Abstract
The expression pattern of regulated genes changes dynamically depending on the developmental stage and the differentiation state of the cell. Transcription factors regulate cellular events at the gene expression level by communicating signals to the general transcription machinery that forms a preinitiation complex (PIC) at class II core promoters. Recent data strongly suggest that PICs are composed of different sets of factors at distinct promoters, reflecting the spatiotemporal profile of gene expression in multicellular organisms. Thus, today it is important to ask the question: how universal are the promoter recognition factors? This review will focus on findings that support the new idea that core promoter recognition by distinct factors is an additional level of transcriptional regulation and that this step is developmentally regulated.
Keywords: embryogenesis, RNA polymerase II, TATA-binding protein (TBP), TBP-like factors (TLFs), TBP-related factors (TRFs), TBP-associated factors (TAFs), TFIID, zygotic transcription
Introduction
RNA polymerase II (Pol II) and a host of other factors, including the general transcription factors (GTFs) TFIIA, -B, -D, -E, -F, -H, work together to form a preinitiation complex (PIC) on core promoters and to allow subsequent transcription initiation (Orphanides et al, 1996). GTFs have been defined biochemically as factors required for correct initiation of Pol II transcription in vitro on a promoter with a classical TATA box and a strong initiator sequence (Roeder, 1996). TFIID, composed of the TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs) (Albright and Tjian, 2000; Tora, 2002, and references therein), was thought to be the only sequence-specific initiation factor recognising a class II promoter and thus to direct PIC assembly on all promoters. Binding of TFIID to the promoter constitutes a critical rate-limiting step at which activators and/or chromatin remodelling factors can control transcription (Orphanides et al, 1996; Hampsey and Reinberg, 1997). It has been widely believed that a PIC is composed of the same set of GTFs at every protein-coding gene promoter. Here we review the findings showing that transcription initiation is also regulated at the level of PIC formation at core promoters, and that many distinct PICs, with varying composition and with distinct promoter recognition factors, play important roles in transcriptional regulation.
GTFs lack diversity
Database searches for paralogue genes encoding GTF subunits have revealed that the components of TFIIB, -E,-F, -H and Pol II are encoded by single-copy genes (Aoyagi and Wassarman, 2000). TFIIA components, which are not typical GTFs, because they are not absolutely required for PIC assembly and subsequent transcription in a purified in vitro transcription system, are encoded by paralogue genes in Drosophila and humans (Aoyagi and Wassarman, 2000, and references therein). However, the fact that Pol II transcription can be initiated at certain promoters with PICs containing only a partial set of the GTFs has challenged the definition of GTFs (Parvin et al, 1994; Usheva and Shenk, 1994).
Diversity in the TBP family members
In contrast to GTF components, a database search for TBP homologues revealed the existence of two additional new gene families in metazoans, which encode factors related to TBPs (Figure 1). TBP-like factor (TLF) has a significantly high similarity to TBP, but groups separately in unrooted phylogenic trees (Dantonel et al, 1999). This factor is also called TBP-related factor 2 (TRF2) (Rabenstein et al, 1999) and TBP-like protein (TLP) (Ohbayashi et al, 1999). The Caenorhabditis elegans genome contains only two TBP-type factors (TBP and TLF) (Figure 1). TLFs and TBPs have well-conserved core domains, which however only share about 60% similarity (Dantonel et al, 1999). The different TLF core domains showed much less conservation during evolution (about 40–45% identity) when compared with the different TBP core domains (about 80% identity). This suggests that the functions of TLFs and TBPs have been subjected to different evolutionary pressures. While TBP function seems to be identical from yeast to humans, TLFs might serve a function(s) that tolerates more diversity.
Figure 1.
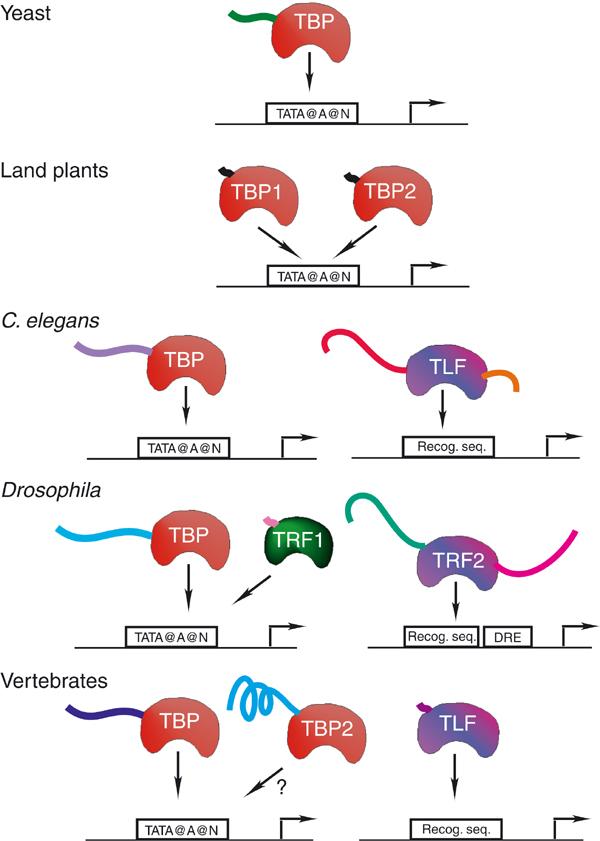
Factors belonging to the TBP family encoded by different genomes are presented. The well-conserved core domains of these factors are displayed as saddle-like shapes. The nonconserved N- and C-terminal domains are shown as ‘noodles', and the lengths of these are proportional. Similar colours reflect high conservation, and different colours mean no or weak sequence homology. The target of these factors, a typical TATA box-containing (TATA@A@N) class II promoter with initiator element (arrow), or a promoter with TATA-less sequences (Recog. seq.), is diagrammed. Drosophila TRF2 (TLF) binds to promoters containing a DNA replication-related element (DRE). ‘?' indicates that binding of vertebrate TBP2 to TATA box-containing promoters has not yet been documented.
A second set of TBP paralogue genes was very recently identified in vertebrate organisms (Veenstra and Wolffe, 2001). These genes encode factors that share a highly homologous core domain with the already known TBPs (92% identity), while they significantly differ from TBPs in their N-terminal domain, hereafter called TBP2s (Figure 1). Drosophila melanogaster has three different TBP-type factors: TBP, TRF2 (TLF) and another TBP-related factor (called TRF1). Based on sequence comparisons, TRF1 was classified in the TBP family in agreement with functional data showing that it is able to bind TATA boxes (Figure 1). Since a large number of plants and metazoans encode TBP paralogue genes, it is likely that these organisms have developed distinct TBP-type factors to facilitate differential gene expression for the regulation of distinct developmental pathways.
Variation in TAFs, TAF paralogues and their expression pattern
Similar to the TBP-type factors, TAFs also have paralogue genes and these TAF-like proteins are often expressed in a cell type- or tissue-specific pattern. The first TAF paralogue seems to have appeared in fission yeast. In this organism, two genes encode TAF5 homologues, both of which are present in the same TFIID complex (Mitsuzawa et al, 2001). Several TAF paralogue genes have been described from the C. elegans, D. melanogaster and human genomes (Aoyagi and Wassarman, 2000; Georgieva et al, 2000; Hiller et al, 2001; Tora, 2002). Interestingly, the D. melanogaster homologues of TAF5 and TAF4, Nohitter (Nht) and Cannonball (Can), respectively, are exclusively expressed in testis and seem to regulate a subset of genes (Hiller et al, 2001). Along the same lines, Drosophila TAF10 homologues (TAF10 and TAF10b) are differentially expressed during Drosophila embryogenesis (Georgieva et al, 2000). A retroposed copy of human TAF1 (TAF1L) and a homologue of TAF7 (TAF7L) are specifically expressed during male germ-cell differentiation (Wang and Page, 2002; Pointud et al, 2003). Mammalian TAF4b, a homologue of TAF4, is expressed tissue specifically in B lymphocytes, in the granulosa cells of the ovary and testis (Dikstein et al, 1996; Freiman et al, 2001). The expression patterns of TAF5L, TAF6L and TAF9L have not yet been investigated in organisms. Given that the general mechanism for the retention of duplicated genes is the divergence in their expression domains, it is likely that these proteins will also show restricted expression patterns. In addition, even core TAFs are expressed at very different levels in distinct cell types or tissues (Perletti et al, 1999; Mohan et al, 2003). Thus, the nonubiquitous expression pattern of several of the TAFs and the tissue-specific expression of certain TAF homologues in metazoans suggest that they cannot be essential components of all PICs formed in every cell of an organism.
Functional differences among distinct TBP-type factors
Crystallographic studies of the core domain of TBP bound to a TATA box revealed that TBP has a saddle-like structure (Burley, 1996). It was argued that TBP is required for the correct initiation of all RNA polymerase (I, II and III)-mediated transcription in eukaryotes. However, with the discovery of other TBP-type factors, it became clear that TBP homologues might play complementing roles in transcriptional regulation. Modelling of the three-dimensional structure of TLFs suggested that they form an asymmetric saddle-shaped core domain and that TLFs, unlike TBP, may bind to DNA sequences other than canonical TATA boxes (Dantonel et al, 1999). This view is supported by transient reporter assays demonstrating that vertebrate TLFs can potentiate transcription from exogenous TATA-less promoters (Ohbayashi et al, 2003).
TBPs and TBP2s mainly differ in their N-terminal domains. It is therefore conceivable that unique domain features contribute to the functional difference of the two TBP-type factors. In agreement, a defined region within the N-terminal domain of human TBP may be involved in specific interactions required for the assembly of functional PICs on TATA-containing, but not on TATA-less, Pol II and III promoters (Lescure et al, 1994). Moreover, deletion of the N-terminal domain from mouse TBP revealed that this part of the protein functions in the regulation of a placental immune defence process (Hobbs et al, 2002). In contrast, in mouse fibroblasts this deletion did not cause changes in gene expression in general (Schmidt et al, 2003). These findings indicate that the activities of the TBP N-terminal domain must either be compensated for by redundant domains or are restricted to the placenta.
Multiple functionally different TFIID complexes
The rather diverse spectrum of TBP- and TAF-type factors allows eukaryotic cells to form multiple promoter recognition complexes, differing in composition and function. Indeed, a number of in vitro studies reported that a single human cell contains TFIID complexes with or without TAF10, which exhibit functionally distinct properties (Brou et al, 1993; Jacq et al, 1994) (Figure 2). TFIID complexes, composed of core TAFs and cell type-specific TAFs, can also be found. TAF4b was shown to be enriched in differentiated human B lymphocytes, and a unique TAF4b-containing TFIID was isolated from these cells (Dikstein et al, 1996) (Figure 2). Later studies suggested that TAF4b has redundant functions in B cells with other related factors, and it was shown that TAF4b mediates transcription of a subset of genes required for proper folliculogenesis in the ovary (Freiman et al, 2001). Moreover, during spermatogenesis, TAF7L-containing TFIID complexes have been identified (Pointud et al, 2003) (Figure 2). Another example is the recent observation that apoptotic stimuli can lead to alternative splicing of TAF6 resulting in TAF6δ, which is part of a TFIID-like complex lacking TAF9 (Bell et al, 2001) (Figure 2). The fact that an inducible TAF isoform is capable of selectively altering gene expression programmes further illustrates that the composition of TFIID in living cells is dynamic and responsive to cellular signals.
Figure 2.
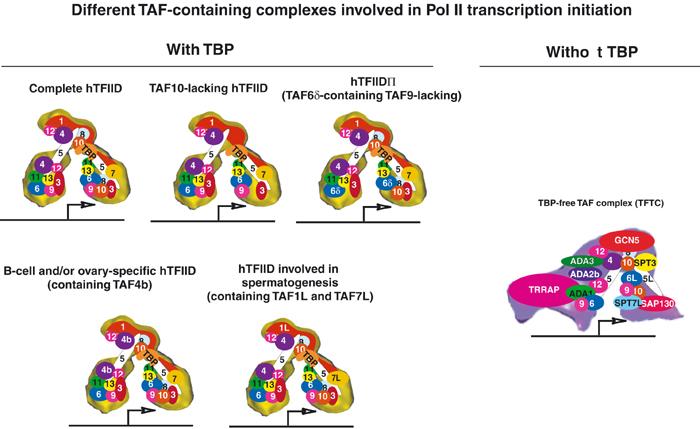
Schematic representation of various TAF-containing complexes isolated from human cells. The target promoter of TAF-containing complexes is diagrammed as a line with an initiator element displayed as an arrow. TFIID complexes are shown at the left with their corresponding TAF composition, and the human TFTC complex is illustrated at the right. Distinct TAFs are represented by different colours and corresponding numbers. Their position in the complexes reflects our present structural knowledge. The HATs in the distinct TAF-containing complexes are shown in red.
The above studies highlight the possibility that the various TFIID complexes have specific roles in (i) recognition of different promoters, (ii) interaction with distinct sets of general transcription factors and (iii) mediating differential responses from transcriptional activators to the PIC. The existence of different TFIID complexes is also consistent with the finding that different TAFs regulate only a limited subset of specific genes.
Alternative TBP-containing complexes play a role in Pol II transcription initiation
Another form of a TBP-containing complex, human B-TFIID or yeast TBP/Mot1 complex, does not contain the classical core TAFs, but instead a single Snf2/Swi2-related ATP-ase, human BTAF1 (formerly TAF170) or yeast Mot1 (Timmers et al, 1992; Poon et al, 1994). Mot1 is an essential regulator of Pol II-dependent transcription in vivo (Davis et al, 1992) and dissociates TBP–DNA complexes in vitro (Adamkewicz et al, 2000). Genome-wide transcriptional profiling analysis suggests that Mot1 positively and negatively affects subsets of yeast genes (Dasgupta et al, 2002; Geisberg et al, 2002). In vitro, human B-TFIID mediates basal Pol II transcription, but does not support stimulation of transcription by factors containing acidic or glutamine-rich activating motifs (Timmers and Sharp, 1991).
Negative cofactor 2 (NC2, also called Dr1/DRAP1) is an evolutionarily conserved regulator, originally identified as an inhibitor of basal transcription, as it binds to TBP and thereby blocks recruitment of TFIIA and TFIIB (Meisterernst et al, 1991; Inostroza et al, 1992). Surprisingly, NC2 stimulates TATA-less transcription in a Drosophila in vitro transcription system (Willy et al, 2000). Moreover, NC2 can directly stimulate activated transcription from TATA-driven promoters, both in vivo and in vitro in yeast (Cang and Prelich, 2002). Indeed, NC2 is required for the expression of many yeast genes and plays both positive and negative roles in vivo (Creton et al, 2002, and references therein). These studies reveal that the NC2/TBP complex is a bifunctional basal transcription factor, which, similar to the Mot1/TBP complex, differentially regulates initiation of Pol II transcription.
Recently, another TBP-containing complex, TAC (TBP-TFIIA-containing complex), was described in embryonic carcinoma cells, but this seems to be absent in differentiated cells (Mitsiou and Stunnenberg, 2000). TAC contains the TFIIAγ subunit and the unprocessed form of TFIIAαβ, and mediates transcription initiation by Pol II in the absence of TAFs.
TAFs are present in multiprotein complexes that lack TBP
Several TAF-containing multiprotein complexes lacking TBP have been isolated from yeast, Drosophila or human cells (Muratoglu et al, 2003, and references therein). The TBP-free TAF-containing complex (TFTC) (Figure 2) was shown to replace TFIID or TBP both on TATA-containing and TATA-less promoters in transcription assays on naked DNA templates in vitro (Wieczorek et al, 1998; Brand et al, 1999). This finding challenged the view that assembly of the PIC was nucleated exclusively by the sequence-specific binding of TBP/TRF1/TLF-containing complexes to promoters. TFTC is composed of a subset of TAFs, TAF homologues, the GCN5 histone acetyltransferase (HAT), TRRAP (a cofactor for Myc and E2F activation), and proteins belonging to the SPT and the ADA family of factors (Figure 2) (Wieczorek et al, 1998; Brand et al, 1999). In agreement with a role of TFTC in transcription initiation, a TFTC-like complex containing the GCN5 HAT was shown to facilitate type I interferon-stimulated transcription in a TBP-independent manner (Paulson et al, 2002). These findings point to an important role of TBP-independent transcription, which may add another level to the complexity of the regulation of eukaryotic gene expression. TAFs have also been described in other TBP-free HAT complexes: the yeast SAGA, the Drosophila TFTC, human PCAF/GCN5 and STAGA complexes (Martinez, 2002; Muratoglu et al, 2003, and references therein). Although these HAT complexes have a similar polypeptide composition as TFTC and have been implicated in the regulation of Pol II transcription at the chromatin level, they have not yet been shown to be involved in promoter recognition. Further studies are required to understand the molecular mechanisms by which TBP-free TAF complexes regulate promoter accessibility, recognition and PIC assembly.
Promoter recognition factors in development
After the discovery of the numerous TBP-type protein- and TAF-containing complexes, the next obvious question is how do these different sets of transcription complexes regulate tissue and developmental stage-dependent transcription. Information on the role of GTFs in multicellular organisms during development is only starting to emerge. The two areas of development in which the role of GTFs has been addressed in some detail are the activation of the zygotic genome in the early embryo and gametogenesis (the latter reviewed recently by Hochheimer and Tjian, 2003). The transition of embryo differentiation by maternally inherited gene products to zygotic gene activation provides a fascinating model for the analysis of the interplay between cell cycle regulation, chromatin assembly, epigenetic reprogramming and assembly of the transcription machinery (Newport and Kirschner, 1982; Stancheva and Meehan, 2000; Vassetzky et al, 2000). In most animal models including Drosophila, C. elegans, zebrafish and Xenopus, zygotic gene activation is preceded by synchronous cell cycles with no gap phases (Newport and Kirschner, 1982; Edgar and Schubiger, 1986), and zygotic gene activity is also delayed in mammals (Bolton et al, 1984; Thompson et al, 1998).
A number of studies explored the mechanisms by which zygotic gene activation is initiated in frog embryos at the mid-blastula transition (MBT). Interestingly, priming TBP binding to DNA allows premature transcription before the MBT, supporting a model that titration of chromatin assembly of histones and other repressive factors in the egg results in the dramatic upregulation of zygotic transcription (Prioleau et al, 1994). Additionally, TBP protein levels were shown to be limiting during pre-MBT and are dramatically upregulated at the initiation of zygotic transcription (Veenstra et al, 1999).
In the mouse embryo, TBP was shown to accumulate in the nucleus at the time when transcription starts, probably by a process involving new protein synthesis (Worrad et al, 1995). Drosophila and human TAFs were also found to localise both in the nucleus and cytoplasm in different tissues and at different time points (Georgieva et al, 2000; Pointud et al, 2003), suggesting that translocation of TAFs may regulate their activity. When somatic cell nuclei were transplanted into Xenopus oocytes, TBP was actively removed from these nuclei by a mechanism involving the chromatin-remodelling nucleosomal adenosine triphosphatase (ISWI), indicating a possible mechanism for intracellular TBP transport between transcriptionally active and inactive states (Kikyo et al, 2000).
TBP is indeed required for initiation of zygotic Pol II transcription in frog, fish and mouse, but not universally (Veenstra et al, 2000; Muller et al, 2001; Martianov et al, 2002). At the same time, TLF loss-of-function phenotypes of embryos revealed a critical role for TLF in zygotic gene activation in C. elegans (Dantonel et al, 2000; Kaltenbach et al, 2000), Xenopus (Veenstra et al, 2000) and zebrafish (Muller et al, 2001). Importantly, TLF- or TBP-depleted embryos both in fish and frogs revealed specific non-overlapping roles for the two factors in regulating distinct subsets of promoters. C. elegans TLF may have repressor as well as transcriptional activator function. Elimination of TLF expression by RNA interference in C. elegans caused not only lack of expression of early patterning genes but also premature ectopic expression of others (Dantonel et al, 2000). Thus, TLFs may positively or negatively regulate Pol II transcription, depending on the developmental stage of the embryo (Figure 3). Surprisingly, TLF mutants in mouse have no embryonic phenotype, but are male sterile (Martianov et al, 2001; Zhang et al, 2001), and a role for mouse TLF in Pol II transcription initiation is yet to be demonstrated. The significant difference between functional requirement for vertebrate TLFs is yet to be understood, especially in the light of the very high degree of conservation among them.
Figure 3.
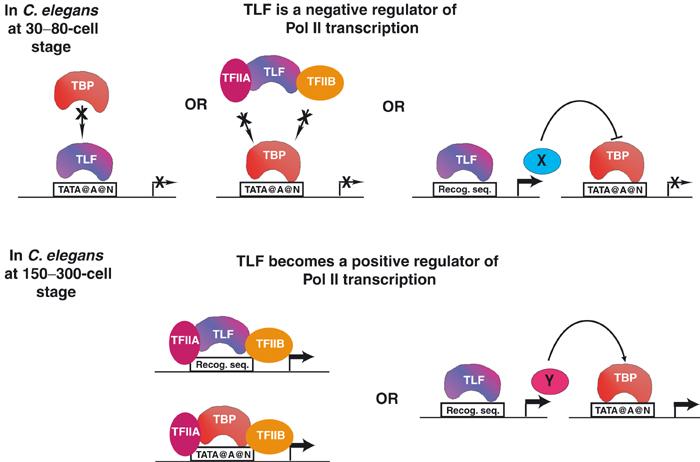
In C. elegans, TLF can function as a negative or a positive regulator of transcription. At the 30–80-cell stage, TLF may function as a repressor, which blocks TBP binding to promoters, or by titrating other GTFs from PIC formation, or by regulating a Pol II repressor factor (X). At the 150–300-cell stage, TLF becomes a positive regulator of Pol II transcription by either forming PICs on promoters where it will find its binding site (Recog. seq.) or by mediating transcription of an essential factor (Y) of Pol II transcription.
TAF10 is required for early development in the mouse in the inner cell mass, but not in the trophoblast, which develops normally in TAF10 knockout (KO) mice, indicating a TAF10-independent mechanism of transcription in this specialised extra-embryonic tissue (Mohan et al, 2003). The KO of TAF4b resulted in infertile female mice because of a defect in folliculogenesis. Gene expression profiling has uncovered a defective inhibin–activin signalling pathway in TAF4b-deficient ovaries (Freiman et al, 2001). These in vivo studies together with the differential expression patterns of TAFs suggest that vertebrate TAFs mediate the transcription of a subset of genes in different tissues or cell types.
Little is known about the regulation of individual promoters by GTFs during zygotic gene activation and subsequent differentiation. The shift from utilisation of a TATA-containing promoter to a TATA-less promoter of the protein translation initiation factor gene (Davis and Schultz, 2000) and those of the microinjected HSV thymidine kinase promoter (Majumder and DePamphilis, 1994) during mouse preimplantation development indicates that different regulatory mechanisms act in promoter recognition in a temporally defined manner. From recent data it seems plausible that distinct complexes are recruited to TATA-containing and TATA-less promoters. It was shown that Drososphila TRF2 (TLF) co-purifies with components of the nucleosome remodelling factor (NURF) complex as well as the DNA replication-related element (DRE)-binding factor DREF, and directs core promoter recognition of the proliferating cell nuclear antigen (PCNA) gene (Hochheimer et al, 2002) (Figure 1). The TATA-box-containing PCNA promoter is bound by TFIID, and a TATA-less promoter of this gene is bound by a TRF2 (TLF)- and DREF-containing complex. Uniquely, Drosophila TRF1 was described as a factor involved in neurogenesis with a restricted expression pattern in the embryo (Crowley et al, 1993). Later, TRF1 was reported to be present in a yet unidentified multiprotein complex (Hansen et al, 1997), and to direct the expression of the embryogenesis-related tudor gene from an alternate promoter (Holmes and Tjian, 2000). TRF1 is also present in the Drososphila TFIIIB complex, and thus was implicated in the regulation of the expression of Pol III genes (Takada et al, 2000). These findings further indicate that the core promoter is an active participant in the regulation of eukaryotic gene expression, which may be regulated by a developmentally changing set of initiation complexes. In spite of the fact that TRF1, TLFs, and possibly also TBP2s, are required for the regulation of a subset of promoters, the exact molecular mechanisms by which these factors nucleate PIC assembly have not yet been elucidated.
Conclusions and future directions
Many studies have documented the diversity of the components of the PICs, but we are still at the very beginning of understanding their biological functions, and how and why they assemble at certain promoters at a given time during development. Further studies focusing on distinct developmental stages or comparing pluripotent and differentiated cell types from different species are required to better understand the regulation of gene expression at the core promoter level. The drawback of the KO and antisense technologies used in different model systems for loss-of-function analyses is that they reveal only the first biological functions in very early steps of development. Conditional mutants with controlled inactivation of TBP-type factors or TAFs will be essential to uncover their specific regulatory roles during later stages of development, establishment and gradual determination of cell lineages as well as differentiation of tissues. Moreover, in vivo visualisation and chromatin immunoprecipitation-based techniques (using intact embryos or tissues) will be very informative to provide a temporal and spatial analysis of promoter occupancy by the diversity of the above-described factors. These experiments can then be complemented with biochemical approaches using extracts or factors prepared from differentiated cells or tissues to understand the exact molecular mechanisms that are relevant to the regulation of genes in cell type- and developmental stage-specific ways. It is very likely that the future results will draw a much more ‘colourful view' (Figure 2) of complexes playing a role in promoter recognition and PIC formation than those found in HeLa cells and yeast, which until recently provided most of our knowledge on PIC assembly.
Acknowledgments
We are very grateful to present and former colleagues for discussions, and apologise to colleagues whose work could only be cited indirectly in this review. We thank H Szutorisz and G Richards for critically reading the manuscript. The present work was supported by funds from INSERM, CNRS, Hôpital Universitaire de Strasbourg, ARC, FRM, AICR and European Community grants.
References
- Adamkewicz JI, Mueller CG, Hansen KE, Prud'homme WA, Thorner J (2000) Purification and enzymic properties of Mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J Biol Chem 275: 21158–21168 [DOI] [PubMed] [Google Scholar]
- Albright SR, Tjian R (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242: 1–13 [DOI] [PubMed] [Google Scholar]
- Aoyagi N, Wassarman DA (2000) Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation. J Cell Biol 150: F45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B, Scheer E, Tora L (2001) Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol Cell 8: 591–600 [DOI] [PubMed] [Google Scholar]
- Bolton VN, Oades PJ, Johnson MH (1984) The relationship between cleavage, DNA replication, and gene expression in the mouse 2-cell embryo. J Embryol Exp Morphol 79: 139–163 [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L (1999) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem 274: 18285–18289 [DOI] [PubMed] [Google Scholar]
- Brou C, Wu J, Ali S, Scheer E, Lang C, Davidson I, Chambon P, Tora L (1993) Different TBP-associated factors are required for mediating the stimulation of transcription in vitro by the acidic transactivator GAL-VP16 and the two nonacidic activation functions of the estrogen receptor. Nucleic Acids Res 21: 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK (1996) The TATA box binding protein. Curr Opin Struct Biol 6: 69–75 [DOI] [PubMed] [Google Scholar]
- Cang Y, Prelich G (2002) Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP). Proc Natl Acad Sci USA 99: 12727–12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creton S, Svejstrup JQ, Collart MA (2002) The NC2 alpha and beta subunits play different roles in vivo. Genes Dev 16: 3265–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley TE, Hoey T, Liu JK, Jan YN, Jan LY, Tjian R (1993) A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature 361: 557–561 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Quintin S, Lakatos L, Labouesse M, Tora L (2000) TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell 6: 715–722 [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Wurtz JM, Poch O, Moras D, Tora L (1999) The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem Sci 24: 335–339 [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT (2002) Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci USA 99: 2666–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JL, Kunisawa R, Thorner J (1992) A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol 12: 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W Jr, Schultz RM (2000) Developmental change in TATA-box utilization during preimplantation mouse development. Dev Biol 218: 275–283 [DOI] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R (1996) Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell 87: 137–146 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G (1986) Parameters controlling transcriptional activation during early Drosophila development. Cell 44: 871–877 [DOI] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R (2001) Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293: 2084–2087 [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Kuras L, Struhl K (2002) Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol Cell Biol 22: 8122–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S, Kirschner DB, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B, Tora L (2000) Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol Cell Biol 20: 1639–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D (1997) Transcription: why are TAFs essential? Curr Biol 7: R44–R46 [DOI] [PubMed] [Google Scholar]
- Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R (1997) Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 91: 71–83 [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT (2001) Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev 15: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs NK, Bondareva AA, Barnett S, Capecchi MR, Schmidt EE (2002) Removing the vertebrate-specific TBP N terminus disrupts placental beta2m-dependent interactions with the maternal immune system. Cell 110: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R (2003) Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev 17: 1309–1320 [DOI] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R (2002) TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420: 439–445 [DOI] [PubMed] [Google Scholar]
- Holmes MC, Tjian R (2000) Promoter-selective properties of the TBP-related factor TRF1. Science 288: 867–870 [DOI] [PubMed] [Google Scholar]
- Inostroza JA, Mermelstein FH, Ha I, Lane WS, Reinberg D (1992) Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70: 477–489 [DOI] [PubMed] [Google Scholar]
- Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L (1994) Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79: 107–117 [DOI] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE (2000) The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell 6: 705–713 [DOI] [PubMed] [Google Scholar]
- Kikyo N, Wade PA, Guschin D, Ge H, Wolffe AP (2000) Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 289: 2360–2363 [DOI] [PubMed] [Google Scholar]
- Lescure A, Lutz Y, Eberhard D, Jacq X, Krol A, Grummt I, Davidson I, Chambon P, Tora L (1994) The N-terminal domain of the human TATA-binding protein plays a role in transcription from TATA-containing RNA polymerase II and III promoters. EMBO J 13: 1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML (1994) TATA-dependent enhancer stimulation of promoter activity in mice is developmentally acquired. Mol Cell Biol 14: 4258–4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I (2001) Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 7: 509–515 [DOI] [PubMed] [Google Scholar]
- Martianov I, Viville S, Davidson I (2002) RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 298: 1036–1039 [DOI] [PubMed] [Google Scholar]
- Martinez E (2002) Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol 50: 925–947 [DOI] [PubMed] [Google Scholar]
- Meisterernst M, Roy AL, Lieu HM, Roeder RG (1991) Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell 66: 981–993 [DOI] [PubMed] [Google Scholar]
- Mitsiou DJ, Stunnenberg HG (2000) TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAalphabeta precursor and the TFIIAgamma subunit. Mol Cell 6: 527–537 [DOI] [PubMed] [Google Scholar]
- Mitsuzawa H, Seino H, Yamao F, Ishihama A (2001) Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J Biol Chem 276: 17117–17124 [DOI] [PubMed] [Google Scholar]
- Mohan IW, Scheer E, Wendling O, Metzger D, Tora L (2003) TAF10 (TAF(II)30) is necessary for TFIID stability and early embryogenesis in mice. Mol Cell Biol 23: 4307–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Lakatos L, Dantonel J, Strahle U, Tora L (2001) TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol 11: 282–287 [DOI] [PubMed] [Google Scholar]
- Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, Cserpan I, Lebedeva L, Nabirochkina E, Udvardy A, Tora L, Boros I (2003) Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol 23: 306–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M (1982) A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30: 687–696 [DOI] [PubMed] [Google Scholar]
- Ohbayashi T, Makino Y, Tamura T (1999) Identification of a mouse TBP-like protein (TLP) distantly related to the Drosophila TBP-related factor. Nucleic Acids Res 27: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T, Shimada M, Nakadai T, Wada T, Handa H, Tamura T (2003) Vertebrate TBP-like protein (TLP/TRF2/TLF) stimulates TATA-less terminal deoxynucleotidyl transferase promoters in a transient reporter assay, and TFIIA-binding capacity of TLP is required for this function. Nucleic Acids Res 31: 2127–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D (1996) The general transcription factors of RNA polymerase II. Genes Dev 10: 2657–2683 [DOI] [PubMed] [Google Scholar]
- Parvin JD, Shykind BM, Meyers RE, Kim J, Sharp PA (1994) Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem 269: 18414–18421 [PubMed] [Google Scholar]
- Paulson M, Press C, Smith E, Tanese N, Levy DE. (2002) IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat Cell Biol 4: 140–147 [DOI] [PubMed] [Google Scholar]
- Perletti L, Dantonel JC, Davidson I (1999) The TATA-binding protein and its associated factors are differentially expressed in adult mouse tissues [In Process Citation]. J Biol Chem 274: 15301–15304 [DOI] [PubMed] [Google Scholar]
- Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I (2003) The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci 116: 1847–1858 [DOI] [PubMed] [Google Scholar]
- Poon D, Campbell AM, Bai Y, Weil PA (1994) Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J Biol Chem 269: 23135–23140 [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, Mechali M (1994) Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 77: 439–449 [DOI] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian R (1999) TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci USA 96: 4791–479610220372 [Google Scholar]
- Roeder RG (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 21: 327–335 [PubMed] [Google Scholar]
- Schmidt EE, Bondareva AA, Radke JR, Capecchi MR (2003) Fundamental cellular processes do not require vertebrate-specific sequences within the TATA-binding protein. J Biol Chem 278: 6168–6174 [DOI] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR (2000) Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev 14: 313–327 [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Takada S, Lis JT, Zhou S, Tjian R (2000) A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 101: 459–469 [DOI] [PubMed] [Google Scholar]
- Thompson EM, Legouy E, Renard JP (1998) Mouse embryos do not wait for the MBT: chromatin and RNA polymerase remodeling in genome activation at the onset of development. Dev Genet 22: 31–42 [DOI] [PubMed] [Google Scholar]
- Timmers HTM, Meyers RE, Sharp PA (1992) Composition of transcription factor B-TFIID. Proc Natl Acad Sci USA 89: 8140–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers HTM, Sharp PA (1991) The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev 5: 1946–1956 [DOI] [PubMed] [Google Scholar]
- Tora L (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev 16: 673–675 [DOI] [PubMed] [Google Scholar]
- Usheva A, Shenk T (1994) TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell 76: 1115–1121 [DOI] [PubMed] [Google Scholar]
- Vassetzky Y, Hair A, Mechali M (2000) Rearrangement of chromatin domains during development in Xenopus. Genes Dev 14: 1541–1552 [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Destree OH, Wolffe AP (1999) Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol 19: 7972–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP (2000) Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290: 2312–2315 [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP (2001) Gene-selective developmental roles of general transcription factors. Trends Biochem Sci 26: 665–671 [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC (2002) Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet 11: 2341–2346 [DOI] [PubMed] [Google Scholar]
- Wieczorek E, Brand M, Jacq X, Tora L (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393: 187–191 [DOI] [PubMed] [Google Scholar]
- Willy PJ, Kobayashi R, Kadonaga JT (2000) A basal transcription factor that activates or represses transcription. Science 290: 982–985 [DOI] [PubMed] [Google Scholar]
- Worrad DM, Turner BM, Schultz RM (1995) Temporally restricted spatial localization of acetylated isoforms of histone H4 and RNA polymerase II in the 2-cell mouse embryo. Development 121: 2949–2959 [DOI] [PubMed] [Google Scholar]
- Zhang D, Penttila TL, Morris PL, Roeder RG (2001) Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech Dev 106: 203–205 [DOI] [PubMed] [Google Scholar]


