Abstract
Translational regulation plays an essential role in development and often involves factors that interact with sequences in the 3′ untranslated region (UTR) of specific mRNAs. For example, Nanos protein at the posterior of the Drosophila embryo directs posterior development, and this localization requires selective translation of posteriorly localized nanos mRNA. Spatial regulation of nanos translation requires Smaug protein bound to the nanos 3′ UTR, which represses the translation of unlocalized nanos transcripts. While the function of 3′ UTR-bound translational regulators is, in general, poorly understood, they presumably interact with the basic translation machinery. Here we demonstrate that Smaug interacts with the Cup protein and that Cup is an eIF4E-binding protein that blocks the binding of eIF4G to eIF4E. Cup mediates an indirect interaction between Smaug and eIF4E, and Smaug function in vivo requires Cup. Thus, Smaug represses translation via a Cup-dependent block in eIF4G recruitment.
Keywords: Cup, Drosophila, eIF4E, Smaug, translation
Introduction
Development of multicellular organisms is directed by regulated gene expression. Although control of gene expression often occurs at the level of transcription, post-transcriptional events play essential roles. For example, translational regulation has been demonstrated to function in the development of a wide range of organisms including Xenopus, Caenorhabditis elegans, Drosophila, and mammals (Wickens et al, 2000). Often, regulation is mediated by trans-acting factors that interact with target mRNAs through cis-acting elements in the transcript's 3′ untranslated region (UTR). These interactions temporally or spatially restrict the translation of target transcripts, which in turn contributes to proper execution of the relevant developmental program.
While a number of sequence-specific RNA-binding proteins have been identified that regulate translation through the 3′ UTR, the molecular mechanisms involved remain poorly understood. Specifically, the molecular interactions that allow proteins bound to the 3′ end of an mRNA to influence the function of the basic translation machinery have in large part not been identified. There are two notable exceptions, both of which involve translational repressors that function through direct or indirect interactions with the translation initiation factor eIF4E. One involves translational repression mediated by Bicoid protein, which recognizes sequences in the 3′ UTR of caudal mRNA and represses translation via its ability to interact with eIF4E (Dubnau and Struhl, 1996; Rivera-Pomar et al, 1996; Niessing et al, 2002). eIF4E is the cap-binding protein that associates with eIF4G, a scaffolding protein that ultimately recruits the 40S ribosomal subunit to the mRNA (Gingras et al, 1999). Both Bicoid and eIF4G recognize eIF4E via a conserved eIF4E-binding motif with the consensus sequence Y-X-X-X-X-L-Φ (where Φ is hydrophobic and X is any amino acid) (Mader et al, 1995; Niessing et al, 2002). Thus, recruitment of Bicoid to an mRNA blocks translation by preventing recruitment of eIF4G.
The second well-characterized mechanism whereby a protein bound to the 3′ end of an mRNA interacts with the basic translation machinery involves cytoplasmic polyadenylation element-binding protein (CPEB) (Hake and Richter, 1994). CPEB, which functions in both oocytes and neurons, interacts with target mRNAs through cis-acting elements known as cytoplasmic polyadenylation elements (Richter, 2000). CPEB bound to RNA interacts with a protein called Maskin, which in turn uses an eIF4E-binding motif to interact with eIF4E (Stebbins-Boaz et al, 1999). Thus, CPEB blocks translation using Maskin as an adaptor protein to interact indirectly with eIF4E, thereby blocking eIF4G recruitment.
The mechanism by which Bicoid and Maskin repress translation is similar to that employed by a conserved group of proteins known as 4E-BPs. 4E-BPs contain an eIF4E-binding motif that allows them to repress translation by blocking the eIF4E/eIF4G interaction (Gingras et al, 1999). They are not known to interact with any RNA-binding proteins but are thought to regulate specifically the translation of transcripts with extensive secondary structure in their 5′ UTRs.
Spatial regulation of translation plays an essential role in Drosophila development. For example, in the early embryo, nanos (nos) mRNA is inefficiently localized to the posterior where it is translated (Wang and Lehmann, 1991; Gavis and Lehmann, 1994; Bergsten and Gavis, 1999). Localized Nos protein represses the translation of hunchback mRNA in the posterior, allowing for development of the posterior of the embryo (Tautz, 1988; Wharton and Struhl, 1991; Gavis and Lehmann, 1992).
The nos 3′ UTR contains three stem/loop structures that function as cis-acting elements, which repress the translation of unlocalized nos mRNA. One stem/loop appears to represent the binding site for an as yet unidentified translational repressor (Crucs et al, 2000). The other two stem/loops are binding sites for a sequence-specific RNA-binding protein known as Smaug (Smg), which functions as a translational repressor (Dahanukar and Wharton, 1996; Smibert et al, 1996, 1999; Dahanukar et al, 1999). These stem/loops are thus referred to as SREs (Smg recognition elements). Smg contains a sterile alpha mating (SAM) domain, and recent work has shown that the Smg SAM domain functions as the protein's RNA recognition domain (Aviv et al, 2003; Green et al, 2003). This domain defines a new family of post-transcriptional regulators, which are conserved from yeast to humans, that function in part through a common mechanism of transcript recognition via their RNA-binding SAM domains.
In an effort to understand how Smg represses translation, we have searched for Smg-binding proteins. Here we show that Smg interacts with a protein known as Cup and that Cup is an eIF4E-binding protein that mediates an indirect interaction between Smg and eIF4E. Cup contains two eIF4E-binding sites, both of which block eIF4G's interaction with eIF4E. Taken together, these results suggest that Smg represses translation through Cup's ability to block the recruitment of eIF4G to target mRNAs. Consistent with this model, we demonstrate that Cup contributes to Smg-mediated translational repression in vivo. Taken together, this work suggests that the use of adaptor eIF4E-binding proteins is likely to be a common mechanism employed in translational regulation mediated by 3′ UTR-binding proteins.
Results
Smg interacts with the Cup protein
To understand the mechanisms that underlie Smg's ability to repress translation, we set out to identify Smg-binding proteins. Our initial work focused on proteins that would interact with amino acids 583–763. This region contains the Smg SAM domain, which is the protein's RNA-binding domain (Aviv et al, 2003; Green et al, 2003). An affinity resin carrying covalently coupled GST-Smg583–763 was mixed with early embryo extracts. After extensive washing, bound proteins were eluted and detected via silver staining following SDS–PAGE. Several proteins were eluted from both the GST-Smg583–763 resin and a resin carrying covalently coupled GST-Smg179–307 (Figure 1A). However, an ∼80 kDa protein and an ∼140 kDa protein were specifically eluted from the Smg583–763 resin. Both proteins were subjected to MALDI-TOF mass spectrometry (performed by Borealis Biosciences Inc.), and while the smaller protein was not identified the larger was identified as Cup, which plays an essential but ill-defined role during oogenesis and early embryogenesis (Schupbach and Wieschaus, 1991; Keyes and Spradling, 1997). To confirm that Cup interacts with Smg583–763, we generated Cup via in vitro translation in rabbit reticulocyte lysate. This protein interacted with GST-Smg583–763, as assayed by capture of Cup on glutathione agarose in the presence of GST-Smg583–763 but not in the presence of GST protein alone or GST-Smg179–307 (Figure 1B). To confirm that the selective capture of Cup by Smg583–763 did not reflect differences in the levels of the different GST fusions captured on the glutathione agarose, we verified that similar levels of each protein were present in the eluates (Figure 1C).
Figure 1.
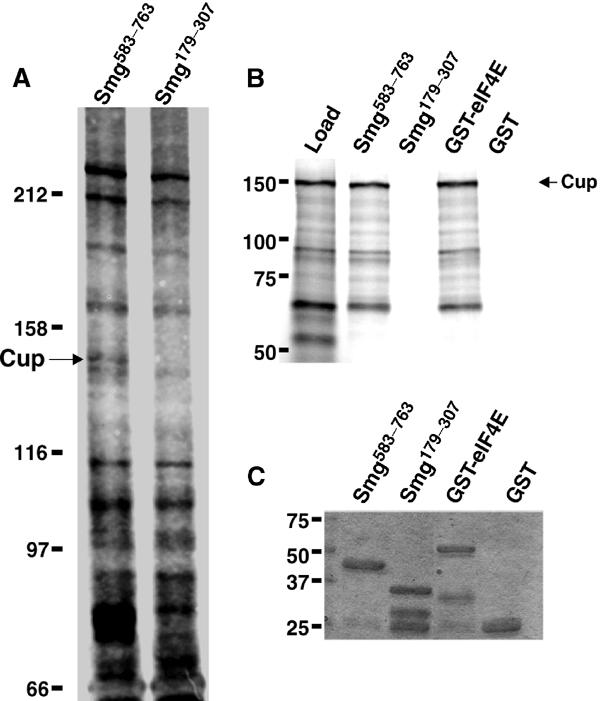
Cup interacts with Smg and eIF4E. (A) Embryo extracts were mixed with beads carrying the indicated GST fusion protein covalently bound to the resin. Bound proteins were resolved via SDS–PAGE and stained with silver. The position of Cup is indicated. (B) 35S-methionine-labeled Cup (load) generated via in vitro translation was mixed with various GST fusion proteins in the presence of glutathione agarose. Equivalent amounts of load and eluates from the indicated GST fusion proteins are shown. (C) Eluates from (B) were stained with Coomassie blue to detect the captured GST fusion proteins.
Cup is an eIF4E-binding protein
Database comparisons revealed that Cup shares a region of similarity with a mammalian protein 4E-T (Dostie et al, 2000). 4E-T is a nucleocytoplasmic shuttling protein that employs an eIF4E-binding motif to transport eIF4E into the nucleus. The similarity between Cup and 4E-T prompted us to test whether Cup is also an eIF4E-binding protein. Figure 1B demonstrates that in vitro-translated Cup interacts with GST-eIF4E.
A search of the Cup amino-acid sequence identified a potential eIF4E-binding motif, matching the Y-X-X-X-X-L-Φ consensus (Mader et al, 1995), from amino acids 342–348. Mutation of this motif within the context of a 203-amino-acid region of Cup (residues 285–487) had little effect on the ability of this region to interact with eIF4E (data not shown), suggesting that this region might contain multiple eIF4E-binding sites. To explore this possibility, we generated seven GST fusion proteins that represent overlapping fragments of residues 285–487, and assayed their ability to capture eIF4E on glutathione agarose (Figure 2A, with results summarized in Figure 2D). We identified three fragments, Cup311–360, Cup335–385, and Cup361–410, that interacted with eIF4E. As Cup311–360 and Cup361–410 do not overlap, Cup contains at least two binding sites for eIF4E. To confirm that the selective capture of eIF4E by the different Cup fragments did not reflect dramatic differences in the levels of the different GST fusions captured on the glutathione agarose, we verified that similar levels of each protein were present in the eluates for all fragments employed in Figure 2.
Figure 2.
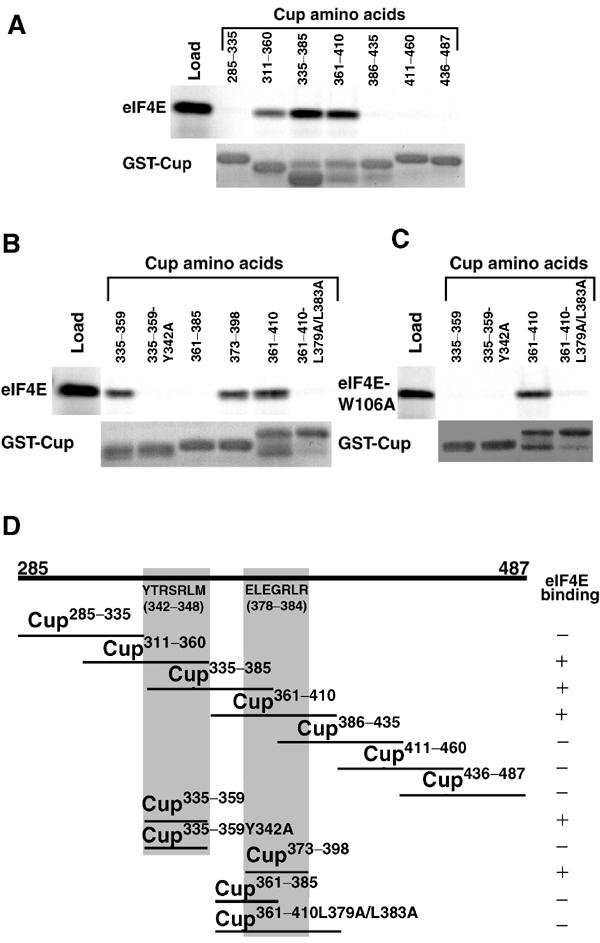
Identification of two eIF4E-binding sites in Cup. (A–C) Labeled eIF4E and eIF4E-W106A were generated via coupled transcription/translation in rabbit reticulocyte lysate in the presence of 35S-methionine and mixed with GST fusion proteins carrying various fragments of Cup in the presence of glutathione agarose. Equivalent amounts of the in vitro-translated eIF4E or eIF4E-W106A (load) and the elutions from the indicated GST-Cup proteins are shown. A fraction of each eluate was stained with Coomassie blue to detect the captured GST fusion proteins (GST-Cup). (D) Schematic representation of Cup amino acids 285–487. The ability of various Cup fragments from this region to interact with eIF4E is indicated. Proteins that interact with eIF4E capture 15–47% of the input eIF4E, while fragments that do not interact capture less than 0.5% of the input. These experiments identified two Cup-binding sites: one located within amino acids 335–369 (outlined in gray), which contains a sequence matching the consensus for an eIF4E-binding motif from amino acids 342–348, and a second site from 373–398 (outlined in gray), which does not contain a sequence matching the consensus. Note that the Y342A change within the consensus site and the L379A/L383A change within the second site block eIF4E binding. In addition, the W106A mutation in eIF4E blocks the interaction of the consensus eIF4E-binding site with eIF4E while having no effect on the interaction with the second site.
To determine if the potential eIF4E-binding motif located from amino acids 342–348 is functional, two smaller GST fusions were generated: one expressing amino acids 335–359 and a mutated version where Y 342 was changed to an A. This change has been shown previously to disrupt the function of other eIF4E-binding motifs (Mader et al, 1995). Figure 2B shows that Cup335–359 interacts with eIF4E while the Y342A mutation blocks this interaction, confirming that this region is indeed a bona fide eIF4E-binding motif.
To map the location of the second binding site, two additional fusion proteins were expressed. Figure 2B shows that Cup373–398 interacts with eIF4E while Cup361–385 does not. Inspection of this region failed to identify an eIF4E-binding motif that matches the consensus. However, simultaneous mutation of L379 and L383 to A residues blocks the capture of eIF4E by Cup361–410 (Figure 2B).
eIF4E-binding motifs interact with a conserved portion of the convex surface of eIF4E, and mutation of a conserved W residue to A within this surface blocks the ability of eIF4E to interact with eIF4E-binding motifs (Ptushkina et al, 1998; Marcotrigiano et al, 1999; Pyronnet et al, 1999). As expected, mutation of this residue in Drosophila eIF4E (W106A) blocked the interaction of the Cup eIF4E-binding motif contained within GST-Cup335–359 (Figure 2C). In contrast, the second Cup eIF4E-binding site (Cup361–410) interacts with eIF4E-W106A. These results therefore demonstrate that the second eIF4E-binding site within Cup interacts with eIF4E through a mechanism that is distinct from that employed by eIF4E-binding motifs.
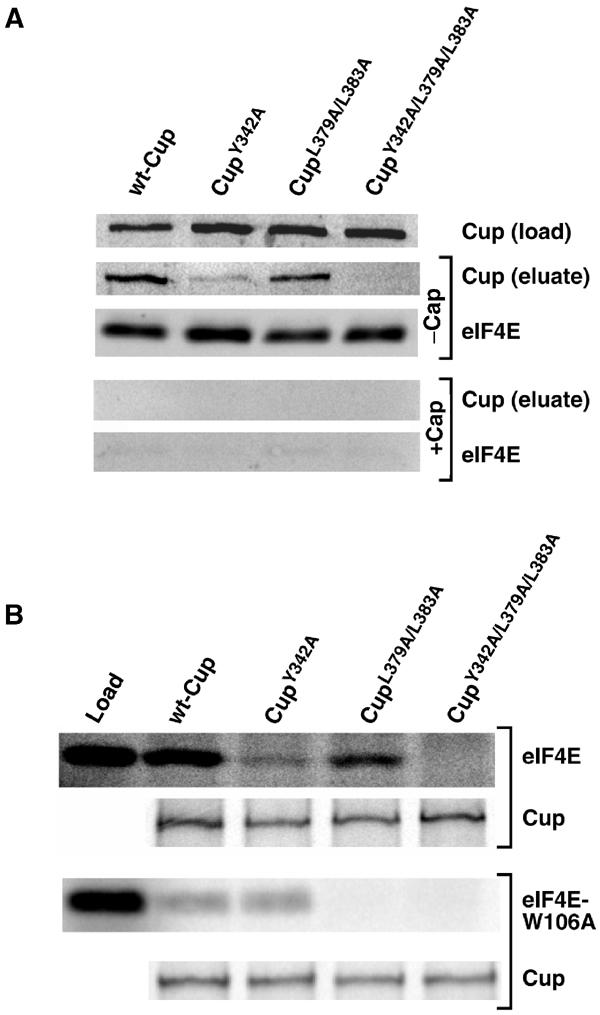
To test the importance of these two binding sites in the ability of full-length Cup to interact with eIF4E, we took two complementary approaches. First we expressed protein A-tagged versions of wild-type and mutant Cup proteins in the Drosophila tissue culture S2 cell line and assayed their ability to interact with endogenous eIF4E. Extracts prepared from transfected cells were mixed with 7m-GTP-sepharose (a cap column), which captures both eIF4E and wild-type Cup (Figure 3A). Addition of excess soluble 7m-GDP to the extract blocked the capture of both eIF4E and Cup, indicating that Cup interacts with the cap column through its interaction with eIF4E (Figure 3A). We found that the Y342A mutation reduced the amount of Cup captured on the cap column, while the L379A/L383A mutation had a more modest effect. Quantification of the amount of L379A/L383A protein captured compared to the amount of wild-type Cup captured in three independent experiments demonstrated a 2.7±0.75-fold decrease consistent with a modest effect on eIF4E binding. Mutation of both binding motifs completely blocked the capture of Cup on the cap column.
Figure 3.
Cup contains two eIF4E-binding sites. (A) Drosophila S2 cells were transfected with plasmids expressing protein A-tagged wild-type and mutant forms of Cup. Extracts prepared from transfected cells were mixed with 7m-GTP-sepharose with and without soluble 7m-GDP (cap). Western blots to detect the indicated Cup proteins in the total cell extracts, which represent 15% of protein used in the immunoprecipitations, as well as Cup and eIF4E in the eluates from captures are shown. (B) Labeled eIF4E or eIF4E-W106A as well as wild-type and mutant FLAG-tagged Cup proteins were generated via coupled transcription/translation in rabbit reticulocyte lysate in the presence of 35S-methionine. The eIF4E or eIF4E-W106A was mixed with each of the Cup proteins and immunoprecipitated using an anti-FLAG antibody and protein G agarose. A sample of the in vitro-translated eIF4E (load) representing 50% of protein used in the captures and the elutions from the immunoprecipitations of the indicated Cup proteins are shown.
To confirm the results obtained using cell culture, we expressed full-length FLAG-tagged wild-type and mutant Cup via in vitro translation. These proteins were assayed for their ability to co-immunoprecipitate in vitro-translated eIF4E using an anti-FLAG antibody. This assay gave results similar to those described above: the Y342A mutation in the consensus motif reduced eIF4E capture, the L379A/L383A double mutation in the second site had a more modest effect, and simultaneous mutation of both binding sites completely blocked the Cup/eIF4E interaction (Figure 3B). We also employed this assay to assess the interaction of full-length Cup with eIF4E carrying the W106A mutation, which blocks the eIF4E/eIF4E-binding motif interaction. The results from Figure 2 demonstrate that the Cup eIF4E-binding motif requires W106 to interact with eIF4E while the second site does not. Thus the interaction of full-length Cup with eIF4E-W106A should be independent of the eIF4E-binding motif and rely only on the second site. Consistent with this model, both wild-type Cup and the Y342A mutant protein interacted with eIF4E-W106A at a low level while the L379A/L383A mutation eliminated the interaction (Figure 3B).
Taken together, our efforts to map the eIF4E-binding sites within Cup suggest that Cup contains one high-affinity eIF4E-binding motif and a second lower affinity site. This second site interacts with eIF4E through a mechanism that is distinct from the one employed by eIF4E-binding motifs.
Cup mediates an interaction between Smg and eIF4E
As described above, Cup interacts with both eIF4E and Smg. To test the possibility that Cup might mediate an indirect interaction between the two proteins, we tested the ability of GST-Smg583–763 to capture in vitro-translated eIF4E on glutathione agarose in the presence of in vitro-translated Cup (Figure 4). If Cup mediates an interaction between Smg and eIF4E, we predict that GST-Smg583–763 would capture eIF4E in the presence of wild-type Cup but not in the presence of mutant Cup, which is unable to interact with eIF4E. We found that while both wild-type Cup and the mutant Cup, which carries mutations in eIF4E-binding sites, were captured to similar extents on the glutathione resins carrying GST-Smg583–763, eIF4E was only captured when wild-type Cup was included. These results therefore demonstrate that Cup mediates an indirect interaction between eIF4E and Smg.
Figure 4.
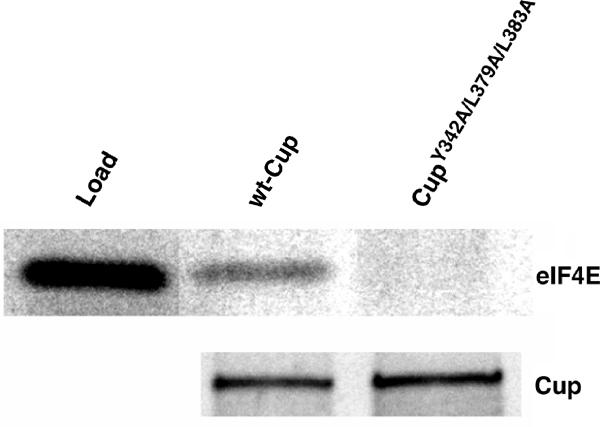
Cup mediates an indirect interaction between Smg and eIF4E. Labeled eIF4E as well as wild-type and mutant Cup proteins were generated via coupled transcription/translation in rabbit reticulocyte lysate in the presence of 35S-methionine. The eIF4E was mixed with either of the Cup proteins in the presence of GST-Smg583–763 and glutathione agarose. A sample of the in vitro-translated eIF4E (load) representing 30% of protein used in the captures and the elutions from the captures in the presence of wild-type Cup or CupY342A/L379A/L383A are shown.
To further assess the molecular interactions described above, we employed co-immunoprecipitation experiments using extracts from early Drosophila embryos. Anti-Smg antibody immunoprecipitates Cup from these extracts while normal rat serum does not (Figure 5). In addition, both an anti-Smg antibody and an anti-Cup antibody immunoprecipitate eIF4E, while failing to immunoprecipitate two irrelevant proteins—Vasa and DDP1. As expected, Cup and eIF4E were not immunoprecipitated with the anti-Smg antibody when extracts were prepared from embryos derived from smg mutant mothers, which lack Smg protein. Co-precipitation of eIF4E with Smg could simply reflect the fact that both are bound to the same mRNA. However, RNase A treatment of extracts had no effect on the amount of either Cup or eIF4E immunoprecipitated with the anti-Smg antibody. Taken together, the results of these co-immunoprecipitation experiments are consistent with Cup interacting with both Smg and eIF4E, thus mediating an interaction between Smg and eIF4E.
Figure 5.
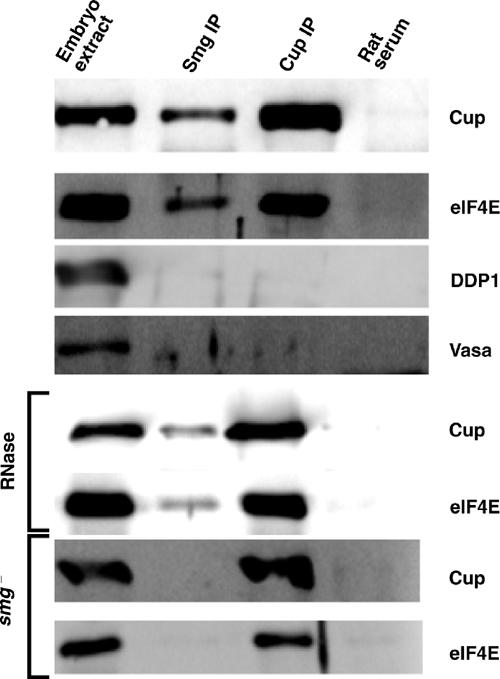
Co-immunoprecipitation of Smg, Cup, and eIF4E. Extracts derived from embryos collected 0–3 h post-egg-laying were immunoprecipitated with an anti-Smg antibody, an anti-Cup antibody, or normal rat serum in the presence of protein G agarose. Western blots to assay for the indicated proteins in crude embryo extracts as well as the indicated immunoprecipitates are shown. Where indicated, immunoprecipitations were performed in the presence of RNase A or employing extracts derived from embryos collected from smg mutant mothers. Embryo extract lanes represent 5% of the material used in the immunoprecipitations.
Cup's eIF4E-binding motifs block the interaction between eIF4E and eIF4G
The interaction between eIF4E and eIF4G is mediated via an eIF4E-binding motif in eIF4G (Mader et al, 1995), and proteins that carry eIF4E-binding motifs block translation by disrupting the eIF4E/eIF4G interaction (Haghighat et al, 1995; Altmann et al, 1997; Gingras et al, 1999). To determine whether the Smg/Cup/eIF4E complex might also block translation, we tested the ability of Cup's eIF4E-binding sites to block eIF4G binding to eIF4E. We recapitulated the eIF4E/eIF4G interaction using eIF4E covalently coupled to sepharose beads. This resin captures a fragment of Drosophila eIF4G (residues 434–804), generated via in vitro translation, which contains the protein's eIF4E-binding motif. As expected, increasing amounts of a peptide of Drosophila eIF4G corresponding to the protein's eIF4E-binding motif blocked eIF4G capture, while a similar peptide carrying a Y to A substitution in the eIF4E-binding motif did not (Figure 6). Similarly, inclusion of increasing amounts of GST fusion protein corresponding to Cup's (Cup335–359) eIF4E-binding motif also blocked eIF4G capture, while the Y to A substituted version of this protein did not. Surprisingly, Cup's second eIF4E-binding site (Cup361–410) also blocked eIF4G capture. This block was specific, as the mutant version of Cup361–410 that fails to interact with eIF4E did not block eIF4G capture. As described above, the second Cup eIF4E-binding site interacts with an eIF4E-W106A while this mutation disrupts binding of eIF4E-binding motifs. Thus, the second Cup eIF4E-binding site must employ a different mechanism to recognize eIF4E. The ability of this site to block eIF4G binding may suggest that, like an eIF4E-binding motif, it interacts with the convex surface of eIF4E and therefore precludes eIF4G binding; however, this interaction does not require W106. Alternatively, the second site interacts with another region of eIF4E and blocks eIF4G binding through an allosteric mechanism.
Figure 6.
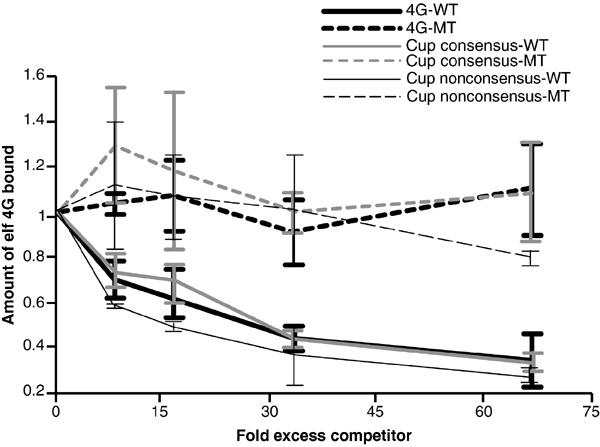
Cup's eIF4E-binding sites block the eIF4E/eIF4G interaction. A labeled fragment of Drosophila eIF4G (residues 434–804), which carries the protein's eIF4E-binding motif, was generated via coupled transcription/translation in rabbit reticulocyte lysate in the presence of 35S-methionine and mixed with a resin carrying covalently coupled eIF4E. Captures were performed in the presence of increasing amounts of either GST-Cup335–359 or GST-Cup335–359Y342A, which correspond to the wild-type and mutant version of the Cup eIF4E-binding motif, respectively, or GST-Cup361–410 or GST-Cup361–410L379A/L383A, which correspond to the wild-type and mutant version of the Cup second eIF4E-binding site, respectively, or a wild-type or mutant version of a 20-mer peptide corresponding to the eIF4G protein's eIF4E-binding motif. The effect of these competitors on eIF4G capture was quantitated and expressed as the fraction of eIF4G captured in the absence of competitors.
Smg-mediated translational repression requires Cup
The above in vitro data argue that the Smg/Cup/eIF4E complex underlies Smg's ability to repress translation. In principle, the existence of cup mutants could allow us to determine the role of Cup in Smg function in vivo. However, Smg functions in early embryos, while Cup plays an essential role during oogenesis, complicating efforts to assay Smg function in the absence of Cup. To circumvent this problem, we developed a quantitative in vivo assay to measure Smg's ability to repress translation. This involved generating an RNA encoding firefly luciferase and carrying three wild-type SREs in the transcript's 3′ UTR (luc3 × SRE+). The 3 × SRE+ element has been shown to mediate translational repression (Smibert et al, 1996, 1999). A control transcript (luc3 × SRE−) was also used where each SRE is point mutated, eliminating Smg binding and hence translational repression. We reasoned that the luc3 × SRE− transcript would be translated upon injection into embryos while the luc3 × SRE+ transcript would be translationally repressed. A similar approach has been employed to study translational repression mediated by Nos protein in the early embryo (Chagnovich and Lehmann, 2001). To control for variables such as the amount of RNA injected, a control RNA encoding Renilla luciferase was co-injected with both luc3 × SRE+ and luc3 × SRE− RNAs. The Renilla and firefly enzymes have distinct substrate requirements, allowing us to assay the activity of both proteins in the same extract. Thus, in each case, we normalized the levels of firefly luciferase to the levels of Renilla luciferase. The normalized data were used to calculate the ratio of luc3 × SRE−/luc3 × SRE+, which is a measure of the efficiency of repression of the luc3 × SRE+ RNA. Figure 7 shows that the luc3 × SRE+ RNA was repressed 12.5-fold in embryos derived from wild-type mothers (column 1), showing that Smg represses translation in an SRE-dependent manner. Embryos from smg mutant mothers translated luc3 × SRE+ and luc3 × SRE− RNAs at similar levels, giving a ratio of 1.17 (column 2). Thus, the injection assay accurately recapitulates Smg-mediated translational repression.
Figure 7.
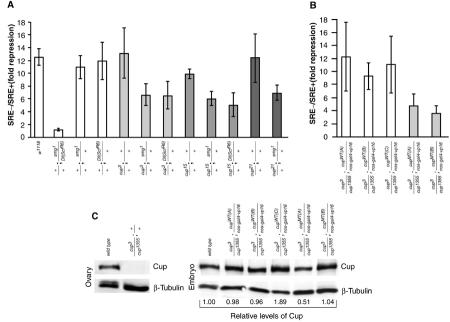
Cup functions in Smg-mediated translational repression. (A,B) Embryos derived from mothers with the indicated genotypes were injected with both Renilla luciferase and luc3 × SRE+ RNAs, or Renilla luciferase and luc3 × SRE− RNAs. Injected embryos were aged, and firefly and Renilla enzyme levels were assayed in embryo extracts. After correcting the levels of firefly enzyme activity using the levels of Renilla activity as a control, the amount of Smg-mediated repression (fold repression) was quantitated by dividing the corrected luc3 × SRE− value for a given genotype by the corresponding corrected luc3 × SRE+ value for that genotype. (C) Ovaries and embryos from mothers with the indicated genotypes were assayed for the levels of Cup and β-tubulin via Western blot. The levels of Cup protein in embryos were quantitated using β-tubulin as a loading control, and the amount of Cup in each sample relative to wild type is indicated.
We then investigated whether reduction of Smg or Cup levels, singly or together, affects the translational repression of the luc3 × SRE+ RNA. The levels of translational repression in embryos from mothers singly heterozygous for smg1, cup3, cup15, or cup21 are similar to those seen in embryos from wild-type mothers (Figure 7). In contrast, translational repression is significantly reduced in embryos from trans-heterozygous mothers that carry one copy of the smg1 allele along with any of the three cup alleles assayed here. We observe similar results when embryos are collected from mothers that are trans-heterozygous for the smg deficiency chromosome Df(ScfR6) and either the cup3 or cup15 alleles. Statistical analysis using the t-test confirmed that the fold repression seen for each trans-heterozygous combination was significantly different from the repression seen with the appropriate singly heterozygous controls (supplementary Table I). These in vivo results suggest that Cup is required for Smg to repress translation and are consistent with our in vitro data, which suggest that Smg-mediated recruitment of Cup to a target mRNA would lead to translational repression.
To obtain further evidence that Cup is involved in Smg function, we employed the GAL4/UAS system to express Cup in the female germ line. Expression of wild-type Cup using the nos-gal4-vp16 driver (Van Doren et al, 1998) rescues the cup mutant phenotype. For example, while cup3/cup1355 females do not lay eggs, expression of wild-type Cup (CupWT) in this mutant background results in females that lay eggs of which 80–90% hatch (supplementary Table II). Expression of CupY342A/L379A/L383A (CupMT), which does not interact with eIF4E, in the cup3/cup1355 mutant background also rescues egg laying, but only 6% of these eggs hatch. We exploited the fact that expression of both CupWT and CupMT protein rescues the ability of cup3/cup1355 females to lay eggs to test directly the role of Cup in Smg function using the injection assay described above. We found that three independent CupWT lines supported wild-type levels of Smg-mediated translational repression while two independent CupMT lines did not (Figure 7B). Statistical analysis confirmed that that the fold repression for each CupWT line was significantly different than the repression for each CupMT line (supplementary Table III). The failure to see complete abrogation of Smg-mediated repression may reflect the fact that cup3 and cup1355 are leaky alleles (Keyes and Spradling, 1997) and thus are likely to provide some Cup activity. Alternatively, Smg may employ both Cup-dependent and Cup-independent mechanisms to repress translation. Despite these caveats, our results demonstrate that wild-type Cup is required for wild-type levels of Smg repression.
We next compared the levels of Cup protein expressed in CupWT and CupMT embryos. Our anti-Cup antibody fails to detect any intact Cup protein in cup3/cup1355 ovaries, indicating that any full-length Cup protein present in transgenic embryos is transgene encoded (Figure 7C). We found that two of the CupWT lines and one of the CupMT lines expressed levels of Cup similar to those seen in wild-type embryos while one CupMT line expressed reduced amounts and one CupWT line overexpressed Cup. Thus the defect in translational repression in CupMT embryo does not result from a decrease in the amount of CupMT protein. Instead these results are consistent with our model, as they strongly suggest that Cup must interact with eIF4E to function in Smg-mediated translational repression.
Discussion
Cup mediates Smg-dependent translational repression by functioning as an eIF4E-binding protein
We present biochemical and genetic evidence that are consistent with Cup functioning as an eIF4E-binding protein that mediates an interaction between Smg and eIF4E. Cup blocks the eIF4E/eIF4G interaction, suggesting that Smg-dependent translational repression of SRE-containing mRNAs results from a Cup-mediated block in the recruitment of eIF4G. Cup's role in Smg function is therefore similar to that played by Maskin in translational repression mediated by CPEB (Stebbins-Boaz et al, 1999). Given that Maskin and Cup are not homologous, this suggests that other undiscovered adaptor eIF4E-binding protein/3′ UTR-binding protein pairs will employ this mechanism to regulate translation.
Cup interacts with eIF4E using both an eIF4E-binding motif and a second site that interacts with eIF4E through a distinct mechanism. Despite this difference, the second site is still able to inhibit the eIF4E/eIF4G interaction in vitro. Further work will be required to assess the significance of this site to Cup function in vivo.
nos translational repression occurs at multiple levels
Our model for Cup suggests that Smg represses translation at the level of initiation. However, the association of repressed nos mRNA with polysomes indicates that translational repression is achieved at a step after initiation (Clark et al, 2000). This apparent contradiction may reflect the fact that repression of nos translation is mediated by at least two trans-acting factors: Smg (Smibert et al, 1996, 1999; Dahanukar et al, 1999) and a yet to be identified factor that functions through sequences in the nos 3′ UTR that are distinct from the SREs (Crucs et al, 2000). Thus, while Smg regulates translation at the level of initiation, additional factors may function at other levels. Similarly, Smg itself may utilize multiple mechanisms to repress nos expression, only one of which is Cup dependent.
Activation of translation
Regulation of translation during development often involves both translational repression and translational activation. The combination of these controls can spatially or temporally restrict the expression of an mRNA, thereby directing the proper development of a cell type or tissue. For example, nos translation is spatially regulated allowing for the proper development of the posterior of the Drosophila embryo. Smg plays an essential role in this process by repressing the translation of unlocalized nos mRNA, while nos mRNA localized to the posterior escapes this repression allowing for the accumulation of Nos protein specifically at the posterior (Gavis and Lehmann, 1994). Given that Smg protein is distributed throughout the embryo, this suggests that Smg function must be over-ridden at the posterior (Dahanukar et al, 1999; Smibert et al, 1999). Cup is also distributed throughout the embryo (Keyes and Spradling, 1997), suggesting that spatial regulation of nos translation may involve disrupting Cup and/or Smg function specifically at the posterior. Osk protein, which is localized to the posterior, is required for nos translation and Osk interacts with Smg (Ephrussi et al, 1991; Kim-Ha et al, 1991; Gavis and Lehmann, 1994; Dahanukar et al, 1999). Thus translational activation could involve Osk binding to Smg thereby blocking Smg function. Interestingly, Cup and Osk interact with the same region of the Smg protein. This might imply that Osk's interaction with Smg could disrupt the Cup/Smg complex and in so doing play a role in activating nos translation at the posterior.
In Xenopus, temporal regulation of translation involves Maskin-mediated repression of target mRNAs in immature oocytes. Upon oocyte maturation, this repression is disrupted resulting in the activation of translation (Stebbins-Boaz et al, 1999; Cao and Richter, 2002). This activation of translation involves a CPEB-mediated increase in the length of the transcript's poly(A) tail and subsequent recruitment of poly(A)-binding protein (PABP) to the message. PABP brings eIF4G to the mRNA, which in turn disrupts the Maskin/eIF4E complex resulting in translational activation. Measurement of the length of the nos poly(A) tail suggests that regulation of nos translation does not involve changes in poly(A) tail length (Sallés et al, 1994; Gavis et al, 1996). Thus, activation of nos translation does not likely involve disruption of the Cup/eIF4E complex through poly(A)-dependent eIF4G recruitment. Taken together, these results also suggest that the use of adaptor proteins such as Cup in translational regulation mediated by sequence-specific RNA-binding proteins is not restricted to mRNAs whose translation is regulated through their poly(A) tail.
Other functions for Smg in the early embryo
Our data demonstrate that the same region of Smg that has previously been shown to function in sequence-specific RNA binding also interacts with Cup (Dahanukar et al, 1999). Our model therefore suggests that this region of the protein would be sufficient to repress translation. However, a transgene that expresses the Smg RNA-binding domain plus a short carboxy-terminal extension fails to rescue the smg mutant phenotype (Dahanukar et al, 1999). These results would suggest that Smg has other essential functions in the early embryo in addition to Cup-dependent translational repression. Our published and unpublished work suggests that Smg induces the degradation of target mRNAs in a process that may be distinct from its ability to repress translation (Smibert et al, 1996; JL Semotok, HD Lipshitz and CA Smibert, unpublished). Perhaps this ability to induce mRNA degradation is essential and requires regions of Smg outside of amino acids 583–763.
Cup function during development
Phenotypic analysis of several cup mutant alleles highlights that Cup is involved in a number of different biological processes during oogenesis and early embryogenesis, including oocyte growth, maintenance of chromosome morphology, and establishment of egg chamber polarity (Schupbach and Wieschaus, 1991; Keyes and Spradling, 1997). However, the molecular mechanisms that underlie Cup function have not been characterized. Our demonstration that Cup is an eIF4E-binding protein suggests that at least some of the defects associated with mutations in the cup gene result from misregulation of translation. Consistent with this possibility is the fact that Cup has been previously shown to interact with Nos protein, which is itself a translational repressor (Verrotti and Wharton, 2000). Genetic experiments suggest that Cup negatively regulates Nos activity during oogenesis, but the molecular mechanisms are not understood. This contrasts Cup's positive effect on Smg-mediated translational repression. Thus Cup might utilize different molecular mechanisms to influence different translational repressors. The pleiotropic nature of the cup mutant phenotype suggests that Cup may serve as an adaptor protein that is utilized by multiple translational repressors to interact with eIF4E.
Cup is homologous to 4E-T, a human nucleocytoplasmic shuttling protein that employs an eIF4E-binding motif to transport eIF4E into the nucleus (Dostie et al, 2000). The similarity between these proteins may suggest that Cup also functions to transport eIF4E into the nucleus. Thus some of the phenotypes associated with cup mutants may be related to a defect in eIF4E shuttling during oogenesis.
The similarity between Cup and 4E-T also suggests that 4E-T might function in translational repression as an adaptor protein that mediates interactions between eIF4E- and 3′ UTR-binding proteins. Specifically, 4E-T could function in translational repression mediated by the human Smg homolog. Similarly, additional RNA-binding proteins that interact with other eIF4E-binding proteins could function to regulate translation spatially or temporally. These protein pairs could control the translation of different mRNAs in various cell types throughout development.
Materials and methods
Affinity purification of Cup
GST-Smg583–763 and GST-Smg179–307 were covalently coupled to CNBr-activated sepharose beads (Amersham Pharmacia Biotech), resulting in a resin with ∼1 mg of protein coupled per milliliter of resin. Embryos collected 0–3 h post-egg-laying from w1118 animals were disrupted in a minimal volume of lysis buffer (150 mM KCl, 20 mM HEPES-KOH (pH 7.4), 1 mM DTT, protease inhibitors (Complete EDTA-free tablets, Roche)). After centrifugation, the supernatant was supplemented with glycerol to a final concentration of 10% v/v and stored at −80°C. A measure of 200 μl of the extract was mixed with 30 μl of the indicated resin for 3 h. Note that for all capture experiments, including GST pull-downs and immunoprecipitations, mixing was performed at 4°C, centrifugation steps were performed for 10 min at 21 000 g at 4°C, captures were washed with the buffer that the captures were performed in, proteins were eluted by boiling in SDS sample buffer, resolved by SDS–PAGE and where appropriate labeled proteins were visualized by phosphorimaging.
GST pull-down assays
All cDNAs are expressed sequence tags (ESTs) generated by the Berkeley Drosophila Genome Project. Cup and eIF4EII proteins were generated from ESTs LD30411 and GH18803, respectively, in rabbit reticulocyte lysate using the TNT-coupled transcription/translation system (Promega) according to the manufacturer's instructions.
GST-tagged proteins (20 μg GST-Smg583–763, 20 μg GST-Smg179–763, 20 μg GST-eIF4EI, or 5 μg GST-Cup fragments) were combined with 10 μl of glutathione agarose, 5 μl in vitro-translated protein and capture buffer (100 mM KCl, 20 mM HEPES-KOH (pH 7.4), 0.025% Tween-20) to a final volume of 100 μl and mixed for 2 h.
To demonstrate a Cup-dependent interaction between Smg and eIF4E, pull-down assays were performed as above, except that 20 μg of GST-Smg583–763 and 10 μl of wild-type or mutant in vitro-translated Cup carrying an amino-terminal FLAG epitope (Sigma) expressed from the pSPUTK vector (Falcone and Andrews, 1991) and 10 μl of in vitro-translated eIF4E were used.
Immunoprecipitation of in vitro-translated proteins
A measure of 10 μl of in vitro-translated FLAG-tagged Cup and 5 μl of in vitro-translated eIF4E or eIF4E-W106A were mixed for 1 h with 1.25 μg of anti-FLAG M2 antibody (Sigma) and 80 μl of capture buffer, centrifuged, and the supernatant was removed to 5 μl of protein G agarose (Roche) and mixed for 2 h.
Immunoprecipitation from the embryo extract
An anti-Cup antibody was raised in rats by Pocono Rabbit Farm & Laboratory (Canadensis, PA) against residues 1–225 of Cup while the anti-Smg antibody is described in Smibert et al (1999). Prior to immunoprecipitation, the embryo extract, prepared as described above, was diluted in an equal volume of dilution buffer (100 mM KCl, 30 mM HEPES-KOH (pH 7.4), 2% Tween-20, protease inhibitors (Complete EDTA-free tablets, Roche)) and centrifuged twice. A volume of 300 μl of the diluted extract was then mixed with antibody or normal rat serum (Sigma) for 1 h, with or without 0.35 μg/μl RNase A, centrifuged, and 250 μl of the supernatant was mixed with 30 μl of protein G resin and mixed for 2 h. Western blots of eluates employed anti-Vasa antibody (Styhler et al, 1998), anti-eIF4E antibody (Lavoie et al, 1996), and anti-DDP1 antibody (H Luo and HD Lipshtiz, unpublished) in addition to the anti-Cup antibody described above.
Tissue culture
For expression in D. melanogaster S2 cell culture, Cup was expressed with an amino-terminal protein A tag using the pRmHa3 expression vector (Bunch et al, 1988). Cells (2 ml) were transfected at a density of 0.5 × 106 cells/ml with 2 μg DNA and 6 μl of Fugene transfection reagent (Roche) according to the manufacturer's instructions and induced with 0.75 mM CuSO4. At 24 h post-induction, cells were lysed in 400 μl cell lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1% NP-40, protease inhibitors (Complete EDTA-free tablets, Roche)) and centrifuged. The supernatant was held on ice for 10 min with or without 0.2 mM soluble 7m-GDP (Sigma). The supernatant was mixed with 7m-GTP-sepharose (Amersham Pharmacia Biotech), and eluates were assayed by Western blot using rabbit anti-eIF4E antibody, which also detected the Cup proteins via their protein A tags.
Competition assay
Drosophila eIF4EI was purified from Escherichia coli as a GST fusion protein, and the GST portion was removed by protease cleavage. This protein was covalently linked to CNBr-activated sepharose beads resulting in a resin with ∼100 μg of eIF4E coupled per milliliter of resin. Wild-type (PSGKKQYDREQLLQLREVKA) and mutant (PSGKKQADREQLLQLREVKA) eIF4G peptides were purchased from Invitrogen. GST-Cup fragments or eIF4G peptides were incubated with 5 μl eIF4E affinity resin for 1 h in capture buffer in a final volume of 100 μl. A region of eIF4G corresponding to amino acids 434–804 was expressed from the pSPUTK vector in rabbit reticulocyte lysate (Promega) labeled with 35S-methionine. A measure of 1 μl of eIF4G translation was added to the protein/resin mixture and mixed for 2 h.
RNA injection assay
smg1 is described by Dahanukar et al (1999) and Df(ScfR6) by Kopp and Duncan (1997). cup alleles (Schupbach and Wieschaus, 1986; Keyes and Spradling, 1997) were maintained as cup/CyO;Ly/TM3 stocks. Wild-type and mutant cup transgenes were engineered by insertion of the cup cDNA into the UASp vector (Rørth, 1998). Transgenic flies were generated by standard methods (Spradling and Rubin, 1982). Western blots of embryos and ovaries used the E7 anti-β-tubulin antibody (Developmental Studies Hybridoma Bank).
Firefly luc3 × SRE+ and luc3 × SRE− RNAs were generated as described by Smibert et al (1999). Renilla luciferase RNA with a 30-nucleotide 3′ poly(A) tail was generated from a PCR product encoding nucleotides 277–1266 of the phRL-null plasmid (Promega). Embryos (30), collected 0–30 min post-egg-laying for heterozygous embryos and 0–60 min post-egg-laying for transgenic embryos, were injected at the anterior with a solution consisting of 5 mM KCl, 0.1 mM KPO4 pH 7.8, 0.2 mg/ml Renilla RNA and either luc3 × SRE+ or luc3 × SRE− RNAs at a concentration of 1 mg/ml. This was carried out at least three times per RNA per genotype. After injection, embryos were incubated at 20°C and harvested 2.5–3.0 h post-egg-laying for heterozygous embryos and 3.0–3.5 h post-egg-laying for transgenic embryos followed by homogenization in 30 μl 1 × passive lysis buffer (Promega) supplemented with 1 mg/ml BSA, 1 mM AEBSF, and 5 mM DTT, and luciferase activities were assayed using the Dual luciferase assay reagent (Promega).
To control for the amount of RNA injected and the efficiency of extract preparation, the relative light unit (RLU) value resulting from the luc3 × SRE+ and luc3 × SRE− RNAs was corrected by dividing by the RLU value from the Renilla signal. Fold repression is expressed as the corrected luc3 × SRE− value divided by the corrected luc3 × SRE+ value. The t-test was performed as described by Dixon and Massey (1957).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Heli Vari for her excellent technical expertise, A Spradling, T Schupach, and R Wharton for fly stocks, and P Lasko, H Luo, and H Lipshitz for antibodies. We also thank H Krause and H Lipshitz for reviewing the manuscript. MRN was supported by an Ontario Graduate Scholarship and CAS is supported by a scholarship from the Canadian Institutes of Health Research. This work was supported by an operating grant from the National Cancer Institute of Canada with funds from the Terry Fox Run.
References
- Altmann M, Schmitz N, Berset C, Trachsel H (1997) A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J 16: 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA (2003) The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 10: 614–621 [DOI] [PubMed] [Google Scholar]
- Bergsten S, Gavis E (1999) Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development 126: 659–669 [DOI] [PubMed] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LSB (1988) Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res 16: 1043–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Richter JD (2002) Dissolution of the maskin–eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J 21: 3852–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnovich D, Lehmann R (2001) Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci USA 98: 11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER (2000) Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr Biol 10: 1311–1314 [DOI] [PubMed] [Google Scholar]
- Crucs S, Chatterjee S, Gavis ER (2000) Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol Cell 5: 457–467 [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP (1999) Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell 4: 209–218 [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Wharton RP (1996) The nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev 10: 2610–2620 [DOI] [PubMed] [Google Scholar]
- Dixon WJ, Massey FJ (1957) Introduction to Statistical Analysis, 2nd edn New York: McGraw-Hill Book Company [Google Scholar]
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N (2000) A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J 19: 3142–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Struhl G (1996) RNA recognition and translational regulation by a homeodomain protein. Nature 379: 694–699 [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R (1991) oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50 [DOI] [PubMed] [Google Scholar]
- Falcone D, Andrews DW (1991) Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol Cell Biol 11: 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R (1992) Localization of nanos RNA controls embryonic polarity. Cell 71: 301–313 [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R (1994) Translational regulation of nanos by RNA localization. Nature 369: 315–318 [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lunsford L, Bergsten SE, Lehmann R (1996) A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122: 2791–2800 [DOI] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Green JB, Gardner CD, Wharton RP, Aggarwal AK (2003) RNA recognition via the SAM domain of Smaug. Mol Cell 11: 1537–1548 [DOI] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N (1995) Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J 14: 5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Richter JD (1994) CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627 [DOI] [PubMed] [Google Scholar]
- Keyes L, Spradling A (1997) The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development 124: 1419–1431 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23–35 [DOI] [PubMed] [Google Scholar]
- Kopp A, Duncan I (1997) Control of cell fate and polarity in the adult abdominal segments of Drosophila by optomotor-blind. Development 124: 3715–3726 [DOI] [PubMed] [Google Scholar]
- Lavoie C, Lachance PE, Sonenberg N, Lasko P (1996) Alternatively spliced transcripts from the Drosophila eIF4E gene produce two different cap-binding proteins. J Biol Chem 271: 16393–16398 [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 15: 4990–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716 [DOI] [PubMed] [Google Scholar]
- Niessing D, Blanke S, Jackle H (2002) Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes Dev 16: 2576–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkina M, von der Haar T, Vasilescu S, Frank R, Birkenhager R, McCarthy JE (1998) Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J 17: 4798–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD (2000) Influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds) pp 785–805. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jäckle H (1996) RNA binding and translational suppression by bicoid. Nature 379: 746–749 [DOI] [PubMed] [Google Scholar]
- Rørth P (1998) Ga14 in the Drosophila female germline. Mech Dev 78: 113–118 [DOI] [PubMed] [Google Scholar]
- Sallés FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S (1994) Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 266: 1996–1999 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E (1986) Maternal-effect mutations altering the anterior–posterior pattern of Drosophila melanogaster. Wilhelm Roux's Arch Dev Biol 195: 302–317 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E (1991) Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Lie YS, Shillinglaw W, Henzel WJ, Macdonald PM (1999) Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA 5: 1535–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM (1996) smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev 10: 2600–2609 [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM (1982) Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347 [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD (1999) Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell 4: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P (1998) vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125: 1569–1578 [DOI] [PubMed] [Google Scholar]
- Tautz D (1988) Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centers. Nature 332: 281–284 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Ruth L (1998) Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8: 243–246 [DOI] [PubMed] [Google Scholar]
- Verrotti A, Wharton R (2000) Nanos interacts with cup in the female germline of Drosophila. Development 127: 5225–5232 [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R (1991) Nanos is the localized posterior determinant in Drosophila. Cell 66: 637–647 [DOI] [PubMed] [Google Scholar]
- Wharton RP, Struhl G (1991) RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67: 955–967 [DOI] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze MW (2000) Translational control of developmental decisions. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds) pp 295–370. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data