Abstract
The detection of thousands of volatile odorants is mediated by several hundreds of different G protein-coupled olfactory receptors (ORs). The main strategy in encoding odorant identities is a combinatorial receptor code scheme in that different odorants are recognized by different sets of ORs. Despite increasing information on agonist–OR combinations, little is known about the antagonism of ORs in the mammalian olfactory system. Here we show that odorants inhibit odorant responses of OR(s), evidence of antagonism between odorants at the receptor level. The antagonism was demonstrated in a heterologous OR-expression system and in single olfactory neurons that expressed a given OR, and was also visualized at the level of the olfactory epithelium. Dual functions of odorants as an agonist and an antagonist to ORs indicate a new aspect in the receptor code determination for odorant mixtures that often give rise to novel perceptual qualities that are not present in each component. The current study also provides insight into strategies to modulate perceived odorant quality.
Keywords: antagonist; calcium imaging; G protein-coupled receptor; mixture; odorant; olfactory; Abbreviations:; EG, eugenol; GPCR, G protein-coupled receptor; HEK293, human embryonic kidney 293; ISF, isosafrole; MIEG, methyl isoeugenol; OR, olfactory receptor
Introduction
A broad range of odorant molecules are recognized by the G protein-coupled olfactory receptor (OR) superfamily (Buck, 1996; Mombaerts, 1999; Firestein, 2001; Touhara, 2002). Studies on the receptive range of ORs have suggested that ORs discriminate subtle differences in the chemical structures of odorants, but also tolerate other molecular features in the ligand-binding site (Zhao et al, 1998; Touhara et al, 1999; Wetzel et al, 1999; Araneda et al, 2000; Kajiya et al, 2001; Gaillard et al, 2002). However, due to the difficulty in expressing and characterizing ORs in a heterologous cell system, detailed pharmacological information has been obtained for only a few ORs. For example, mOR-EG, a mouse OR encoded by MOR174-9 gene (Zhang and Firestein, 2002), has been shown to recognize eugenol (EG) (a major odorant in clove oil), vanillin (a vanilla flavor), and other structurally similar odorants (Figure 1A) (Kajiya et al, 2001).
Figure 1.
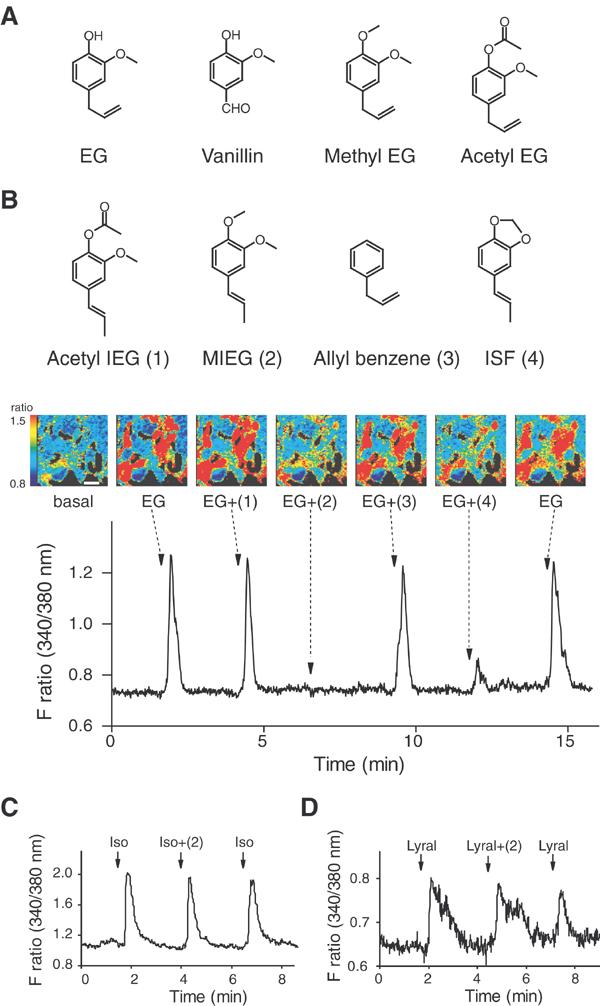
Inhibition of odorant-induced Ca2+ responses by odorants in HEK293 cells. (A) mOR-EG agonists. See Kajiya et al. (2001) for other mOR-EG agonists. EG, eugenol. (B) Inhibition of EG-induced Ca2+ increases in mOR-EG-expressing HEK293 cells by MIEG and ISF. EG (100 μM) was applied for 15 s at the times indicated by arrows with or without various odorants (1 mM) (±(1)–(4)). (Upper panel) Odorant compounds structurally similar to EG that were used for screening. Acetyl IEG (1), acetyl isoeugenol; MIEG (2), methyl isoeugenol; ISF (4), isosafrole. (Middle panel) Pseudocolored images of HEK293 cells at six representative time points of odorant applications. A change in fluorescent ratio intensities before and after application of each stimulus is shown in pseudocolored images, in which red equals the greatest change in fluorescent ratio. Scale bar, 30 μm. (Bottom panel) A representative Ca2+- response profile. (C) Effect of MIEG on isoproterenol-induced Ca2+ increases via endogenous β-adrenergic receptor in HEK293 cells. Isoproterenol (3 μM) was applied for 10 s at the times indicated by arrows with or without 1 mM MIEG (±(2)). (D) Effect of MIEG on lyral-induced Ca2+ increases in MOR23-expressing HEK293 cells. Lyral (1 mM) was applied for 15 s at the times indicated by arrows with or without 1 mM MIEG (±(2)).
The encoding of an odorant quality is determined by a combination of ORs, which is set for each odorant (Malnic et al, 1999; Kajiya et al, 2001). A receptor code for an odorant mixture, therefore, is expected to be the sum of the codes for its components. However, theoretical and psychophysical experiments showed that the perceived magnitude of an odorant mixture was neither additive nor a simple average of its components, but instead fell between these limits (Baker, 1964; Johns and Woskow, 1964; Cain and Drexler, 1974). This observation has been designated as masking (i.e. modification of perceived odor) or counteraction (i.e. reduction of odor intensity). Recent behavioral and psychophysical studies demonstrated that mixing some odorants led to the emergency of novel perceptual qualities that were not present in each component, suggesting that odorant mixture interactions occurred at some levels in the olfactory system (Jinks and Laing, 2001; Wiltrout et al, 2003).
Numerous electrophysiological studies in various vertebrate and invertebrate species also suggested that peripherally initiated mixture suppression resulted in less response to the mixture than that expected from simple additivity among the components (Ache, 1989; see the other chapters in Laing et al, 1989). Using a metabolic marker, 2-deoxyglucose, Bell et al, (1987) observed odorant mixture suppression predominantly in presynaptic receptor axons in the olfactory bulb, providing evidence that odorant mixture interaction begins at the peripheral neurons. It is, therefore, conceivable that odorants compete to bind the receptor sites and activate or antagonize olfactory neurons, resulting in a nonadditive receptor code for an odorant mixture.
Based on these observations, we hypothesized that odorants could activate ORs as agonists and also antagonize OR responses to odorants. Since ORs belong to the G protein-coupled receptor (GPCR) superfamily as the most common target of therapeutic drugs (Lefkowitz, 2000; Howard et al, 2001; Young and Trask, 2002), we reasoned that a standard medicinal chemistry approach could be applied to screen antagonist(s) that blocked a given OR. We adopted a Ca2+ imaging odorant-response assay in HEK293 cells expressing an OR and the promiscuous G protein, Gα15 (Krautwurst et al, 1998; Katada et al, 2003), for a high-throughput ligand screening system. We herein report the identification of odorants that inhibit EG response of mOR-EG, and the determination of their pharmacological properties as competitive antagonists in HEK293 cells and in single olfactory neurons that express mOR-EG. We further demonstrate antagonism between odorants at the level of the olfactory epithelium, and propose a concept of antagonism-based modulation of receptor codes for odorants.
Results
Screening of antagonistic odorants
Stimulation of mOR-EG-expressing HEK293 cells with its cognate ligand EG elicits an increase in intracellular Ca2+ level by virtue of Gα15 coupling to mOR-EG followed by an inositol phosphate-mediated signalling cascade (Kajiya et al, 2001; Katada et al, 2003). Odorant cocktails containing EG (100 μM) and a series of odorants were sequentially applied to screen odorant(s) that inhibited the EG response in HEK293 cells expressing mOR-EG and Gα15. Among approximately 500 tested odorants, we found that EG-induced Ca2+ responses were significantly inhibited by methyl isoeugenol (MIEG) or isosafrole (ISF) (Figure 1B). In the presence of 1 mM MIEG, Ca2+ responses to EG stimulation (100 μM) were almost completely inhibited (the average response was 8.4±2.1% (±s.e.) of that finally elicited by EG alone in 21 cells), while a residual Ca2+ response was observed for the ISF inhibition at the same concentration (the average response was 50.9±4.9% of that finally elicited by EG alone in 21 cells). Other odorants including structurally similar odorants (i.e. allyl benzene and acetyl isoeugenol; Figure 1B) showed no inhibitory activity. No Ca2+ response was observed when MIEG or ISF was applied alone even at 1 mM concentration, suggesting that these compounds were not an agonist or a partial agonist for mOR-EG (data not shown).
Ca2+ responses elicited by isoproterenol via endogenous β-adrenergic receptors were not affected by MIEG (Figure 1C). The average response to isoproterenol in the presence of MIEG was 117.8±5.6% of that finally elicited by isoproterenol in 26 cells. These results exclude the possibility that the inhibitory activity is due to nonspecific effects on downstream signalling via Gα15. In HEK293 cells that coexpressed MOR23 and Gα15, Ca2+responses evoked by lyral, an agonist of MOR23, were not inhibited by MIEG (Figure 1D). The average response to lyral in the presence of MIEG was 115.7±13.9% of that finally elicited by lyral in 10 cells. These results demonstrate that the antagonistic activity of MIEG is not a nonspecific event to any OR family.
Pharmacological characterization of antagonists
To determine whether the inhibition by MIEG or ISF was derived from specific competition at the ligand-binding site, dose–response properties were examined using different concentrations of MIEG and ISF (Figure 2A). EG-mediated Ca2+ responses were inhibited in a dose-dependent manner with IC50 values of 66 μM for MIEG and 742 μM for ISF (Figure 2B). It is of note that the IC50 value for MIEG was similar to the EC50 value for EG (46 μM) (Kajiya et al, 2001). The EG response was rescued when EG concentrations were increased at the fixed amount of MIEG (Figure 2C). The EG response curve was shifted to the right in the presence of MIEG (Figure 2D). These results suggest that MIEG is a competitive antagonist for mOR-EG with an appropriate order of potency in a HEK293 heterologous expression system.
Figure 2.
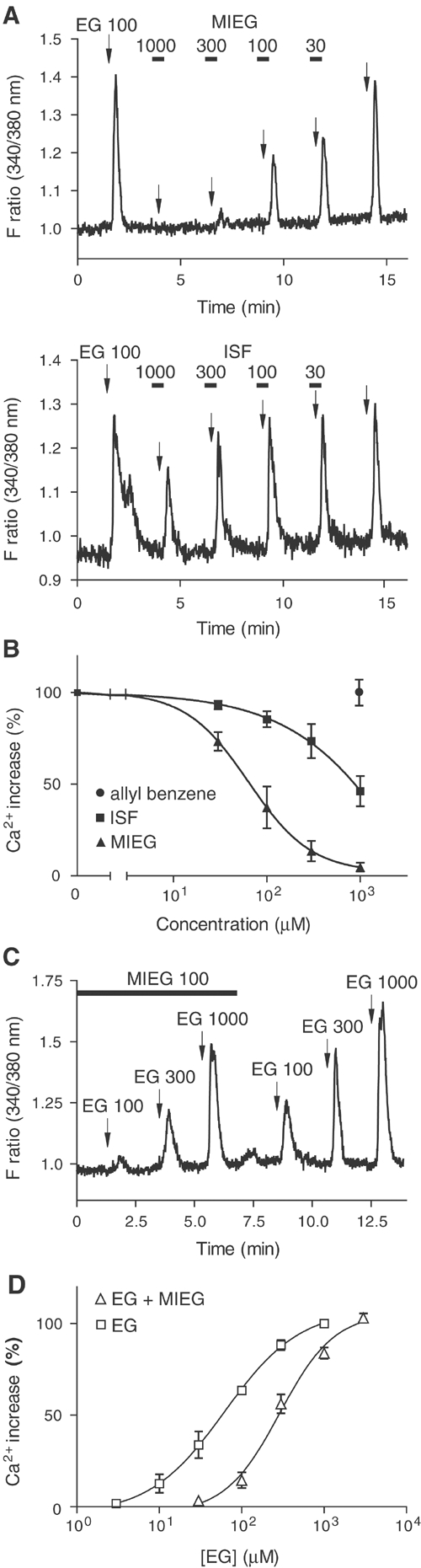
Pharmacological characterization of the antagonists for mOR-EG in HEK293 cells. (A) Dose-dependent inhibition of EG-induced Ca2+ increases by MIEG and ISF. In all, 100 μM EG (EG 100) was applied to mOR-EG-expressing HEK293 cells at the times indicated by arrows. Black bars indicate the periods during which 1000, 300, 100, and 30 μM MIEG or ISF were included in the buffer. The antagonist (MIEG or ISF) was applied 10 s prior to and during EG application. (B) Dose-dependent inhibition curves of mOR-EG by MIEG, ISF or allyl benzene as a percentage of the response to 100 μM EG. Each point represents the mean ± s.e. of five to six responding cells. •, allyl benzene; ▪, ISF; ▴, MIEG. (C) EG responses in the absence or presence of MIEG. In all, 100, 300, and 1000 μM EG were applied to the cells at the times indicated by arrows. The black bar indicates the period during which 100 μM MIEG was included in the buffer. (D) Dose–response curves of Ca2+ responses of mOR-EG to EG in the absence or presence of 100 μM MIEG. The data are shown as a percentage of the response to 1 mM EG without MIEG. Each point represents the mean ±s.e. of eight responding cells. △, EG response in the presence of 100 μM MIEG; □, EG response in the absence of MIEG.
Antagonism of odorant responses in single olfactory neurons
We next asked whether the antagonism of the EG response by MIEG occurred in the olfactory neurons expressing mOR-EG. Olfactory neurons suitable for odorant response recordings were isolated from the mouse olfactory epithelium located on the upper septum and roof (zone 1 among four distinct spatial receptor zones (Vassar et al, 1993)) where mOR-EG is expressed (Kajiya et al, 2001). Since OR-mediated cAMP increases trigger Ca2+ influx through a cyclic nucleotide-gated channel, the isolated olfactory neurons were subjected to Fura 2-based Ca2+ imaging to record responses to odorant stimulations (Schild and Restrepo, 1998; Touhara et al, 1999). In all, 300 μM EG was sequentially applied to the olfactory neurons in the absence or presence of 300 μM MIEG (Figure 3A). MIEG was applied 10 s prior to and during the second EG application so that the response to MIEG alone could be monitored at the times indicated by dotted lines in Figure 3A. The neurons in Figures 3A-3 and 4 showed the response to MIEG alone (dotted lines), while the neuron in Figure 3A-2 showed no response to MIEG but responded to the second EG application.
Figure 3.
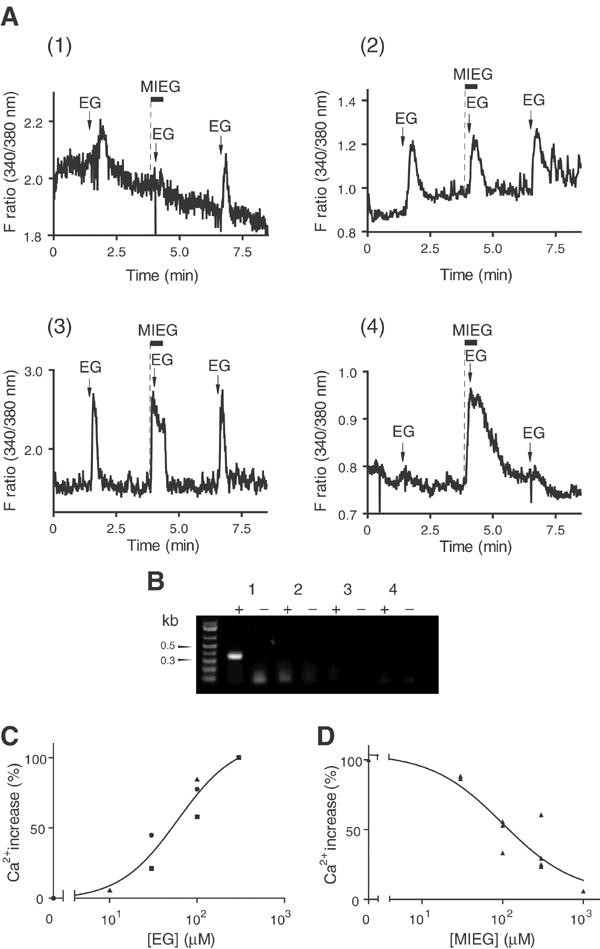
Ca2+ responses of mouse olfactory neurons to EG or MIEG followed by single-cell RT–PCR. (A) Representative Ca2+-response profiles of single olfactory neurons that responded to EG or MIEG. Responses were induced by repetitive application of 300 μM EG at the times indicated by arrows. The black bars indicate the periods during which 300 μM MIEG was applied. MIEG was applied 10 s prior to (dotted line) and during EG application. Four representative response profiles were shown: (1), EG response was antagonized by MIEG; (2), EG response was not antagonized by MIEG; (3), both EG and MIEG were responsive; (4), MIEG, but not EG, was responsive. (B) mOR-EG expression in single olfactory neurons. RT–PCR was performed using specific primers designed from mOR-EG, and the products were separated on 1.2% agarose gel. Plus or minus signs indicate RT–PCR experiment with or without reverse transcriptase using the same RNA preparation. Lanes 1–4 correspond to representative single-cell RT–PCR experiments of the neurons whose response profiles are shown in Figs. A1–4, respectively. (C) Dose–response curves of Ca2+ responses to EG in mOR-EG-expressing neurons. The data are shown as a percentage of the response to 300 μM EG. The data were obtained from three neurons that expressed mOR-EG, based on single-cell RT–PCR experiments. EG responses in the same neuron are shown by the same symbol: •, EG 30, 100, 300 μM; ▪, EG 30, 100, 300 μM; ▴, EG 10, 100, 300 μM. (D) Dose-dependent inhibition of the EG response by MIEG in mOR-EG-expressing neurons. The data are shown as a percentage of the response to 300 μM EG in the absence of MIEG. The dose-inhibition curve was obtained from the data of nine neurons that expressed mOR-EG, based on single-cell RT–PCR experiments.
Figure 4.
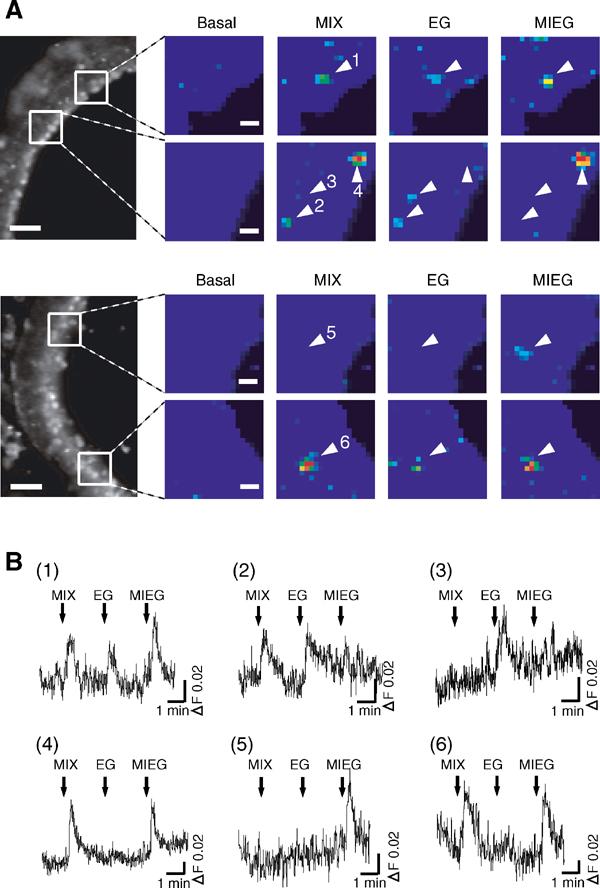
In situ Ca2+ imaging of the olfactory epithelium slices responding to EG, MIEG, or a mixture of EG and MIEG. (A) Pseudocolored images of Ca2+-measurement at 300 μM EG (EG), 300 μM MIEG (MIEG), and a mixture of 300 μM EG and 300 μM MIEG (MIX) (scale bar, 10 μm). A change in fluorescent ratio intensities before and after application of each stimulus is shown in pseudocolored images, in which red equals the greatest change in fluorescent ratio. The locations of images in the olfactory epithelium are shown on the left (scale bar, 50 μm). Arrowheads represent responding neurons. (B) Ca2+ response profiles of olfactory neurons shown by arrowheads in Figure 4A. A mixture of MIEG and EG (MIX), EG alone (EG), and MIEG alone (MIEG) were sequentially applied to the slice for 15 s at the times indicated by arrows. The data are representative of response profiles of cells from five recordings.
Of ∼3000 viable olfactory neurons, we could identify a total of 95 neurons that responded to EG. EG responses in 20 of the 95 neurons were antagonized by MIEG (representative data shown in Figure 3A-1). EG responses in 60 of the 95 cells were not inhibited by MIEG (representative data shown in Figure 3A-2). In 15 of the 95 neurons, the responses were evoked by both EG and MIEG (representative data shown in Figure 3A-3). The magnitudes of EG responses in each type of neuron were not significantly different. Thus, the average ΔF ratio upon EG stimulation was 0.39±0.07 for the type of neurons in Figure 3A-1, 0.39±0.04 for the type of neurons in Figure 3A-2, and 0.41±0.10 for the type of neurons in Figure 3A-3. In the neurons that showed inhibition of EG responses by MIEG (Figure 3A-1), 300 μM MIEG robustly inhibited the responses to 29.3±7.2% of that finally elicited by EG alone. In the neurons that were not antagonized by MIEG (Figure 3A-2), the average amplitude of EG response in the presence of MIEG was 100.3±1.4% of that finally elicited by EG alone. These results suggest that there exist at least three physiologically distinct populations of olfactory neurons that recognize EG in zone 1. We also found cells that responded to MIEG but not to EG (Figure 3A-4). Therefore, MIEG activated OR neurons as an agonist, but inhibited EG responses of other neurons as an antagonist.
Receptor gene amplification from single olfactory neurons
Next, 43 single cells of the 95 EG responsive neurons described above were subsequently picked in a microcapillary tube, and were subjected to single-cell RT–PCR (Touhara et al, 1999) using primers specific to mOR-EG. The mOR-EG fragment (∼380 bp) was amplified from six out of 12 single-isolated neurons in which inhibition of EG response by MIEG was observed (Figure 3B). The results clearly demonstrated that endogenous mOR-EG in olfactory neurons was antagonized by MIEG. The mOR-EG fragment was not amplified from the rest of the six cells that exhibited the antagonism by MIEG. It is, therefore, tempting to speculate that there exist other OR(s) whose activity is antagonized by MIEG. No mOR-EG expression was detected from the neurons whose EG responses were not antagonized by MIEG (0 out of 23 cells), or the neurons that responded to both EG and MIEG (0 out of eight cells) (Figure 3B). These results suggest that EG is recognized by multiple ORs that comprise two arrays of receptors: one type that exhibits no inhibition by MIEG, and another type, including mOR-EG, that is antagonized by MIEG. In mOR-EG-expressing neurons, the EC50 value for EG was about 52 μM (Figure 3C), and the IC50 value for MIEG was approximately 119 μM (Figure 3D). These values are in good agreement with those obtained in heterologous HEK293 cells. ISF exhibited weak antagonistic activity in mOR-EG-expressing neurons (data not shown).
Antagonism between odorants in the olfactory epithelium
We next examined whether the antagonism between EG and MIEG could be visualized by in situ Ca2+ imaging using an intact coronal slice of the olfactory epithelium of a newborn mouse (Omura et al, 2003). EG (300 μM), MIEG (300 μM), and a mixture of equal amounts of EG and MIEG (300 μM each) were applied to the slice preparation. As with the dissociated olfactory neurons, cells with various response profiles were identified (Figure 4). There were neurons that responded only to EG (Figures 4A-2, and B-2), only to MIEG (Figures 4A-4, 6, and B-4, 6), or to both EG and MIEG (Figures 4A-1, and B-1). These three types of neurons correspond to neurons shown in Figures 3A-2, A-4, and A-3, respectively. We also found cells that responded to EG but did not respond to the mixture of EG and MIEG (Figures 4A-3, and 4B-3), demonstrating the antagonism of EG responses by MIEG. This type of neuron corresponds to the neuron shown in Figure 3A-1. There also existed cells that responded to MIEG but did not respond to the mixture (Figures 4A-5, and B-5), showing the inhibition of MIEG responses by EG.
The proportion of various types of neurons with different response profiles in the slice imaging was consistent with that obtained in single-neuron recording experiments. Thus, we found a total of 17 neurons that responded to EG or MIEG in five recording areas (one recording area: 437 × 328 μm): eight cells responded only to EG, six cells responded only to MIEG, and three cells responded to both MIEG and EG. The Ca2+ responses in two of the eight cells, which responded only to EG, were completely blocked in the presence of MIEG, the response to the mixture in one of the eight cells was significantly reduced to 58.3% of that elicited by EG alone, and the responses in five of the eight cells were not affected by MIEG. The response in one of the six cells, which responded only to MIEG, was eliminated in the presence of EG, and five of the six cells showed no inhibition by EG.
Consequently, odorant mixture suppression resulted in a less number activated by the odorant mixture (14 cells) than the total number of EG- or MIEG-responsive neurons (17 cells) (14/17=82%, n=5 recordings). These results demonstrate that antagonism between odorants occurs in the olfactory epithelium when the agonist is mixed with an equal amount of the antagonist that exhibits an approximate order of potency in comparison with that of the agonist. Antagonism between odorants at the level of ORs partly accounts for a mechanism underlying odorant mixture suppression in the peripheral neuronal coding of an odorant mixture.
Discussion
In this study, we provided molecular and cellular evidence for the antagonism of OR activities between odorants. Inhibition of odorant responses of a given OR by different odorants was demonstrated in a heterologous OR-expression system and in single olfactory neurons that expressed the OR. Antagonism between odorants was also observed at the level of the olfactory epithelium. Our data indicate that odorants act as an agonist and an antagonist to ORs. Therefore, when antagonism occurs between components in an odorant mixture, the OR repertoire activated by the odorant mixture is not simply the sum of receptor codes for its components (Figure 5). The current study describes a molecular aspect in the odor-recognition mechanism in the olfactory sensing system that always perceives odorants as a mixture in real life.
Figure 5.
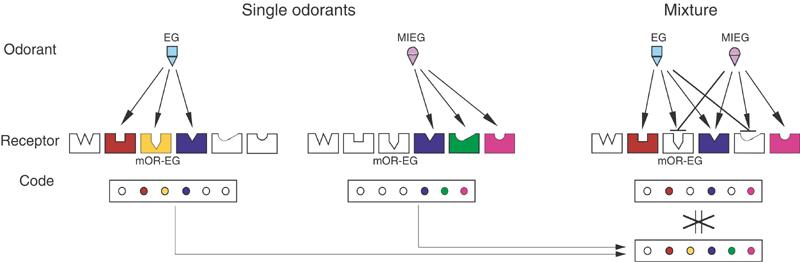
Schematic diagram of receptor coding for single odorants and an odorant mixture. Each odorant activates different but sometimes overlapping subsets of ORs, which define an identity code for an odorant. When the odorants are mixed, a code for the mixture is expected to be the sum of coding patterns of its components. However, when antagonism between the odorants occurs, a smaller number of receptors are activated than the additive number of activated receptors by single odorants. For example, EG is an agonist for the red, yellow, and blue receptors, but an antagonist for the green receptor. MIEG is an agonist for the blue, green, and pink receptors, but an antagonist for the yellow receptor that corresponds to mOR-EG.
Our results reminisce behavioral and psychophysical experiments, in that the perceived intensity or quality of an odorant mixture is not the sum of its components, which is called odorant mixture synergism, when it is greater, or mixture suppression, when it is less than expected (Laing et al, 1989). Although there is no simple way to correlate the observation at a perception level with that at a receptor level, the peripheral events investigated in this study may partly account for a mechanism underlying odorant mixture suppression that occurs when odorants are mixed. When the olfactory epithelium encounters an odorant mixture, stimulation of a combinatorial repertoire of ORs results in the creation of a complex spatial odor map, which is eventually transmitted to the higher cortical areas of the brain where a perception is constructed (Zou et al, 2001). Therefore, a receptor code resulting from OR antagonism between odorants in the peripheral neurons is likely maintained and expressed at the behavioral or perceptual level such that an odorant mixture exhibits a novel perceptual quality that is not present in the components.
It appears that peripheral mixture suppression is the result of molecular events at multiple levels. Odorant mixture interaction at the peripheral level could be derived from (1) OR antagonism as shown in this study, (2) inhibition at the second messenger transduction level (Spehr et al, 2002), or (3) suppression of the inward membrane current (Kurahashi et al, 1994). The possibility of antagonism at the level of ORs has also been suggested for other mammalian ORs (Araneda et al, 2000; Spehr et al, 2003). Our pharmacological analyses of receptor antagonism in HEK293 cells and single olfactory neurons that expressed a defined OR clearly demonstrated that odorant mixture suppression occurred at the receptor level.
In the course of examining the antagonism between EG and MIEG, we found at least three physiologically distinct types of olfactory neurons that responded to EG. It is an intriguing question as to what type of an OR is expressed in the individual neuron. Our preliminary results suggest that ORs, which are structurally related to mOR-EG, are expressed in these cells (unpublished results). Further characterization of these ORs will enable us to investigate molecular and structural information about ligand specificity, and will also allow for the identification of amino-acid residues that are involved in the ligand recognition.
The concept of antagonism has been well established for other GPCRs, at which almost 50% of the pharmaceuticals appear to be targeted. Deducing a structure–activity relationship from profiles of agonists and antagonists of ORs will definitely stimulate the field of drug discovery. Having a repertoire of agonists and antagonists, a combination of computational and mutational strategies will enable us to predict a ligand-binding site and to elucidate molecular bases for ligand discrimination. Further, examining an antagonist-bound form that represents the inactive state will provide insight into a molecular basis for agonist-induced conformational changes of GPCRs.
Extensive analysis of ligand specificity has also been carried out for the rat I7 odorant receptor, showing that the I7 receptor recognized octanal as a primary agonist, and its structurally similar odorant, citral, behaved as a partial agonist or antagonist (Araneda et al, 2000). Most recently, an inhibitor for human OR17-4 was identified to be undecanal, which possessed an aldehyde group as a common functional group with the ligands, bourgenonal and cyclamal (Spehr et al, 2003). These reports and the current study clearly demonstrated that antagonists tend to be structurally related to the agonists, as is often the case for other GPCRs.
Natural fragrant materials usually contain structurally similar odorants (Arctander, 1960). Thus, antagonism between odorants likely happens when we perceive the fragrance of an essential oil or extracts of natural origin. We also experience that odorant quality sometimes changes dramatically by mixing a small amount of a particular odorant, which is empirically well recognized among perfumers. It is of great interest to examine whether such combinations of odorants prove to be an agonist–antagonist relationship for some ORs. Therefore, the concept of antagonism between odorants provides a strategy to control perceived odor quality of malodors or perfumes, which is potentially applicable to odor-modulation technologies (Gilbert and Firestein, 2002).
In conclusion, detailed pharmacological characterization of an OR revealed a previously unexplored strategy utilized for the receptor code scheme in the olfactory sensing system. The fact that odorants possess dual functions as an agonist and an antagonist to ORs provides complexity in the encoding mechanism of an odorant mixture at the receptor level. Emerging evidence on odorant–OR interaction by diverse approaches in pharmacology, medicinal chemistry, and structural biology will eventually converge on fully understanding chemical sensing in the olfactory system. Together with the previous electrophysiological and biochemical studies, molecular evidence for the peripheral OR antagonism provides one of the likely explanations for odorant mixture interactions leading to novel perceptual qualities of odorant mixtures. The concept of antagonism between odorants offers a basis for a receptor-targeted approach to block the perception of specific classes of smells, and eventually to design pleasing scents or flavors. In this context, it is provocative to address that the current study provides opportunities to control perceived odor quality specifically and noninvasively, which is eventually applicable to odor-modulation technologies with implications for a better quality of life.
Materials and methods
Odorant solutions for antagonist screening
Odorants utilized in this study were kindly provided by T Hasegawa Co. Ltd and Takasago International Corp., or purchased from Wako Co. Ltd (Tokyo, Japan) or Sigma-Aldrich (Tokyo, Japan). Isoproterenol was purchased from Sigma-Aldrich (Tokyo, Japan). Odorants were kept at 4°C and their purities were checked by thin-layer chromatography before experiments, since we have experienced that many odorants decomposed during storage under air. Odorants were diluted to the indicated concentrations in Ringer's solution (in mM: 140 NaCl, 5.6 KCl, 2 CaCl2, 2 MgCl2, 2 sodium pyruvate, 9.4 glucose, and 10 HEPES, pH 7.4) before experiments. Approximately 500 odorants were randomly divided into groups that consisted of four different odorants in each group. The odorant cocktails, which contained 100 μM EG and four different odorants at 300 μM each, were sequentially applied to mOR-EG-expressing HEK293 cells for initial screening of antagonistic odorants. Each odorant at 1 mM concentration was then individually tested for antagonistic activity to 100 μM EG response. To prevent artifactual responses, odorant solutions were prepared at total concentrations of less than 1.5 mM.
Ca2+ imaging in HEK293 cells
Ca2+ imaging for mOR-EG-expressing HEK293 cells was performed essentially as described previously (Katada et al, 2003). Briefly, 60–70% confluent HEK293 cells were transfected with 2.0 μg of pME18S-tagged-mOR-EG and 1.5 μg of pME18S-Gα15 by using Lipofectamine 2000 (Invitrogen) (2.5 μl per 1 μg DNA). At 24–28 h post-transfection, the transfected cells were loaded with 4 μM Fura 2/AM for 30 min at 37°C. Odorant solutions were sequentially applied to the cells using a peristaltic pump at a flow rate of 1.5 ml/min for 15 s with a 2.5 min wash with Ringer's solution between stimulus applications. Intracellular Ca2+ levels were monitored using an AQUA COSMOS Ca2+-imaging system (Hamamatsu Photonics). When inhibitory odorants were screened (Figure 1), each odorant was coapplied with EG. In order to perform quantitative pharmacological studies (Figure 2), an antagonist was applied 10 s prior to and during the EG application.
Ca2+imaging of mouse olfactory neurons
Olfactory neurons for Ca2+ imaging were isolated from the olfactory epithelium of 3–4-week-old BALB/c CrSlc mice (Japan SLC, Hamamatsu, Japan) on Cell-TAK (Collaborative Research, Bedford, MA)-coated glass-base dishes, and Fura-2-based Ca2+ responses were recorded as described before (Touhara et al, 1999). Odorant solutions were sequentially applied for 10 s with a 2.5 min wash between stimuli. An antagonist (MIEG) was applied 10 s prior to and during the agonist (EG) application (Figure 3) so that MIEG responses of olfactory neurons could be distinguished from EG responses.
In situ Ca2+ imaging of mouse olfactory epithelium slices
In situ Ca2+ imaging in mouse olfactory epithelium slices was performed as described previously (Omura et al, 2003). Briefly, coronal slices (50–70 μm) of the mouse olfactory epithelium from 3–4-day-old BALB/c CrSlc mice were prepared using Microslicer DTK-3000 (DoSaKa EM). After the slice was loaded with 10 μM Fura-PE3/AM (Calbiochem) for 1 h, Ca2+ responses were recorded under 150 × magnification for simultaneous monitoring of a 437 × 328 μm area. Fluorescent images were acquired at ∼1.2 s intervals with a resolution of 164 × 123 pixels, resulting in a pixel size of 2.7 μm. The ratio analysis was carried out with a series of 4 × 4 pixel regions through the entire epithelium layer as described (Omura et al, 2003). Odorant solutions were sequentially applied for 15 s with a 2.5 min wash between stimuli. In order to examine odor mixture interactions, EG and MIEG were applied to the epithelium slice together as a mixture.
Single-cell RT–PCR
Single odorant-responsive neurons were subjected to RT–PCR analysis using a slightly modified procedure from that described previously (Touhara et al, 1999). After an odorant-responsive olfactory neuron was isolated in a microglass capillary tube, the crude RNA was purified through a Micro-to-Midi RNA Purification System (Invitrogen). Amplification of the mOR-EG fragment was performed with the two-round PCR protocol: the first amplification using a mOR-EG specific primer (5′-CTCCATTGTTGCTCCCAAGATGCTGGTAAATCTAG-3′) and a NotI-adaptor primer designed from d(T)18-NotI that was used for the first-strand cDNA synthesis, and the second amplification using a set of mOR-EG specific primers (5′-ATTGGGATCCTATGCTTGGGGGGTGG-3′ and 5′-GGTATGCCTGGAGTTCTTGGAGTTAGGT-3′). Both amplifications were performed using AmpliTaq Gold (2.5 units/50 μl) according to the following schedule: 10 min treatment at 95°C followed by 40 PCR cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min by using a GeneAmp PCR Cycler (Applied Biosciences).
Acknowledgments
We thank T Hasegawa Co Ltd and Takasago International Corp for odorant compounds, and H Watanabe for valuable discussions. This work was supported in part by grants from MEXT and PROBRAIN Japan. KT is a recipient of grants from Uehara Memorial Foundation, Kato Memorial Bioscience Foundation, The Naito Foundation, and The Mochida Memorial Foundation.
References
- Ache BW (1989) Central and peripheral bases for mixture suppression in olfaction; a crustacean model. In Laing DG, Cain WS, McBride RL , Ache BW (eds). Perception of Complex Smell and Tastes. Sydney, Australia: Academic Press pp. 101–114 [Google Scholar]
- Araneda RC, Kini AD, Firestein S (2000) The molecular receptive range of an odorant receptor. Nat Neurosci 3: 1248–1255 [DOI] [PubMed] [Google Scholar]
- Arctander S (1960) Perfume and Flavor Materials of Natural Origin. Arctander S (ed). Elizabeth, N.J.: Arctander, Pub.
- Baker RA (1964) Response parameters including synergism– antagonism in aqueous odor measurement. Ann NY Acad Sci 116: 495–503 [DOI] [PubMed] [Google Scholar]
- Bell GA, Laing DG, Panhuber H (1987) Odour mixture suppression: evidence for a peripheral mechanism in human and rat. Brain Res 426: 8–18 [DOI] [PubMed] [Google Scholar]
- Buck LB (1996) Information coding in the vertebrate olfactory system. Annu Rev Neurosci 19: 517–544 [DOI] [PubMed] [Google Scholar]
- Cain WS, Drexler M (1974) Scope and evaluation of odor counteraction and masking. Ann NY Acad Sci 237: 427–439 [DOI] [PubMed] [Google Scholar]
- Firestein S (2001) How the olfactory system makes sense of scents. Nature 413: 211–218 [DOI] [PubMed] [Google Scholar]
- Gaillard I, Rouquier S, Pin JP, Mollard P, Richard S, Barnabe C, Demaille J, Giorgi D (2002) A single olfactory receptor specifically binds a set of odorant molecules. Eur J Neurosci 15: 409–418 [DOI] [PubMed] [Google Scholar]
- Gilbert AN, Firestein S (2002) Dollars and scents: commercial opportunities in olfaction and taste. Nat Neurosci 5 Suppl: 1043–1045 [DOI] [PubMed] [Google Scholar]
- Howard AD, McAllister G, Feighner SD, Liu Q, Nargund RP, Patchett LHTVdPaAA (2001) Orphan G-protein-coupled receptors and natural ligand discovery. Trends Pharm Sci 22: 132–140 [DOI] [PubMed] [Google Scholar]
- Jinks A, Laing DG (2001) The analysis of odor mixtures by humans: evidence for a configurational process. Physiol Behav 72: 51–63 [DOI] [PubMed] [Google Scholar]
- Johns FN, Woskow MH (1964) On the intensity of odor mixtures. Ann NY Acad Sci 116: 484–494 [DOI] [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K (2001) Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci 21: 6018–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Nakagawa T, Kataoka H, Touhara K (2003) Odorant response assays for a heterologously expressed olfactory receptor. Biochem Biophys Res Commun 305: 964–969 [DOI] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reed RR (1998) Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95: 917–926 [DOI] [PubMed] [Google Scholar]
- Kurahashi T, Lowe G, Gold GH. (1994) Suppression of odorant responses by odorants in olfactory receptor cells. Science 265: 118–120 [DOI] [PubMed] [Google Scholar]
- Laing DG, Cain WS, McBride RL, Ache BW (1989) Perception of Complex Smell and Tastes. Sydney, Australia: Academic Press [Google Scholar]
- Lefkowitz RJ (2000) The superfamily of heptahelical receptors. Nat Cell Biol 2: E133–136 [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors. Cell 96: 713–723 [DOI] [PubMed] [Google Scholar]
- Mombaerts P (1999) Seven-transmembrane proteins as odorant and chemosensory receptors. Science 286: 707–711 [DOI] [PubMed] [Google Scholar]
- Omura M, Sekine H, Shimizu T, Kataoka H, Touhara K (2003) In situ Ca2+ imaging of odor responses in a coronal olfactory epithelium slice. NeuroReport 14: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Schild D, Restrepo D (1998) Transduction mechanisms in vertebrate olfactory receptor cells. Physiol Rev 78: 429–466 [DOI] [PubMed] [Google Scholar]
- Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H (2003) Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299: 2054–2058 [DOI] [PubMed] [Google Scholar]
- Spehr M, Wetzel CH, Hatt H, Ache BW (2002) 3-Phosphoinositides modulate cyclic nucleotide signaling in olfactory receptor neurons. Neuron 33: 731–739 [DOI] [PubMed] [Google Scholar]
- Touhara K (2002) Odor discrimination by G protein-coupled olfactory receptors. Microsc Res Tech 58: 135–141 [DOI] [PubMed] [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T (1999) Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci USA 96: 4040–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R (1993) Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74: 309–318 [DOI] [PubMed] [Google Scholar]
- Wetzel CH, Oles M, Wellerdieck C, Kuczkowiak M, Gisselmann G, Hatt H (1999) Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J Neurosci 19: 7426–7433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltrout C, Dogra S, Linster C (2003) Configurational and nonconfigurational interactions between odorants in binary mixtures. Behav Neurosci 117: 236–245 [DOI] [PubMed] [Google Scholar]
- Young JM, Trask BJ (2002) The sense of smell: genomics of vertebrate odorant receptors. Hum Mol Genet 11: 1153–1160 [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S (2002) The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5: 124–133 [DOI] [PubMed] [Google Scholar]
- Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S (1998) Functional expression of a mammalian odorant receptor. Science 279: 237–242 [DOI] [PubMed] [Google Scholar]
- Zou Z, Horowitz LF, Montmayeur JP, Snapper S, Buck LB (2001) Genetic tracing reveals a stereotyped sensory map in the olfactory cortex. Nature 414: 173–179 [DOI] [PubMed] [Google Scholar]


