Abstract
Eucaryotic gene expression requires chromatin-remodeling activities. We show by time-course studies that transcriptional induction of the yeast glucose-regulated SUC2 gene is rapid and shows a striking biphasic pattern, the first phase of which is partly mediated by the general stress transcription factors Msn2p/Msn4p. The SWI/SNF ATP-dependent chromatin-remodeling complex associates with the promoter in a similar biphasic manner and is essential for both phases of transcription. Two different histone acetyltransferases, Gcn5p and Esa1p, enhance the binding of SWI/SNF to the promoter during early transcription and are required for optimal SUC2 induction. Gcn5p is recruited to SUC2 simultaneously with SWI/SNF, whereas Esa1p associates constitutively with the promoter. This study reveals an unusual transcription pattern of a metabolic gene and suggests a novel strategy by which distinct chromatin remodelers cooperate for the dynamic activation of transcription.
Keywords: biphasic, Esa1, Gcn5, stress, SWI/SNF
Introduction
Programming the expression of the genome is essential for the cellular response to a variety of signals that regulate metabolism, cell growth, differentiation, and development. Defined sets of genes are induced at specific developmental stages or upon environmental changes. The transcription of such genes must start at the right time and continuously integrate both extracellular stimuli and the cellular outputs in response to these stimuli. Accordingly, transcription of some genes occurs immediately, while transcription of others occurs days after exposure to a stimulus. The duration of transcription may be short or prolonged, and the transcription pattern may be constant or cyclical.
Transcription by RNA polymerase II (pol II) requires the concerted action of a large number of proteins that must be recruited to the target promoter. Each of the steps leading to formation of a transcript must contend with the repressive structure of chromatin, the basic unit of which is the nucleosome, which consists of 146 bp of DNA wrapped around an octamer of the four histones (Luger et al, 1997). The conformation of nucleosomes and the modification state of histones, both of which impact higher-order chromatin structure, are believed to be major determinants of localized chromatin structure. Importantly, the activity of a gene is largely dictated by the chromatin structure in which it resides (Wolffe, 1998), which can be modulated by enzymes that reversibly remodel chromatin.
Chromatin-remodeling enzymes, which are often part of large protein complexes, have been grouped into two major categories—ATP-dependent chromatin remodelers and covalent histone modifiers. ATPase remodelers, such as the yeast SWI/SNF complex, induce conformational changes in nucleosomes by altering DNA–histone interactions (Vignali et al, 2000; Martens and Winston, 2003); histone modifiers catalyze post-translational modifications of histones. Histone acetylation, the first modification shown to correlate strongly with transcriptional competence (Allfrey et al, 1964; Struhl, 1998), is controlled by the antagonistic activities of histone acetyltransferases (HATs) and deacetylases (HDACs) (Khochbin et al, 2001; Roth et al, 2001).
Local increases in chromatin accessibility can be achieved by the recruitment of chromatin-remodeling complexes by gene-specific transcription factors or components of the general transcription apparatus (Peterson and Workman, 2000; Vignali et al, 2000; Cosma, 2002; Sharma et al, 2003). Gene transcription often requires both ATP-dependent chromatin-remodeling complexes and HATs, and recent studies have underscored the critical importance of the interplay between these two types of activities in the regulation of transcription. For example, yeast SWI/SNF is required globally for transcription-associated histone acetylation during mitosis when chromatin is condensed (Krebs et al, 2000); conversely, histone acetylation may facilitate the affinity with which SWI/SNF binds to chromatin (Hassan et al, 2001, 2002; Agalioti et al, 2002). Studies of several promoters that are induced during differentiation or development have revealed that individual ATP-dependent and HAT remodeling enzymes are recruited temporally and that the order can vary at different promoters (Cosma et al, 1999; Krebs et al, 1999; Agalioti et al, 2000; Fry and Peterson, 2001; Reinke et al, 2001; Cosma, 2002; Soutoglou and Talianidis, 2002). Nevertheless, similar kinetic studies have not yet been carried out in genes whose induction occurs more rapidly (e.g., genes involved in stress response or metabolism); such genes may employ different strategies to establish active chromatin structure.
Here, we analyze by time course the transcriptional induction of the yeast glucose-regulated SUC2 gene in response to acute glucose limitation. We discovered that induction of SUC2 is rapid and, unexpectedly, proceeds in two distinct phases, the first of which is a stress response. SWI/SNF is essential for both phases of gene induction and associates with the SUC2 promoter in a biphasic manner. Moreover, both the Gcn5p and Esa1p HATs facilitate the association of SWI/SNF with the promoter for optimal SUC2 induction. Gcn5p is recruited to the promoter concurrently with SWI/SNF, whereas Esa1p associates constitutively with SUC2. Our study suggests a novel strategy by which distinct chromatin remodelers cooperate in activation of dynamic transcription.
Results
Acute glucose limitation induces biphasic transcription of a glucose-repressible gene
When limiting, the preferred carbon sources glucose and fructose can be derived from hydrolysis of other sugars (e.g., raffinose) by secreted invertase, which is encoded by the SUC2 gene. Transcription of SUC2 is therefore repressed by glucose and fructose and induced when glucose and fructose fall below certain threshold levels (Carlson, 1999; Herwig et al, 2001).
To better understand the transcriptional regulation of SUC2, we have studied its induction by an abrupt change of carbon source. We first monitored SUC2 RNA levels in a time course following a rapid shift of yeast cells from glucose to raffinose (containing low glucose) medium. To our surprise, SUC2 RNA accumulated rapidly and in two distinct phases: a short first phase in which transcript levels peaked at 45 min after induction and a prolonged second phase in which RNA reached steady-state levels at 2 h (Figure 1A). Shifting to a medium containing only low glucose (no raffinose) resulted in a similar biphasic pattern, except that SUC2 RNA levels dropped to noninducing levels by 4 h (Figure 1A; data not shown). These results suggest that the biphasic transcription of SUC2 is primarily a response to low glucose and that raffinose is required for the maintenance of SUC2 transcription, as was observed previously (Ozcan et al, 1997; Recht and Osley, 1999; Geng et al, 2001).
Figure 1.
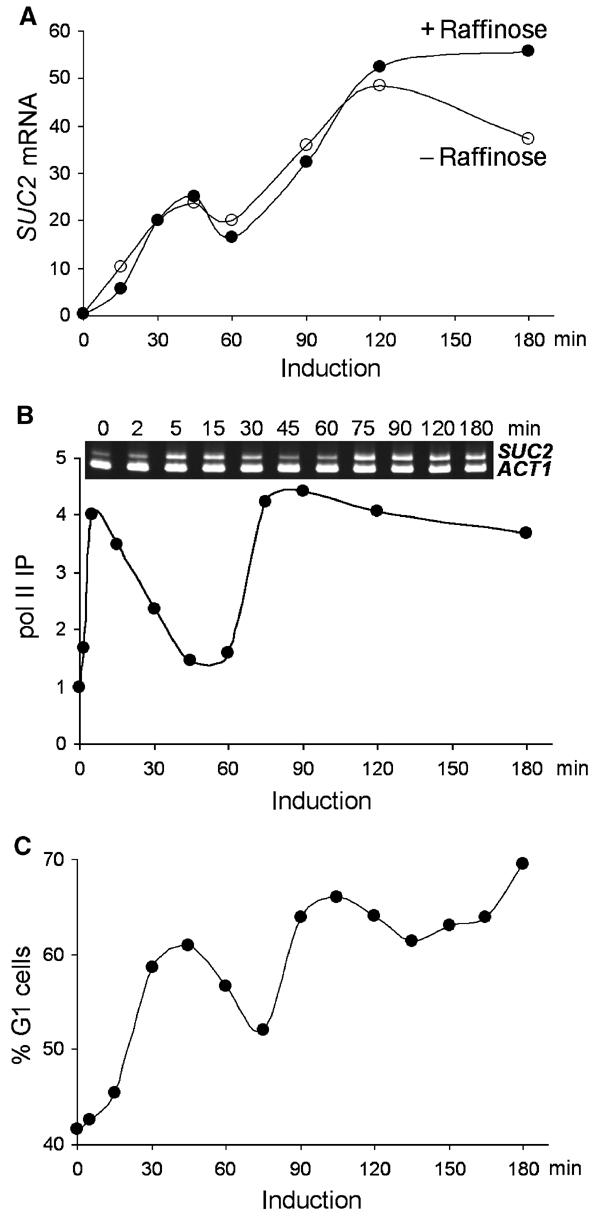
Biphasic induction of SUC2. Wild-type (BLY1) cells were grown in glucose (2%) medium to early logarithmic phase, and then quickly shifted to low-glucose (0.05%) medium containing raffinose (2%) unless noted. (A) Biphasic accumulation of SUC2 mRNA. SUC2 transcripts were quantified by real-time PCR in glucose-repressed cells (time 0) and cells induced in low-glucose media with (+ raffinose) or without (− raffinose) raffinose for the indicated times. The levels of SUC2 transcripts were presented as percentages of ACT1 transcripts. (B) Dynamic recruitment of pol II to SUC2. ChIP analysis using anti-pol II CTD antibody was performed on crosslinked chromatin prepared from glucose-repressed or raffinose-induced cells. IP efficiencies of the SUC2 UAS sequence (nucleotides −154 to +45 relative to the translational start site) at different time points were determined by real-time PCR quantitation and presented as the fold increases relative to that at time 0. A semiquantitative multiplex PCR analysis of the precipitated DNA (upper panel) is also shown. A sequence of the ACT1 promoter was co-amplified as an internal control. The PCR products were separated on an 8% polyacrylamide gel and stained with ethidium bromide. (C) Fluctuation of the G1 (unbudded) cell fractions. Cells were withdrawn from a raffinose-induced culture and fixed immediately in 3.7% formaldehyde. The mitotic index was determined microscopically and presented as the percentages of G1 cells.
To correlate the SUC2 mRNA levels with active SUC2 transcription, we measured the amount of pol II present at the SUC2 promoter in chromatin immunoprecipitation (ChIP) experiments. The level of pol II crosslinking at the SUC2 promoter (TATA box) increased four-fold immediately following the carbon source shift, and its presence at the promoter also showed a biphasic pattern that slightly preceded that of the SUC2 RNA (Figure 1B). As a control, the levels of pol II associated with the ACT1 promoter did not vary significantly during SUC2 induction.
To probe the more general cellular response to acute glucose limitation, we determined the fraction of unbudded cells (cells in G1) in a time course following the carbon source shift. The transition from G1 to S phase is tightly controlled by the availability of carbon source, and glucose deprivation causes G1 arrest (Alberghina et al, 1998; Newcomb et al, 2003). Remarkably, the pattern of cells in G1 following the carbon source shift was also biphasic (Figure 1C). The beginning of each phase correlated with the binding of pol II to the SUC2 promoter (Figures 1C and B). We interpret the accumulation of cells in G1 during limiting glucose as an indication that cells are experiencing a shortage of metabolic glucose. The biphasic accumulation of cells in G1 following acute glucose limitation suggests a dynamic change in cellular glucose metabolism, which likely underlies the biphasic induction of SUC2.
Distinct chromatin-remodeling activities associate with the SUC2 promoter
Under glucose-repressing conditions, the SUC2 promoter is packaged into an array of evenly positioned nucleosomes, which are remodeled in an SWI/SNF-dependent manner upon gene induction (Hirschhorn et al, 1992; Matallana et al, 1992; Gavin and Simpson, 1997; Wu and Winston, 1997; Geng et al, 2001). SWI/SNF is essential not only for the initiation but also for the maintenance of SUC2 transcription (Biggar and Crabtree, 1999; Sudarsanam et al, 1999). Gcn5p, a histone H3/H2B acetyltransferase and component of both the SAGA and ADA complexes (Vignali et al, 2000), has also been implicated in maximally inducing SUC2 (Pollard and Peterson, 1997; Biggar and Crabtree, 1999; Recht and Osley, 1999; Sudarsanam et al, 1999), but the mechanism by which Gcn5p stimulates SUC2 transcription has not been elucidated.
Since transcription can be regulated at multiple steps, we tested whether the biphasic induction of SUC2 results from the temporally regulated recruitment of SWI/SNF and/or Gcn5p. In parallel, we also assessed the role of a second histone acetyltransferase, Esa1p, which specifically acetylates histones H4 and H2A and is the catalytic subunit of the NuA4 HAT complex (Allard et al, 1999; Vogelauer et al, 2000; Suka et al, 2001). Esa1p is essential for cell cycle progression and is required for transcription of ribosomal protein genes and several other genes (Allard et al, 1999; Clarke et al, 1999; Galarneau et al, 2000; Reid et al, 2001). We measured the crosslinking of Snf5p (a core subunit of SWI/SNF), Gcn5p, and Esa1p to the SUC2 promoter in ChIP experiments using specific polyclonal antibodies against each protein. Crosslinking of both Snf5p and Gcn5p to the SUC2 upstream activating sequence (UAS) increased seven- and six-fold, respectively, within 5 min following the carbon source shift (Figure 2A, upper panel) and displayed biphasic patterns similar to that of pol II, although slightly less Snf5p and Gcn5p were crosslinked to SUC2 during the second phase. In contrast, the binding of Esa1p increased two-fold within the first 5 min but decreased quickly to preinduction levels.
Figure 2.
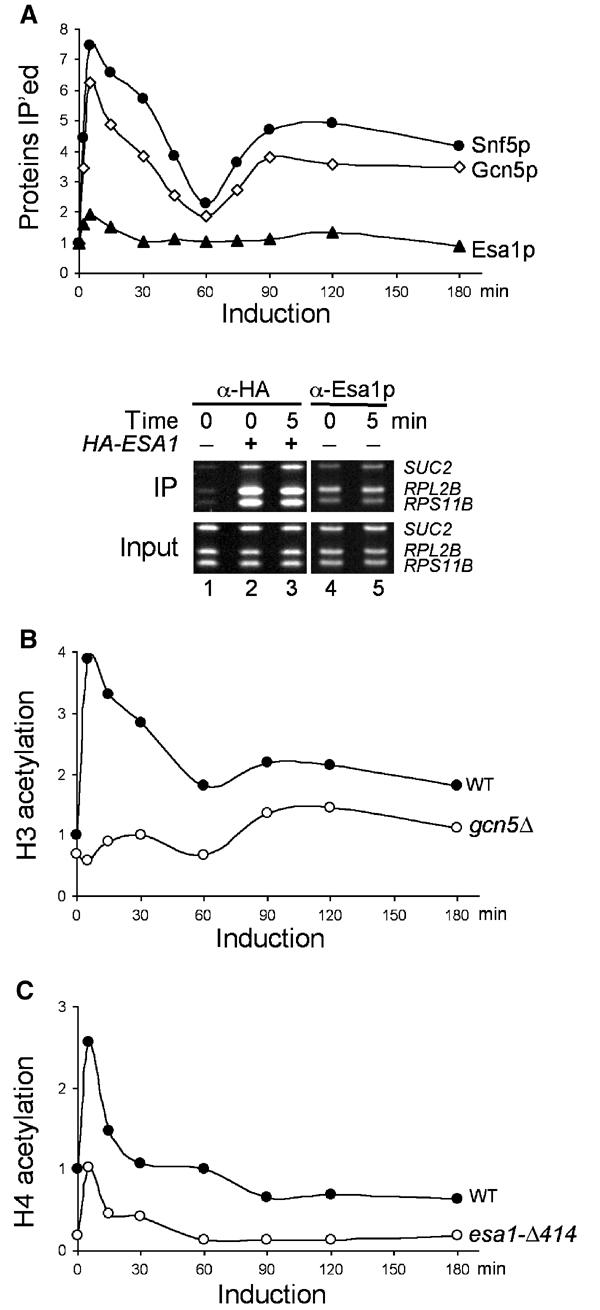
Snf5p and histone acetyltransferase activities associate with the SUC2 promoter. (A) The crosslinking of Snf5p, Gcn5p, and Esa1p to the SUC2 UAS DNA was analyzed by ChIP in raffinose-induced wild-type (BLY1) cells using polyclonal antibodies against each protein and the levels (IP efficiencies) presented as fold over preinduction levels (upper panel). ChIP experiments using anti-HA or anti-Esa1p antibodies were performed in HA-ESA1 (BLY431) and ESA1 (BLY1) cells that were glucose repressed (time 0) or derepressed for 5 min. The SUC2 UAS and the promoter sequences of two ribosomal genes were amplified by PCR from the precipitated (IP) and input DNAs and stained with ethidium bromide after separation on 8% polyacrylamide gels (lower panel). (B) The acetylation of histone H3 at SUC2 UAS in wild-type (BLY1) and gcn5Δ (BLY417) cells was compared by ChIP using an antibody against diacetylated H3 (K9 and K14). The levels of acetylation (IP efficiencies) were shown relative to wild-type preinduction levels. (C) The levels of acetylation of H4 at SUC2 UAS in wild-type (BLY1) and esa1-Δ414 (BLY457) cells were compared by ChIP as in (B) using an antibody against tetra-acetylated H4 (K5, K8, K12, and K16).
To confirm that preinduction levels of Esa1p were significantly higher than background at SUC2, we repeated ChIP assays in a strain expressing HA-tagged Esa1p (Figure 2A, lower panel). The SUC2 UAS and flanking sequences were precipitated by anti-HA antibody much more efficiently from tagged than from untagged strains under glucose-repressing conditions (compare lanes 1 and 2; see also Figure 3C, lower panel). In addition, ChIP signals for SUC2 increased slightly 5 min following the shift to low glucose (compare lanes 2 and 3). Esa1p also associated strongly with the promoters of two ribosome protein genes, RPL2B and RPS11B, consistent with a previous report (Reid et al, 2001). Importantly, similar patterns of Esa1p association with SUC2, RPL2B, and RPS11B were observed in ChIP experiments carried out with anti-Esa1p antibody (compare lanes 2 and 3 with lanes 4 and 5), thus validating the use of anti-Esa1p antibody in this study. We conclude that a significant amount of Esa1p associates constitutively with the SUC2 promoter.
Figure 3.
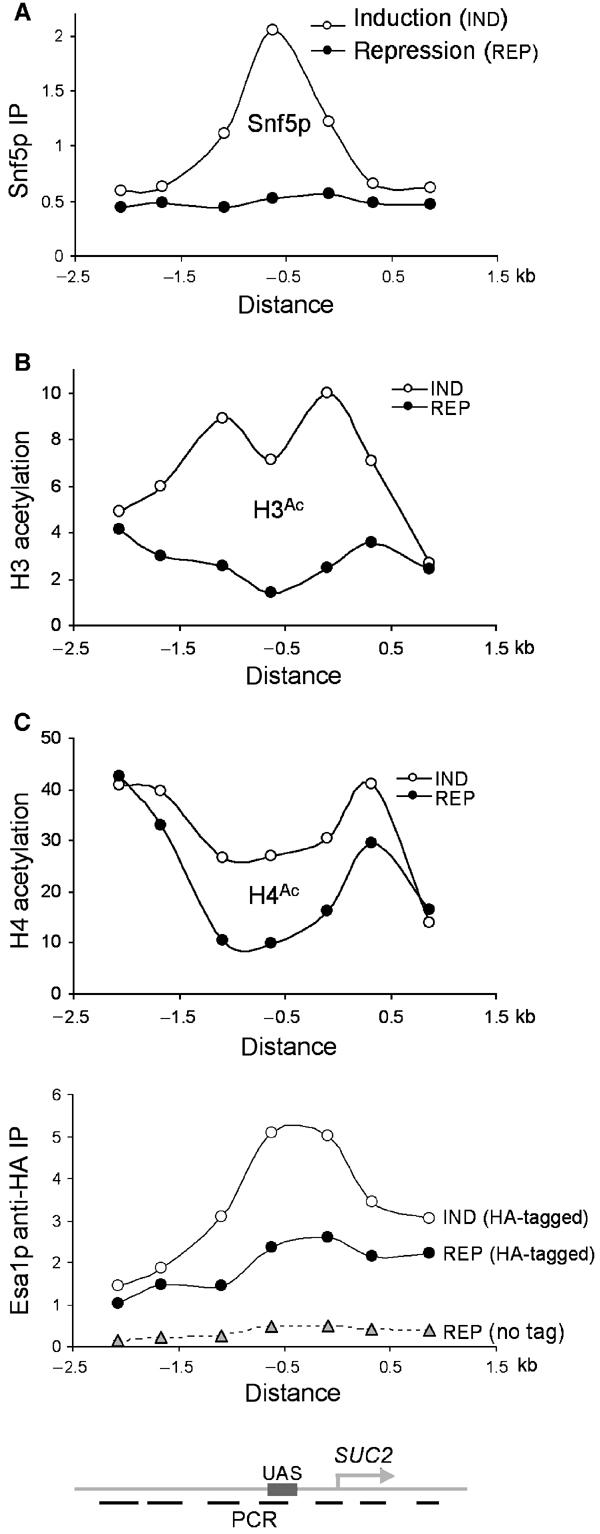
SUC2 promoter-specific association of chromatin remodelers or remodeling activities. (A) Crosslinking of Snf5p to the different SUC2 sequences (shown schematically below (C)) was determined by measuring [α-32P]-dCTP-labeled multiplex PCR products of the precipitated and input DNAs from the same experiment for Figure 2A. IP efficiencies of SUC2 sequences were shown relative to that of a subtelomeric sequence (Vogelauer et al, 2000). The distribution pattern of Snf5p, 5 min following induction, is shown. The distances are relative to the translational start site. The binding of Snf5p to SUC2 following gene induction is under-represented for the sequences tested, as the levels of Snf5p crosslinking to the subtelomeric sequence following SUC2 induction also increased slightly. The distribution of H3 (B) and H4 (C, upper panel) acetylation at SUC2 was determined and presented as in (A). The distribution of Esa1p (C, lower panel) was similarly assayed from anti-HA ChIP experiments, except that IP efficiencies were shown in arbitrary units for comparison between HA-tagged and untagged (no tag) strains.
To correlate the presence of Gcn5p and Esa1p at SUC2 with their histone acetyltransferase activities, we compared the histone acetylation levels in wild-type and gcn5 or esa1 mutant cells. In the wild type, the overall H3 acetylation at the SUC2 UAS increased four-fold immediately following induction and correlated well with Gcn5p cross-linking at SUC2 throughout the experiment (Figure 2B). In gcn5Δ cells, the increase in H3 acetylation was largely abolished (Figure 2B), suggesting that Gcn5p is the major HAT responsible for H3 acetylation during SUC2 activation.
The overall H4 acetylation at SUC2 increased transiently following the carbon shift in the wild type (Figure 2C). In the conditional esa1-Δ414 mutant, the basal levels of H4 acetylation and the induced increase in H4 acetylation were both significantly reduced compared to the wild type, suggesting that Esa1p contributes to both the basal and induced H4 acetylation at SUC2.
To address the specificity of the association of these chromatin-remodeling activities with SUC2, we assessed the crosslinking of Snf5p, Esa1p, and acetylated H3 and H4 at different DNA sequences along the SUC2 locus. At 5 min (and at other time points) following induction, Snf5p crosslinking increased over the entire SUC2 promoter region and peaked at the UAS sequence (Figure 3A; data not shown). Thus, Snf5p is specifically recruited to the SUC2 promoter following the carbon source shift, presumably as SWI/SNF (Geng et al, 2001).
The highest increase in H3 acetylation occurred at the SUC2 promoter, although significant acetylation was also detected at regions upstream and downstream of the SUC2 promoter 5 min following induction (and at other time points) (Figure 3B; data not shown). These results suggest that Gcn5p is also specifically recruited to the SUC2 promoter but appears to be distributed more widely than Snf5p.
The increase in H4 acetylation 5 min following SUC2 induction was restricted to the same regions as H3 acetylation (Figure 3C, upper panel). Consistent with this finding, the parallel increase in Esa1p's association with SUC2 also occurred primarily at the SUC2 promoter region (Figure 3C, lower panel). Under repressing conditions, however, Esa1p appeared to associate globally with the SUC2 locus, with a marginally higher affinity for the promoter region. In conclusion, SWI/SNF and Gcn5p were dynamically and specifically recruited to SUC2 following gene induction, whereas Esa1p associated globally with SUC2 under repressing conditions and was specifically but transiently recruited upon gene induction.
SWI/SNF, Gcn5p, and Esa1p differentially regulate the dynamic induction of SUC2
To investigate the roles of SWI/SNF, Gcn5p, and Esa1p in regulating the dynamic transcription of SUC2, we tested how loss-of-function mutations in the chromatin-remodeling factors affect SUC2 mRNA levels at various times following induction. Little SUC2 RNA was synthesized throughout induction in snf5Δ cells (Figure 4A), demonstrating that SWI/SNF is critical for both phases of SUC2 induction.
Figure 4.
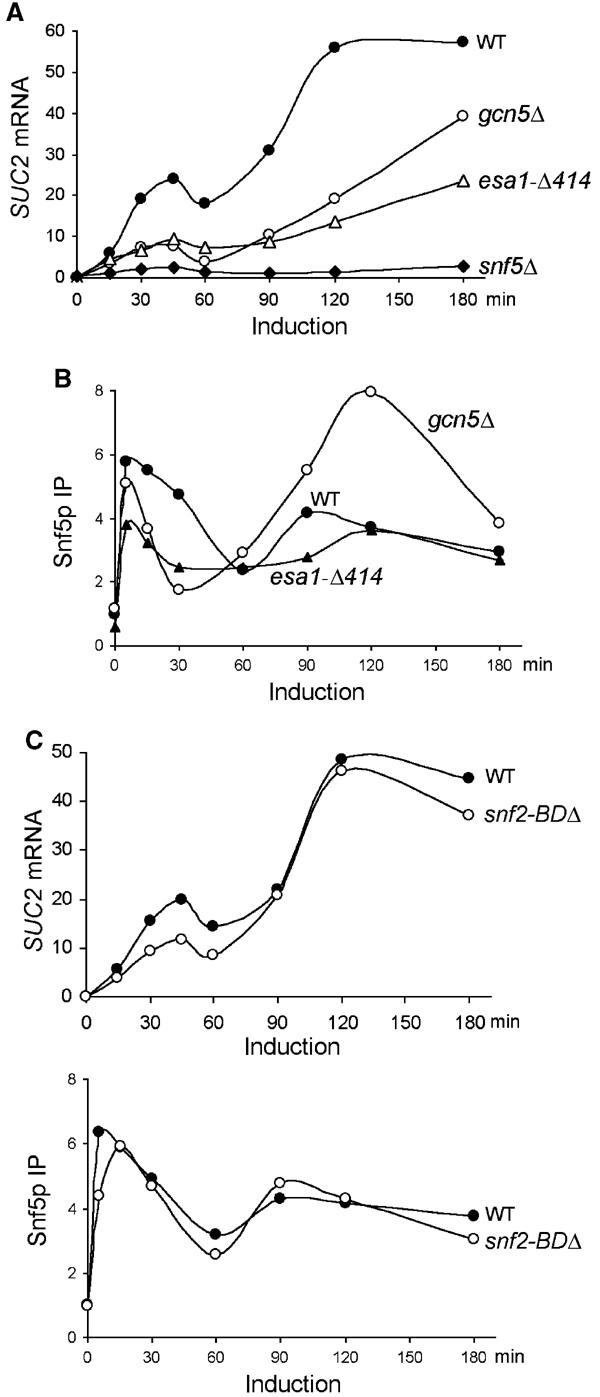
Snf5p, Gcn5p, and Esa1p differentially control SUC2 transcription. (A) The levels of SUC2 mRNA in wild-type (BLY1), gcn5Δ (BLY417), esa1-Δ414 (BLY457), and snf5Δ (BLY3) cells were assayed as in Figure 1A. (B) The crosslinking of Snf5p to the SUC2 promoter was assayed as in Figure 2A. IP efficiencies were shown relative to the wild-type preinduction levels. (C) The levels of SUC2 mRNA (upper panel) and the crosslinking of Snf5p to the SUC2 promoter (lower panel) in wild-type (BLY1) and snf2-BDΔ (BLY663) cells were assayed as in (A) and (B), respectively. These data are representative of three independent time courses.
In gcn5Δ cells, SUC2 transcript levels reached 75% of the wild-type levels after 3 h of induction, consistent with previous studies (Biggar and Crabtree, 1999; Sudarsanam et al, 1999). However, only 25–35% of the wild-type levels of SUC2 mRNA were achieved during the first 2 h of induction in the gcn5 mutant (Figure 4A), indicating that Gcn5p plays a more important role in activating SUC2 transcription than previously appreciated (Biggar and Crabtree, 1999; Recht and Osley, 1999; Sudarsanam et al, 1999). These results demonstrate that Gcn5p accelerates the kinetics of SUC2 induction. A similar role for Gcn5p has been observed at PHO5 (Barbaric et al, 2001).
The esa1-Δ414 mutation reduced SUC2 transcription to the same extent as the gcn5Δ mutation in the first phase of induction and caused a greater reduction in SUC2 transcription during the second phase (Figure 4A). These results suggest that Esa1p is important for SUC2 transcription throughout induction and thus provide physiological significance for the constitutive presence of Esa1p and its HAT activity at SUC2 (Figure 2A and C).
The important, but nonessential, roles of Gcn5p and Esa1p in the transcriptional activation of SUC2 prompted us to test whether Gcn5p or Esa1p stimulates SUC2 transcription by facilitating SWI/SNF chromosomal binding. In gcn5Δ cells, Snf5p was crosslinked to the SUC2 promoter as well as in the wild type 5 min following induction, demonstrating that the initial SWI/SNF association was independent of Gcn5p (Figure 4B). However, the efficient binding of SWI/SNF to the SUC2 promoter during the first phase of induction was lost soon thereafter, as indicated by the rapid decrease in crosslinking of Snf5p between 5 and 30 min. Interestingly, during the second phase of induction, SWI/SNF associated with SUC2 even more efficiently in gcn5 cells than in wild-type cells (Figure 4B). Higher-than-wild-type binding of SWI/SNF to the RNR3 promoter was also observed in gcn5 cells 2 h after induction by DNA damage, although its association with chromatin was not tested at earlier time points (Sharma et al, 2003). Snf5p crosslinking to SUC2 was also significantly reduced in the esa1-Δ414 mutant during early gene induction (0–60 min), but reached wild-type levels by 2 h (Figure 4B). These results show that both Gcn5p and Esa1p are necessary for maximal binding of SWI/SNF to the SUC2 promoter early during gene induction but not at later stages of transcription, suggesting compensation by other mechanisms (see Discussion). Conversely, we found that Gcn5p and Esa1p associate with SUC2 independently of SWI/SNF (data not shown). We conclude that Gcn5p and Esa1 stimulate SUC2 transcription partly by facilitating the association of SWI/SNF with the promoter.
The bromodomain is a conserved protein motif that recognizes and binds to acetylated histones (Zeng and Zhou, 2002). To test the role of the Snf2p bromodomain (the only bromodomain in SWI/SNF) in SWI/SNF recruitment to SUC2, we measured SUC2 mRNA levels and Snf5p's association with SUC2 in cells lacking the Snf2p bromodomain. In contrast to gcn5Δ and esa1-Δ414 mutations, deletion of the Snf2p bromodomain primarily affected the first phase of SUC2 transcription, reducing SUC2 RNA levels by less than two-fold (compare Figure 4A and C, upper panel). Importantly, Snf5p's association with SUC2 was only minimally affected in the snf2-BDΔ mutant (Figure 4C, lower panel). These results suggest that the Snf2p bromodomain plays little role in mediating the HAT-facilitated SWI/SNF association with SUC2. The more dramatic effect of an Snf2p bromodomain deletion on SWI/SNF binding to SUC2 reported previously (Hassan et al, 2002) may reflect differences in experimental reagents or conditions.
AMPK/Snf1 and cAPK co-regulate SWI/SNF and Gcn5p occupancy and SUC2 transcription
Two major signaling pathways transmit the glucose signal in yeast cells. One pathway involves the Snf1 kinase, the yeast homolog of the mammalian AMP-dependent protein kinase (AMPK). Snf1p is activated by low glucose, and is essential for the transcriptional induction of glucose-repressed genes, including SUC2 (Carlson, 1999). The other pathway involves the cAMP-dependent protein kinase (cAPK), which controls cell growth in response to nutrient availability and acts antagonistically to Snf1 in nutrient response (Cannon and Tatchell, 1987; Thompson-Jaeger et al, 1991; Thevelein and de Winde, 1999), although its role in regulating SUC2 transcription is unresolved (Hubbard et al, 1992).
To test the potential roles of Snf1 and cAPK in regulating the dynamic recruitment of SWI/SNF and Gcn5p to the SUC2 promoter, we monitored Snf5p and Gcn5p promoter occupancy in the snf1K84R and bcy1Δ mutants. The snf1K84R mutation specifically abolishes the Snf1 kinase activity (Celenza and Carlson, 1989), and deletion of the BCY1 gene, which encodes the regulatory subunit of cAPK, causes constitutive cAPK activity (Cannon and Tatchell, 1987; Toda et al, 1987). snf1K84R completely abolished the limiting glucose-induced increase in crosslinking of both Snf5p and Gcn5p (Figure 5A (upper panel) and B). We conclude that the snf1K84R mutation's abrogation of both Snf5p and Gcn5p recruitment is a selective rather than a general event, as Snf5p and Gcn5p recruitment to another target promoter, RNR3 (Sharma et al, 2003), was not dramatically affected by snf1K84R (Figure 5A, lower panel; data not shown).
Figure 5.
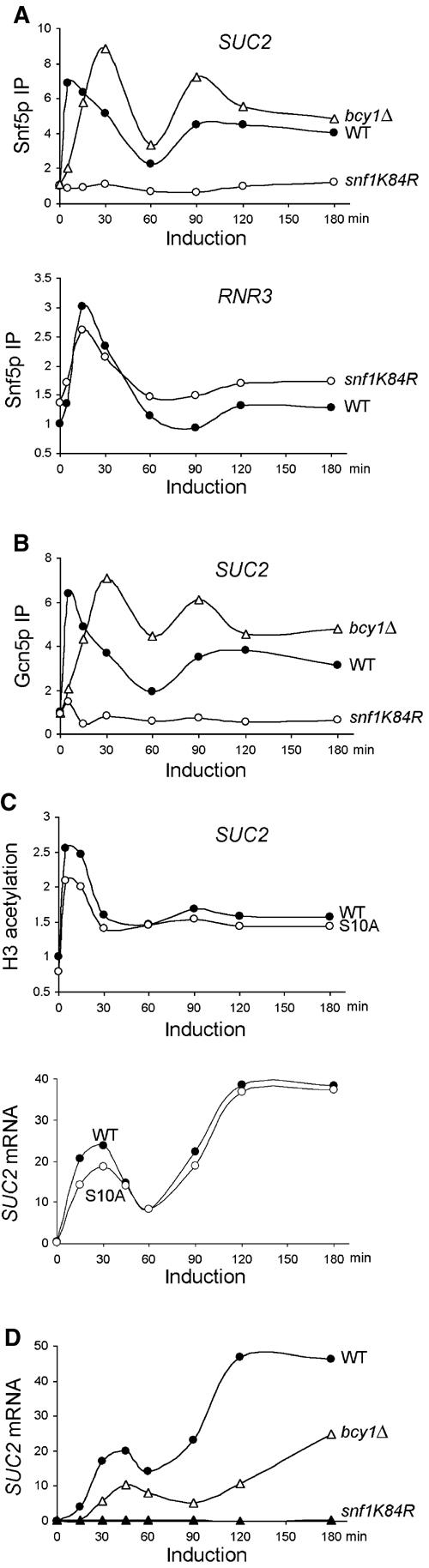
AMPK/Snf1 and cAPK regulate both recruitment of Snf5p and Gcn5p and transcription of SUC2. The crosslinking of Snf5p (A) and Gcn5p (B) to the SUC2 or RNR3 promoters and the levels of SUC2 mRNA (D) were measured in wild-type (BLY1), bcy1Δ (BLY553), and snf1K84R (BLY463) cells as in Figure 4. (C) H3 acetylation (upper panel) and SUC2 mRNA levels (lower panel) were assayed in wild-type (BLY484) and H3 S10A mutant (BLY485, in which serine 10 was changed to alanine) cells as in Figures 2B and 4A, respectively. These data are representative of at least two independent time courses each.
Deletion of BCY1 delayed the promoter association of both Snf5p and Gcn5p in the first phase of SUC2 induction, so that the crosslinking of both proteins peaked at 30 min rather than at 5 min following induction (Figure 5A (upper panel) and B). Notably, at later time points, higher levels of crosslinking of both proteins were detected in bcy1Δ cells compared to wild-type cells. These results demonstrate that Snf1 kinase activity is essential for the recruitment of SWI/SNF and Gcn5p to SUC2 in response to limiting glucose, and suggest that downregulation of cAPK activity is important for their early recruitment.
Gcn5p-dependent H3 acetylation at the INO1 and HO promoters requires phosphorylation of H3 serine 10. Moreover, Snf1 is required for H3 phosphorylation at INO1 (Lo et al, 2001). To test whether Snf1 similarly controls Gcn5p's HAT activity at SUC2, we measured SUC2 H3 acetylation in the same mutant (H3 S10A) in which H3 acetylation at both INO1 and HO promoters was diminished (Lo et al, 2001). By contrast, we found that the S10A mutation only slightly reduced the basal levels of H3 acetylation at the SUC2 UAS, but did not alter the magnitude of increase in H3 acetylation following induction (Figure 5C, upper panel), suggesting that S10 phosphorylation is not a prerequisite for H3 acetylation at SUC2. Moreover, the H3 S10A mutation only slightly reduced SUC2 mRNA levels and Snf5p binding to SUC2 during the early phase of transcription (Figure 5C, lower panel; data not shown). Therefore, it is unlikely that Snf1 plays a prominent role in regulating Gcn5p's H3-acetylating activity via phosphorylation of H3 serine 10 at SUC2.
Consistent with the essential role of Snf1 in controlling the recruitment of SWI/SNF and Gcn5p to SUC2, no SUC2 transcripts were detected in the snf1K84R mutant throughout gene induction (Figure 5D). Despite the overall increased efficiency in SWI/SNF and Gcn5p crosslinking to SUC2 in bcy1Δ cells compared to wild type, the SUC2 mRNA levels in the mutant were actually reduced two-fold, suggesting that cAPK also negatively regulates SUC2 transcription at steps other than promoter recruitment of SWI/SNF and Gcn5p.
Stress response transcription factors Msn2p/Msn4p function specifically in early SUC2 induction
Several lines of evidence suggest that Msn2p and Msn4p, two functionally redundant transcriptional factors involved in multiple stress responses, function in the dynamic induction of SUC2. First, the early induction of SUC2 resembles a stress response in that SWI/SNF, Gcn5p, and pol II are recruited to the promoter within 2 min (Figures 2A and 1B). Second, acute glucose deprivation activates Msn2p/Msn4p transcriptional activity, presumably through downregulation of cAPK (Gorner et al, 2002). Third, MSN2 was isolated as a multicopy suppressor of the SUC2 expression defects in both snf1 and snf5 temperature-sensitive mutants (Estruch and Carlson, 1993; our unpublished data). Finally, Msn2p and Msn4p bind to the SUC2 promoter DNA in vitro (Estruch and Carlson, 1993).
Like bcy1Δ, deletion of MSN2/MSN4 delayed the recruitment of both Snf5p and Gcn5p to SUC2 (Figure 6A and B). Moreover, SUC2 mRNA levels in the msn2Δ msn4Δ mutants were two-fold lower than in the wild type in the first phase of gene induction. Interestingly, in the second phase of induction, SUC2 mRNA accumulated more rapidly and reached higher levels in the mutant than in the wild type (Figure 6C), consistent with the parallel increases in promoter association of both Snf5p and Gcn5p (Figure 6A and B). We infer from these results that Msn2p/Msn4p act specifically in the early phase of SUC2 induction.
Figure 6.
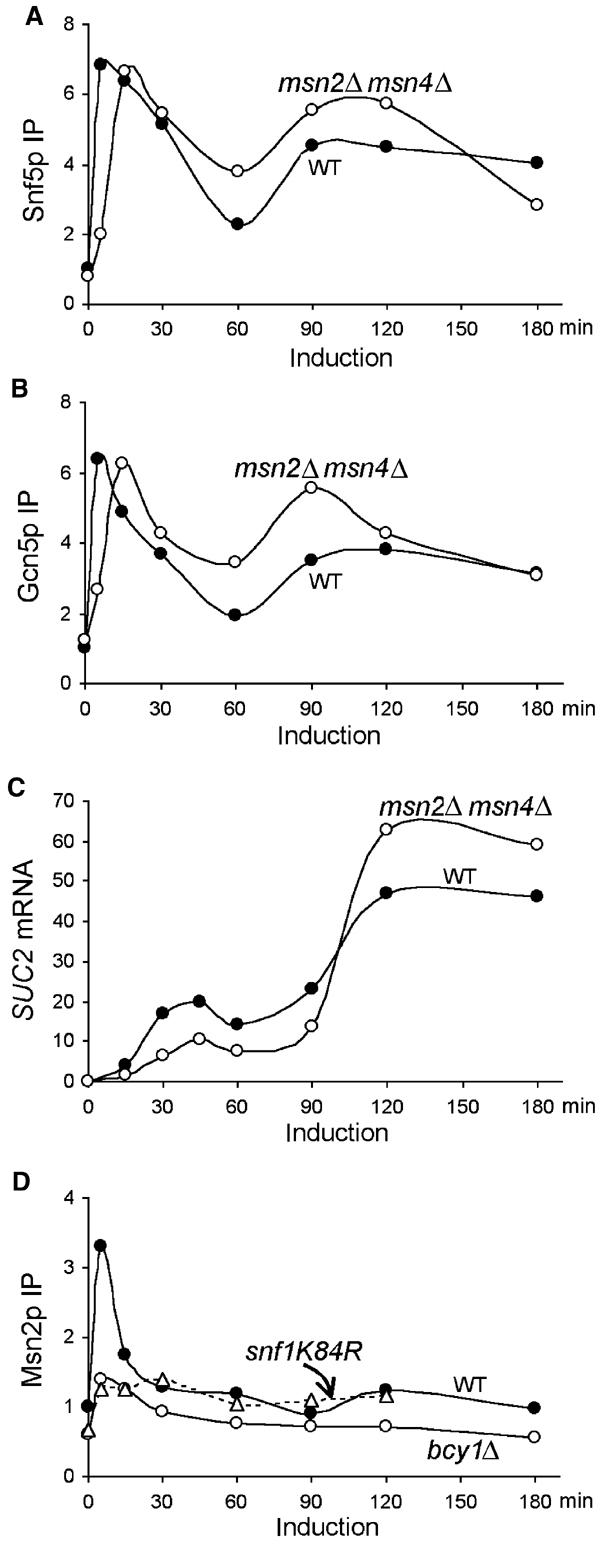
Msn2p/Msn4p are involved in early SUC2 induction. The crosslinking of Snf5p (A) and Gcn5p (B) to the SUC2 promoter and levels of SUC2 mRNA (C) in the msn2Δmsn4Δ mutant (BLY559) were measured in the same experiments as in Figure 5. (D) The crosslinking of Msn2p to the SUC2 UAS in wild-type (BLY1), bcy1Δ (BLY553), and snf1K84R (BLY463) cells was measured by ChIP using polyclonal anti-Msn2p antibody, and the levels (IP efficiencies) were shown relative to wild-type preinduction levels. The data in (A–C) are representative of three independent time courses, and those in (D) of two independent time courses.
To substantiate the involvement of Msn2p/Msn4p in the first phase of SUC2 transcription, the association of Msn2p with SUC2 chromatin was measured by ChIP using a polyclonal anti-Msn2p antibody. We found that Msn2p crosslinking at the SUC2 promoter increased more than three-fold 5 min following gene induction but quickly returned to repression levels within 30 min and did not increase further throughout the induction (Figure 6D). Additionally, the rapid recruitment of Msn2p to the SUC2 promoter was essentially abolished in the bcy1Δ mutant (Figure 6D), consistent with the ability of cAPK to inhibit the nuclear localization of Msn2p (Gorner et al, 2002). Since the nuclear localization of Msn2p and Msn4p is regulated in the same fashion (Gorner et al, 2002), we assume that Msn4p associates with SUC2 similarly.
Interestingly, further analysis of the association of Msn2p with the SUC2 promoter in snf5Δ and snf1K84R mutants suggested that Msn2p binding to SUC2 requires the presence of SWI/SNF and the Snf1 kinase activity. Like in the bcy1Δ mutant, the induced increase in Msn2p crosslinking to the SUC2 promoter was largely abolished in the snf1K84R mutant (Figure 6D). Identical results were obtained in the snf5Δ mutant (data not shown). The effects of snf5Δ and snf1K84R on Msn2p association with the SUC2 promoter are unlikely to be caused by defective expression of MSN2 or the failure of Msn2p to enter the nucleus, as SWI/SNF does not regulate MSN2 transcription (Holstege et al, 1998) and snf1Δ does not block the nuclear localization of Msn2p (Mayordomo et al, 2002). Together, these results suggest a role for Msn2p/Msn4p in the early induction of SUC2. Although we cannot rule out the possibility that Msn2p and Snf5p interact indirectly, we favor the idea that Msn2p and SWI/SNF function cooperatively for promoter association to initiate SUC2 transcription.
Discussion
Here, we present evidence that SUC2 transcription proceeds in a biphasic fashion. Different chromatin remodelers (SWI/SNF, Gcn5p, and Esa1p) play distinct roles and cooperatively activate SUC2 transcription. Our study underscores the importance of applying time-course approaches in exploring the interplay of transcription factors in transcription.
Biphasic transcriptional induction in nutrient starvation
Although raffinose, a substrate of the SUC2 gene product, is required for continual SUC2 transcription, it does not appreciably affect the accumulation pattern of SUC2 transcripts during the first 2 h of induction (Figure 1A). This result suggests that the biphasic pattern of SUC2 transcription is not due to feedback from SUC2 transcription itself, but rather reflects dynamic cellular changes in glucose metabolism. Therefore, it is likely that transcription of other glucose-regulated genes proceeds in a similar dynamic fashion, although this remains to be proven. Interestingly, we noticed that both nitrogen and amino-acid starvation also induce biphasic transcription from a large set of genes (Gasch et al, 2000). Thus, biphasic transcriptional induction could be a general response to the stress of nutrient starvation (Gasch et al, 2000; Causton et al, 2001; Gorner et al, 2002).
Why is the transcriptional induction biphasic? We suggest that the initial stress response triggers a transient repression of energy-consuming processes such as translation, transcription, and cell cycle progression (Martinez-Pastor and Estruch, 1996; Alberghina et al, 1998; Ashe et al, 2000; Gasch et al, 2000; Causton et al, 2001; Newcomb et al, 2003) (Figure 1C). As a result, the cell compensates for the sudden shortage of energy or other nutrients and quickly restores the balance between energy production and consumption. However, this balance is then upset when the cell resumes energy-consuming processes once it has corrected the stress-induced damages. The biphasic transcriptional repression that parallels transcriptional induction during amino-acid or nitrogen starvation (Gasch et al, 2000) further supports the idea that a general mechanism is responsible for the biphasic pattern.
The two phases of SUC2 transcription may initiate independently, resembling the cyclical transcription of the estrogen-induced human cathespin D gene, in which the same group of transcription factors is recruited for each cycle (Shang et al, 2000). At SUC2, the two phases seem to communicate. For example, cells increase recruitment of SWI/SNF and Gcn5p to SUC2 after early phase transcription is reduced by deletions of BCY1 or MSN2/MSN4, and the second phase of recruitment of SWI/SNF increases in the gcn5 mutant. We suggest that the cell senses the overall response in the first phase and adjusts the levels of the following responses accordingly. Nevertheless, it remains possible that factors recruited during the first phase, or stable modifications of histones (e.g., methylation) made in the first phase, persist and contribute to later phase(s) of transcription.
Distinct modes of recruitment of chromatin-remodeling enzymes
The data presented here strongly suggest, although do not prove, that SWI/SNF and Gcn5p are recruited to the SUC2 promoter concurrently, rather than in a temporal order as has been defined at cell cycle-regulated or differentiation-induced promoters (Cosma et al, 1999; Shang et al, 2000; Agalioti et al, 2002; Soutoglou and Talianidis, 2002). A recent study suggests that the two remodeling factors are also recruited in parallel to osmotic stress-responsive promoters (Proft and Struhl, 2002). We hypothesize that different strategies for transcriptional regulation underlie the different modes of recruitment. For example, genes involved in stress response or metabolism respond to signals that sense different aspects of the cell's status simultaneously. The concurrent association of these chromatin-remodeling activities assures a rapid on–off switch for gene activity. In contrast, genes required for cell cycle progression, differentiation, or development sense and integrate various events that occur in sequence; the sequential recruitment of chromatin remodelers may function as checkpoints for subsequent transcriptional events to ensure that a gene will be activated at the right time.
Notably, Esa1p associates constitutively with SUC2 to promote rapid and efficient transcriptional induction. In contrast to this mode of action, Reid et al (2001) have shown that Esa1p is specifically targeted to ribosomal protein genes to activate transcription, and that levels of promoter-associated Esa1p correlate with levels of gene transcription. We propose a new role for Esa1p, based on our results and by analogy to the role of nontargeted HDACs in rapidly turning off gene transcription (Vogelauer et al, 2000), in which Esa1p functions globally to turn on rapidly gene transcription by maintaining chromatin fluidity via acetylation of histone H4. Future studies examining the kinetics of transcription are needed to test this model.
Dynamic interplay of chromatin-remodeling activities
Despite differences in their temporal association with the SUC2 promoter, Gcn5p and Esa1p are equally important for optimal SUC2 transcription. This may be explained, in part, by our finding that both proteins facilitate SWI/SNF's binding to the SUC2 promoter (Figure 4B). Our results further support the notion that HATs enhance the interaction of SWI/SNF with chromatin (Hassan et al, 2001, 2002; Agalioti et al, 2002), which could explain the ability of Gcn5p to increase nucleosome remodeling in vivo (Gregory et al, 1998, 1999; Syntichaki et al, 2000; Sharma et al, 2003). In addition, we showed that the Snf2p bromodomain plays at most only a minor role in SWI/SNF's association with SUC2 (Figure 4C). Therefore, other mechanism(s) must exist. One possibility is that the local HAT-modifiable structure of chromatin plays an important role in mediating HATs-facilitated SWI/SNF–chromatin association.
Interestingly, SWI/SNF binding to SUC2 is impaired by esa1 and gcn5 mutations only in the first phase of induction; wild-type or even higher levels of SWI/SNF binding are achieved at later stages of induction. One explanation is that the HAT mutants compensate by generating stronger signals (e.g., a more active Snf1 kinase) to recruit more SWI/SNF to SUC2 during later gene induction. This compensatory mechanism may also involve other factors that normally play redundant roles with the Gcn5p and Esa1p HATs. Regardless of the mechanism, an important finding from this study is that the effects of loss of HAT functions on the binding of SWI/SNF to chromatin in vivo differ temporally during the transcription of a single gene. Such roles for HATs could easily be missed in studies examining only steady-state transcription. Dynamic time-course studies therefore provide a more unambiguous examination of the interplay of these activities (and other factors) throughout the transcription of a gene.
Materials and methods
Yeast strains and media
Saccharomyces cerevisiae strains used in this study are in the S288c genetic background and are listed in Table I. Mutant strain constructions are described in Supplementary data.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| BLY1 | MATα his3-Δ200 lys2-801 ura3-52 | This lab |
| (=MCY829) | ||
| BLY3 | MATα ade2-101 his3-Δ200 ura3-52 snf5-Δ2 | This lab |
| BLY417 | MATα his3-Δ200 lys2-801 gcn5∷HIS3 | This study |
| BLY431 | MATα his3-Δ200 lys2-801 ura3-52 HA3-ESA1 | This study |
| BLY457 | MATα his3-Δ200 lys2-801 ura3-52 esa1-Δ414 | This study |
| BLY463 | MATa his3-Δ200 leu2-3,112 ura3-52 snf1K84R | M Carlson |
| (=MCY2693) | ||
| BLY553 | MATα his3-Δ200 lys2-801 ura3-52 bcy1∷URA3 | This study |
| BLY484 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1 hhf1) | CD Allis |
| (=JHY86) | Δ(hht2 hhf2) pJH18[CEN ARS TRP1 HHT2 HHF2] | |
| BLY485 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1 hhf1) | CD Allis |
| (=JHY87) | Δ(hht2 hhf2) pJH15[CEN ARS TRP1 hht2-3(S10A) HHF2] | |
| BLY559 | MATα his3-Δ200 lys2-801 ura3-52 msn2∷URA3 msn4∷HIS3 | This study |
| BLY663 | MATα his3-Δ200 lys2-801 ura3-52 snf2-BDΔ | This study |
YEP medium contains 1% yeast extract (Difco) and 2% bacto-peptone (Difco). To induce the SUC2 gene, early logarithmic phase (OD600 0.5–0.6) cells grown in YPD medium (YEP+2% dextrose) were collected onto a filter by filtration, washed with water, and resuspended immediately in YPR medium (YEP+0.05% dextrose+2% raffinose) unless noted. All cells were cultured at 30°C.
RNA analysis
Yeast cells were frozen on dry ice following collection. Total RNA was isolated with hot phenol (Ausubel et al, 1995) and reverse-transcribed with oligo (dT)12–18 (Amersham) and Omniscript reverse transcriptase (Qiagen). The cDNA was quantified by real-time PCR analysis with primers amplifying ACT1 (+323 to +613) or SUC2 (−1 to +120) sequences. The ACT1 transcript serves as a control. All the experiments were performed with at least two independent RNA preparations.
Chromatin immunoprecipitation
ChIP was carried out essentially as described previously (Geng et al, 2001), except that the IP buffer contains 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, and 0.1% sodium deoxycholate. The following amounts of antibodies were used in a typical IP reaction: 3 μl anti-Snf5p (Geng et al, 2001), 2 μl anti-Pol II CTD (unphosphorylated; BabCO), 10 μl anti-Gcn5p or anti-Esa1p (Santa Cruz), 8 μl anti-HA (Santa Cruz), 6 μl anti-Msn2p (gift of F Estruch), or 1.5 μl anti-diacetyl-H3 (acetylated at K9 and K14) or anti-tetra-acetyl-H4 (acetylated at K8, K12, K14, and K16) (Upstate Biotechnology). The amounts of chromatin inputs varied with individual antibodies and were within the linear ranges in which the amount of DNA precipitated was proportional to the input. Inputs and precipitated DNAs were measured for specific sequences by real-time PCR or semiquantitative multiplex PCR. IP efficiencies were calculated by dividing IP signals by the corresponding input signals. All ChIP experiments were performed with at least two independent chromatin preparations. Primers were designed to amplify the following SUC2 sequences: (−2239 to −1900), (−1817 to −1506), (−1228 to −960), UAS (−748 to −498), (−208 to 24), (−150 to +45), (+225 to +432), and (+780 to +960). Other primers amplify a 150-bp subtelomeric sequence (Vogelauer et al, 2000) or promoter sequences of ACT1 (−290 to −117), RPL2B (−439 to −232), RPS11B (−421 to −253), or RNR3 (−450 to −288).
Mitotic index
Yeast cells were fixed in 3.7% formaldehyde in the culture medium. After resuspension in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) and sonication, the single (unbudded), small-budded, and large-budded cells were determined microscopically. Experiments were carried out with three independent cultures.
PCR
Real-time PCR was performed with a DNA Engine Opticon system (MJ Research) and SYBR Green (Sigma). PCR reactions (15 μl) were set up in triplicate for each DNA sample with 0.5 U of HotStarTaq DNA polymerase (Qiagen). Relative quantitation was achieved from standard curves prepared from serial dilutions of genomic DNA. For most experiments, the cycle threshold (c(t)) was set at a fluorescence intensity of 0.0046 on baseline-subtracted data graphs (fluorescence versus cycle number). The values obtained from triplicate reactions were averaged, with errors <20%.
Multiplex PCR was conducted in reactions similar to those for real-time PCR except that they contained multiple primer pairs. After 27–29 cycles of amplification, the PCR products were separated on 8% polyacrylamide gels and stained with ethidium bromide; in some experiments, the PCR products were labeled by [α-32P]-dCTP (Amersham) and quantified with a phosphorImager (Molecular Dynamics). PCR amplifications of both input DNA and ChIP-ed DNA were carried out in predetermined linear ranges.
Supplementary Material
Supplementary Materials and Methods
Acknowledgments
We thank CD Allis, M Carlson, L Pillus, K Struhl, K Tatchell, and G Thireos for providing yeast strains or plasmids, and F Estruch for anti-Msn2p antibody. We also thank B Chai and J Huang for construction of strains BLY457 and BLY663, respectively, and BR Cairns, C Parada and Laurent lab members J-M Hsu, B Chai, and J Huang for useful comments on the manuscript. This work was supported by Public Health Service grant GM56700 from the NIH.
References
- Agalioti T, Chen G, Thanos D (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392 [DOI] [PubMed] [Google Scholar]
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniastis T, Thanos D (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667–678 [DOI] [PubMed] [Google Scholar]
- Alberghina L, Smeraldi C, Ranzi BM, Porro D (1998) Control by nutrients of growth and cell cycle progression in budding yeast, analyzed by double-tag flow cytometry. J Bacteriol 180: 3864–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA 51: 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB (2000) Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11: 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1995) Current Protocols in Molecular Biology. Boston, MA: John Wiley and Sons, Inc. [Google Scholar]
- Barbaric S, Walker J, Schmid A, Svejstrup JQ, Horz W (2001) Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J 20: 4944–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR (1999) Continuous and widespread roles for the Swi–Snf complex in transcription. EMBO J 18: 2254–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JF, Tatchell K (1987) Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol 7: 2653–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M (1999) Glucose repression in yeast. Curr Opin Microbiol 2: 202–207 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1989) Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol 9: 5034–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L (1999) Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP (2002) Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell 10: 227–236 [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311 [DOI] [PubMed] [Google Scholar]
- Estruch F, Carlson M (1993) Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol 13: 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CJ, Peterson CL (2001) Chromatin remodeling enzymes: who's on first? Curr Biol 11: r185–r197 [DOI] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault A, Zhang Y, Heliot L, Allard S, Savard J, Lane W, Stillman D, Cote J (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigentic control of transcription. Mol Cell 5: 927–937 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin IM, Simpson RT (1997) Interplay of yeast global transcriptional regulators Ssn6p–Tup1p and Swi–Snf and their effect on chromatin structuire. EMBO J 16: 6263–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Cao Y, Laurent BC (2001) Essential roles of Snf5p in Snf–Swi chromatin remodeling in vivo. Mol Cell Biol 21: 4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schuller C (2002) Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Lui L, Berger SL, Horz W (1998) Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell 1: 495–505 [DOI] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W (1999) Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J 18: 6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL (2001) Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Herwig C, Doerries C, Marison I, Von Stockar U (2001) Quantitative analysis of the regulation scheme of invertase expression in Saccharomyces cerevisiae. Biotechnol Bioeng 76: 247–258 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F (1992) Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6: 2288–2298 [DOI] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Yang XL, Carlson M (1992) Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics 130: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D (2001) Functional significance of histone deacetylase diversity. Curr Opin Genet Dev 11: 162–166 [DOI] [PubMed] [Google Scholar]
- Krebs JE, Fry CJ, Samuels ML, Peterson CL (2000) Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102: 587–598 [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo M-H, Allis CD, Peterson CL (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev 13: 1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Tolga NC, Emre Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL (2001) Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2 A resolution. 389: 251–260 [DOI] [PubMed]
- Martens JA, Winston F (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Estruch F (1996) Sudden depletion of carbon source blocks translation, but not transcription, in the yeast Saccharomyces cerevisiae. FEBS Lett 390: 319–322 [DOI] [PubMed] [Google Scholar]
- Matallana E, Franco L, Perez-Ortin JE (1992) Chromatin structure of the yeast SUC2 promoter in regulatory mutants. Mol Gen Genet 231: 395–400 [DOI] [PubMed] [Google Scholar]
- Mayordomo I, Estruch F, Sanz P (2002) Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J Biol Chem 277: 35650–35656 [DOI] [PubMed] [Google Scholar]
- Newcomb LL, Diderich JA, Slattery MG, Heideman W (2003) Glucose regulation of Saccharomyces cerevisiae cell cycle genes. Eukaryot Cell 2: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Vallier LG, Flick JS, Carlson M, Johnston M (1997) Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 13: 127–138 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev 10: 187–192 [DOI] [PubMed] [Google Scholar]
- Pollard KJ, Peterson CL (1997) Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol 17: 6212–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K (2002) Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Recht J, Osley MA (1999) Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi/Snf in yeast. EMBO J 18: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J, Lyer V, Brown P, Struhl K (2001) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Reinke H, Gregory PD, Horz W (2001) A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol Cell 7: 529–538 [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD (2001) Histone acetyltransferases. Annu Rev Biochem 70: 81–120 [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, Direnzo J, Lazar MA, Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843–852 [DOI] [PubMed] [Google Scholar]
- Sharma VM, Li B, Reese JC (2003) SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev 17: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295: 1901–1904 [DOI] [PubMed] [Google Scholar]
- Struhl K (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12: 599–606 [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Cao Y, Wu L, Laurent BC, Winston F (1999) The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J 18: 3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G (2000) The Gcn5 bromodomain co-ordinates nucleosome remodeling. Nature 404: 414–417 [DOI] [PubMed] [Google Scholar]
- Thevelein JM, De Winde JH (1999) Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 33: 904–918 [DOI] [PubMed] [Google Scholar]
- Thompson-Jaeger S, Francois J, Gaughran JP, Tatchell K (1991) Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics 129: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Scott JD, Mcmullen B, Hurwitz M, Krebs EG, Wigler M (1987) Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol 7: 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL (2000) ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M (2000) Global histone acetylation and deacetylation in yeast. Nature 408: 495–498 [DOI] [PubMed] [Google Scholar]
- Wolffe AP (1998) Chromatin Structure and Function. San Diego, CA: Academic Press [Google Scholar]
- Wu L, Winston F (1997) Evidence that Snf–Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res 25: 4230–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhou MM (2002) Bromodomain: an acetyl-lysine binding domain. FEBS Lett 513: 124–128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods


