline-resistant (Tetr) and tetracycline-susceptible (Tets) strains with a mob-specific probe. Approximately 500 ng of genomic DNA from 20 tetracycline-resistant and 10 tetracycline-sensitive isolates was spotted onto a nylon membrane in the arrangement indicated and hybridized with the 728-bp mob-specific probe.
Conjugative transfer of a tet(W) element from mob+ A. pyogenes isolates.
Each of the five mob+ strains, BBR1, JGS190, 3, 4, and 98-1508, was used in filter mating experiments with a kanamycin-resistant recipient strain, JGS478 (Table 4). Small numbers of putative tetracycline-resistant, kanamycin-resistant transconjugants were obtained with BBR1, JGS190, or 98-1508 as the donor. The frequency of conjugative transfer for these strains was very low (Table 4), and the possibility existed that colonies with resistance to both tetracycline and kanamycin could have arisen through spontaneous mutation. However, mock conjugations involving only donor or recipient cultures did not yield tetracycline-resistant, kanamycin-resistant colonies. Genomic DNA from transconjugants resulting from matings with each of the donors were probed in DNA dot blots with probes specific for the kanamycin resistance gene of the recipient and the tet(W) gene of the donor. DNA from each putative transconjugant hybridized with both probes, confirming that these transconjugants result from the transfer of tet(W) from the donor strain to JGS478 (data not shown). In addition, each of these transconjugants also hybridized to a mob-specific gene probe (data not shown), indicating that mob is part of the transferred genetic element.
TABLE 4.
Conjugation frequencies of Tet W determinants among A. pyogenes strains
| Donor | Recipient | Conjugation frequencya |
|---|---|---|
| BBR1 | JGS478 | 2.5 × 10−10 |
| JGS190 | JGS478 | 2.2 × 10−10 |
| 3 | JGS478 | <7.6 × 10−11 |
| 4 | JGS478 | <4.2 × 10−11 |
| 98-1508 | JGS478 | 9.0 × 10−11 |
| JGS603 | JGS610 | 5.4 × 10−9 |
| JGS605 | JGS610 | 4.4 × 10−9 |
Number of transconjugants per donor cell.
Transconjugants resulting from BBR1 × JGS478 (JGS603) and JGS190 × JGS478 (JGS605) crosses were used as donors in conjugation experiments with JGS610, an erythromycin-resistant derivative of 3274 (Table 4). Both transconjugants were able to transfer tetracycline resistance to JGS610 at frequencies approximately 10-fold higher than that of the original donor strains.
DISCUSSION
In the United States, tetracyclines, predominantly chlortetracycline and oxytetracycline, are fed extensively to food animals (36, 37), so commensal organisms like A. pyogenes are constantly exposed to these antibiotics. Tetracyclines tend to be fed on a long-term basis for growth promotion in pigs (36), whereas they are fed on a shorter-term basis to cattle, primarily for the prevention of certain diseases upon entry of cattle into feedlots (37). These data may partially explain the high prevalence of tetracycline resistance among porcine isolates of A. pyogenes (34). It is now recognized that bacteria commensal on animals provide a reservoir of antibiotic resistance genes that are frequently mobile and capable of being transferred to other bacteria entering the host.
In this paper, we report the cloning of the tetracycline resistance determinant from two bovine isolates of A. pyogenes, a frequent commensal organism on the mucous membranes of domestic animals. The predicted tetracycline resistance protein from A. pyogenes is greater than 80% identical to TetW of the obligately anaerobic rumen bacterium B. fibrisolvens, and thus by the criteria of Levy et al. (19), the A. pyogenes determinant was also classified as Tet W. Tet W determinants have previously been described only in obligately anaerobic bacteria isolated from the rumen of sheep and cattle, pig feces, or the human gut (2, 5, 28). A. pyogenes is commensal on the ruminal wall of cattle (22) and has been isolated from the stomach of pigs (B. H. Jost, K. W. Post, J. G. Songer, and S. J. Billington, unpublished data), and it may have acquired the determinant from obligately anaerobic rumen or gastric bacteria. The tet(W) genes of the obligate anaerobes diverge in DNA sequence by less than 1% over the coding region, consistent with proposed recent intergeneric transfer of the determinant within the gastrointestinal environment (5, 28). The A. pyogenes tet(W) gene diverges by approximately 8% from these sequences, but is still highly homologous, consistent with its potential inheritance from the normal flora anaerobes. The greater divergence of the tet(W) gene of A. pyogenes may be due to a longer residence of this gene in A. pyogenes. However, inconsistent with this hypothesis is the fact that less than half of the base changes between the B. fibrisolvens and the A. pyogenes tet(W) sequences are A/T to G/C transitions. The percent G+C content of the A. pyogenes tet(W) gene and the associated mob gene are lower than those of A. pyogenes housekeeping genes, suggestive of an origin outside of A. pyogenes. A similar argument has been used to suggest that tet(W) did not originate in B. fibrisolvens, which has a much lower percent G+C content. However, several of the gastrointestinal species that carry tet(W) have G+C contents in the range of 50 to 55% (28).
The presence of Tet W in the facultative anaerobe A. pyogenes gives this determinant a life outside the gastrointestinal tract of animals and humans and establishes a bridge between the tet(W)-carrying anaerobic bacteria found in different animal hosts. While A. pyogenes is likely capable of survival in the environment, it is still relatively fastidious and is generally found associated with animals. However, even on the respiratory mucosa, A. pyogenes will interact with a different set of microbiological flora from that present in the gastrointestinal tract. The ability of A. pyogenes to transfer the Tet W determinant to other bacteria may play a role in the dissemination of this determinant. Recent studies of the distribution of tet(W) genes suggest that while they contribute significantly to the resistance of the gastrointestinal flora of animals (2), the ability of these genes to be disseminated even in effluent is limited (9). Despite this fact, tet(W) genes were detected in food components and even antibiotics fed to swine (2).
The region upstream of the A. pyogenes tet(W) gene is similar to a region upstream of the tet(M) gene of Tn916, which is involved in transcriptional attenuation of the tet(M) gene (31). This observation has previously been made for the B. fibrisolvens tet(W) gene, although no inducibility data were described (5). The tetracycline resistance of strain BBR1 was not inducible by previous exposure to tetracycline. This is also true of the resistances of the other tetracycline-resistant isolates examined in this study (34). Therefore, it is unlikely that these upstream sequences are involved in tet(W) induction. Other ribosomal protection tet genes, such as tet(O), also have upstream sequences similar to the tet(M) upstream region, although these determinants are expressed constitutively (39). However, for tet(O), this upstream region is required for full expression of the gene (39).
The association of tet(W) with a functional mob gene is also indicative of the mobile nature of this determinant. The mob gene encodes a protein of the Mob/Pre family of gram-positive mobilization proteins, which function in mobilization by single-stranded cleavage at an oriT sequence. The tet(W)-associated Mob protein is most similar to the TnpZ proteins of the clostridial mobilizable transposons Tn4451 and Tn4453a (11, 20), and its associated oriT shares identity of 22 of 24 bp to the oriT of these transposons. The Mob protein appears capable of acting in trans at its cognate oriT to effect mobilization, at least in E. coli. While the direct effect of Mob on the transmissibility of tet(W) was not addressed in this study, the transfer of tet(W) and mob, from at least some of the mob+ tetracycline-resistant A. pyogenes strains, is consistent with the presence of these genes on a mobile genetic element. The exact nature of the genetic element is not yet known, since mob genes can be present on either mobilizable or conjugative plasmids or transposons. These strains have previously been examined for the presence of plasmids, and the only plasmid identified was the small, broad-host-range plasmid pAP1, which lacks conjugative or mobilization functions, in BBR1 and JGS190 (7). However, the techniques used in those studies may not have allowed the identification of large plasmids. Preliminary sequence data from one end of pJGS279 indicated the presence of the putative housekeeping gene rluC, which encodes pseudouridylate synthase (data not shown). The presence of a housekeeping gene on pJGS279 suggests that the insert from this recombinant plasmid is of chromosomal, rather than plasmid, origin. A chromosomal location for tet(W) is consistent with the low transfer efficiency, since this adds additional rate-limiting steps that could affect conjugative transfer, such as excision, and in the case of nonreplicating DNA, chromosomal insertion. The tet(W) genes of B. fibrisolvens strains 1.23 and 1.230 have been reported to be capable of being transferred by conjugation (27), although the frequency of conjugative transfer was considerably higher than was observed in this study. The B. fibrisolvens element also appears to be chromosomally located, although no additional sequence information is available to determine if it is an element similar to that described here. The lack of transconjugants observed with strains 3 and 4 as donors may reflect a defect in some part of the transfer process, or, given the low transfer frequency observed with the other three mob+ strains, the transfer frequency could be below the level of detection in the assays performed. Interestingly, onward transfer of the element from transconjugants appeared to be more efficient than the initial transfer event. This result may reflect transfer into a strain with the same background, since JGS478 and JGS610 are both derivatives of 3274. Recent evidence suggests that some strains of A. pyogenes carry different restriction-modification mechanisms (S. J. Billington and B. H. Jost, unpublished data), which may ultimately affect transfer frequencies.
The presence of tet(W) in each of the tetracycline-resistant isolates examined in this study suggests that Tet W is a widespread determinant of tetracycline resistance in A. pyogenes. However, since no other determinants were examined in this study, it is possible that some or all of these isolates may carry more than one tetracycline resistance determinant. Despite the fact that all tetracycline-resistant A. pyogenes isolates carry tet(W), only 25% of these strains carry the mob gene, which appears to be part of the genetic element carrying tet(W). Incidentally, the majority of mob+ isolates are of bovine origin, and none of the 12 tetracycline-resistant isolates of porcine origin carries this gene. This difference may reflect the way in which bovine and porcine isolates have inherited this gene, with bovine isolates primarily receiving tet(W) on the element described in this study, but with porcine isolates receiving tet(W) by an alternative route, perhaps through a separate genetic element.
Acknowledgments
We thank Hien Trinh and Dawn Bueschel for excellent technical assistance and Stefani Gilbert for construction of the A. pyogenes BBR1 λGEM12 library.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. (ed.) 1994. Current protocols in molecular biology, vol. 1. Greene Publishing Associates and John Wiley and Sons, Inc., New York, N.Y.
- 4.Bannam, T. L., P. K. Crellin, and J. I. Rood. 1995. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site specific recombinase excises a circular transposon molecule. Mol. Microbiol. 16:535-551. [DOI] [PubMed] [Google Scholar]
- 5.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 6.Billington, S. J., B. H. Jost, W. A. Cuevas, K. R. Bright, and J. G. Songer. 1997. The Arcanobacterium (Actinomyces) pyogenes hemolysin, pyolysin, is a novel member of the thiol-activated cytolysin family. J. Bacteriol. 179:6100-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billington, S. J., B. H. Jost, and J. G. Songer. 1998. The Arcanobacterium (Actinomyces) pyogenes plasmid pAP1 is a member of the pIJ101/pJV1 family of rolling circle replication plasmids. J. Bacteriol. 180:3233-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, G. R., and M. M. Chengappa. 1991. Essentials of veterinary bacteriology and mycology, 4th ed. Lea and Febiger, Philadelphia, Pa.
- 9.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crellin, P. K., and J. I. Rood. 1998. Tn4451 from Clostridium perfringens is a mobilizable transposon that encodes the functional Mob protein, TnpZ. Mol. Microbiol. 27:631-642. [DOI] [PubMed] [Google Scholar]
- 12.Davison, J., M. Heusterspreute, N. Chevalier, H.-T. Vinh, and F. Brunel. 1987. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51:275-280. [DOI] [PubMed] [Google Scholar]
- 13.Guérin-Faublée, V., J. P. Flandrois, E. Broye, F. Tupin, and Y. Richard. 1993. Actinomyces pyogenes: susceptibility of 103 clinical animal isolates to 22 antimicrobial agents. Vet. Res. 24:251-259. [PubMed] [Google Scholar]
- 14.Hoie, S., K. Falk, and B. M. Lium. 1991. An abattoir survey of pneumonia and pleuritis in slaughter weight swine from 9 selected herds. IV. Bacteriological findings in chronic pneumonic lesions. Acta Vet. Scan. 32:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonsson, P., S.-E. Olsson, A.-S. Olofson, C. Fälth, O. Holmberg, and H. Funke. 1991. Bacteriological investigations of clinical mastitis in heifers in Sweden. J. Dairy Res. 58:179-185. [DOI] [PubMed] [Google Scholar]
- 16.Jost, B. H., J. G. Songer, and S. J. Billington. 1999. An Arcanobacterium (Actinomyces) pyogenes mutant deficient in production of the pore-forming cytolysin pyolysin has reduced virulence. Infect. Immun. 67:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jost, B. H., J. G. Songer, and S. J. Billington. 2001. Cloning, expression, and characterization of a neuraminidase gene from Arcanobacterium pyogenes. Infect. Immun. 69:4430-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lechtenberg, K. F., T. G. Nagaraja, H. W. Leipold, and M. M. Chengappa. 1988. Bacteriologic and histologic studies of hepatic abscesses in cattle. Am. J. Vet. Res. 49:58-62. [PubMed] [Google Scholar]
- 19.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyras, D., and J. I. Rood. 2000. Transposition of Tn4451 and Tn4453 involves a circular intermediate that forms a promoter for the large resolvase, TnpX. Mol. Microbiol. 38:588-601. [DOI] [PubMed] [Google Scholar]
- 21.Nagaraja, T. G., S. B. Laudert, and J. C. Parrott. 1996. Liver abscesses in feedlot cattle. I. Causes, pathogenesis, pathology, and diagnosis. Comp. Cont. Edu. Pract. Vet. 18:S230-S241, S256. [Google Scholar]
- 22.Narayanan, S., T. G. Nagaraja, N. Wallace, J. Staats, M. M. Chengappa, and R. D. Oberst. 1998. Biochemical and ribotypic comparison of Actinomyces pyogenes and A. pyogenes-like organisms from liver abscesses, ruminal wall, and ruminal contents of cattle. Am. J. Vet. Res. 59:271-276. [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 25.Reddy, C. A., C. P. Cornell, and A. M. Fraga. 1980. Chemically defined growth medium for Corynebacterium pyogenes. Am. J. Vet. Res. 41:843-845. [PubMed] [Google Scholar]
- 26.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott, K. P., T. M. Barbosa, K. J. Forbes, and H. J. Flint. 1997. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63:3405-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 30.Speer, B. S., L. Bedzyk, and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su, Y. A., P. He, and D. B. Clewell. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trinh, H. T., S. J. Billington, A. C. Field, J. G. Songer, and B. H. Jost. 2002. Susceptibility of Arcanobacterium pyogenes to tetracyclines, macrolides and lincosamides. Vet. Microbiol. 85:353-359. [DOI] [PubMed] [Google Scholar]
- 35.Turner, G. V. 1982. A microbiological study of polyarthritis in slaughter pigs. J. S. Afr. Vet. Assoc. 53:99-101. [PubMed] [Google Scholar]
- 36.U.S. Department of Agriculture. 1996. Swine '95. II. Reference of 1995 U.S. grower/finisher health & management practices. USDA APHIS VS CEAH. National Animal Health Monitoring System, Fort Collins, Colo.
- 37.U.S. Department of Agriculture. 2000. III. Health management and biosecurity in U.S. feedlots 1999. No. N336.1200. USDA APHIS VS CEAH. National Animal Health Monitoring System, Fort Collins, Colo.
- 38.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 39.Wang, Y., and D. E. Taylor. 1991. A DNA sequence upstream of the tet(O) gene is required for full expression of tetracycline resistance. Antimicrob. Agents Chemother. 35:2020-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura, H., A. Kojima, and M. Ishimaru. 2000. Antimicrobial susceptibility of Arcanobacterium pyogenes isolated from cattle and pigs. J. Vet. Med. B 47:139-143. [DOI] [PubMed] [Google Scholar]