Abstract
Competing models for the coordination of processive stepping in kinesin can be tested by introducing a roadblock to prevent lead head attachment. We used T93N, an irreversibly binding mutant monomer, as a roadblock, and measured the rates of nucleotide-induced detachment of kinesin monomers or dimers with and without the T93N roadblock using microflash photolysis combined with stopped flow. Control nucleotide-induced monomer (rK340) unbinding was 73.6 s−1 for ATP and 40.5 s−1 for ADP. Control ADP-induced dimer (rK430) unbinding was 18.6 s−1. Added 20 mM Pi slowed both monomer and dimer unbinding. With the roadblock in place, lead head attachment of dimers is prevented and ATP-induced trail head unbinding was then 42 s−1. This is less than two-fold slower than the stepping rate of unimpeded rK430 dimers (50–70 s−1), indicating that during walking, lead head attachment induces at most only a slight (less than two-fold) acceleration of trail head detachment. As we discuss, this implies a coordination model having very fast (>2000 s−1) ATP-induced attachment of the lead head, followed by slower, strain-sensitive ADP release from the lead head.
Keywords: kinesin, kinesin mechanism, processivity, tubulin
Introduction
A key problem in the kinesin field is the coordination mechanism that links the actions of the two heads of a kinesin molecule as it walks along a microtubule (MT). Remarkably, single-molecule in vitro assays appear to recapitulate the broad features of the processive walking mechanism of native kinesin in vivo. In vitro under a range of loads, including very low loads, kinesin stepping remains tightly coupled to ATP consumption, with each 8 nm step consuming one ATP molecule (Hua et al, 1997; Schnitzer and Block, 1997; Coy et al, 1999; Nishiyama et al, 2002). To achieve this, the mechanochemical actions of the two heads must be tightly linked. Recent attention has focused on the control of lead head attachment by the trail head, and there is firm evidence that MT-activated ADP release from the lead head is dramatically accelerated by an ATP-dependent conformational change of the trail head (Hackney, 1994a; Ma and Taylor, 1997a; Gilbert et al, 1998; Crevel et al, 1999; Hackney, 2002) with a correlated shift in the mobility of the neck linkers that join the heads to the tail (Rice et al, 1999; Case et al, 2000). One coordination rule, that governing leading head attachment, thus emerges. The corresponding rules governing trail head detachment are less clear. Yet if long processive runs and a tight ATPase coupling ratio are to be maintained, trailing head detachment must be timed relative to leading head attachment such that detachment occurs neither too early (resulting in termination of the processive run of steps) nor too late (resulting in a brake on progress).
Concerning the control of trail head detachment, three points at least are controversial. Firstly, which chemical state(s) of the trail head detach(es) from MT? Candidates are the K.ADP.Pi and K.ADP states. Second, how does the direction and magnitude of strain experienced by the trail head influence the rate constant(s) for its detachment? Third, do the trailing heads of kinesin dimers behave differently from isolated kinesin monomers, and if so, how? One reason for the uncertainties is that unbinding rate constants for kinesins are difficult to measure. Ma and Taylor (1995) estimated 12 s−1 for the ATP-induced unbinding of K379 human kinesin dimers. Ma and Taylor (1997b) estimated rate constants for the unbinding of single human kinesin (K332) heads of 40–50 s−1 for ATP and 65–90 s−1 for ADP. Jiang and Hackney (1997) estimated 46 and 52 s−1 for a Drosophila kinesin head (DKH357) construct. Signals were noisy and a slow process was assigned as an artefact. Gilbert et al (1995) obtained 13 s−1 for the rate of ATP-induced unbinding of human K401, predominantly a dimer, but noted a slower phase in their data, assigned as an artefact. Moyer et al (1998) measured 21–22 s−1 for ATP-induced unbinding of Drosophila K340; an artefactual slow process was again noted. Hackney (2002) points out that this monomer lacks part of the neck linker, and consequently dissociates unusually slowly. In general, the light-scattering signals associated with kinesin unbinding from MT are inevitably small (because the proportional change in the diameter of the scattering rodlike polymers is small), and additional artefacts can arise in the stopped flow as MTs initially orient in the direction of flow, and then rerandomise their orientation once flow has stopped.
Other approaches to measure unbinding rate constants have been taken. Rosenfeld et al (2002) have developed an assay based on rhodamine excimer attached to the neck linkers of human kinesin dimers, and estimated 50–60 s−1 by this method and by turbidity for ATP-induced trail head detachment in dimers. Howard and colleagues have analysed a Drosophila kinesin dimer with one head deleted, but both neck linkers present, and found ATP-induced unbinding at 0.3 s−1 (Hancock and Howard, 1998). Yajima et al (2002) measured the residence times of kinesin dimers tagged with a fluorescent actin filament, and estimated 2 s−1 for the detachment rate constant in ADP. In both these latter cases, the kinesin is attached to a large tag with a low diffusion coefficient, which might lead to underestimation of the detachment rate (Vugmeyster et al, 1998). In addition to all this, there is a potentially large effect of molecular crowding (mutual interference between neighbours on the MT lattice) on the unbinding rate constant, which is difficult to quantify.
To try to tease out the rules governing trail head detachment, and thereby distinguish between competing models for head–head coupling, we took two new approaches. Firstly, we used microflash photolysis in combination with conventional stopped-flow to measure unbinding rate constants. In flash photolysis of caged nucleotides, nucleotide concentration can transiently be increased using a laser flash in the absence of flow. Scattering artefacts due to flow are thereby eliminated, giving a substantial improvement in signal to noise, and access therefore to experimental regimes where the MT lattice is sparsely populated with kinesin, or where relatively low amounts of kinesin are caused to unbind. Secondly, we used T93N, a P-loop mutant of rat kinesin that binds irreversibly, as a roadblock (an obstacle to impede progress) to prevent forward stepping by processive kinesin dimers. As we show, this allowed us to measure the rate of nucleotide-induced trail head unbinding under conditions where the lead head could not attach. The roadblock experiment tests whether trail head detachment is accelerated by lead head attachment: if it is, a roadblock will cause the trail head to pause for some time before detaching. If not, the trail head will unbind rapidly. We measured the rates of ATP-induced and ADP-induced unbinding of processive dimers and of monomers of rat kinesin, the effect of added phosphate, and the effect of steric blockade of forward stepping using T93N. We found evidence that K.ADP.Pi is strong binding, that K.TRAPPEDADP is the detaching state, and that detachment of K.TRAPPEDADP trail heads is accelerated approximately two-fold by lead head attachment. The data imply a coordination mechanism in which ATP-induced lead head attachment is very fast, and subsequent ADP release from the lead head is slower and strain dependent.
Results
In flash-photolysis experiments, ATP or ADP were released at concentrations between 5 and 150 μM, and caused an approximately exponential decrease in light scattering (Figure 1A and B). We will assume throughout that this essentially monophasic reduction in light scattering reports purely the unbinding of kinesin from MT. The light-scattering transients were fitted to single exponentials, which were used to determine kobs. kobs increased with the concentration of released nucleotide up to 150 μM, the maximum accessible concentration. The data deviated slightly from single exponential behaviour. The factors limiting available nucleotide concentration and the origins of the deviation from exponentiality are discussed in Supplementary data. In the process of this work, we confirmed the earlier finding of Higuchi et al (1997) that cATP (caged ATP) or cADP competitively inhibit the binding of ATP or ADP to kinesin. To compensate, inhibition constants were determined, and a correction was applied (see Materials and methods and Supplementary data). In monomer experiments, the inhibition constants were 432±30 and 383±40 μM for cATP and cADP, respectively. In dimer experiments, the inhibition constants were 477±33 and 394±43 μM for cATP and cADP, respectively (Table I).
Figure 1.

Nucleotide-induced kinesin dissociation from MTs. (A, B) Light-scattering traces obtained by flash photolytic release of nucleotide. (A) rK340 monomers (0.8 μM) were dissociated from MT (0.4 μM) by flash photolytic release of 60.0 μM ATP or 66.3 μM ADP (as indicated). Single exponential fits to the traces (solid lines superimposed) gave kobs values of 21.2 and 10.6 s−1 for the ATP and ADP cases, respectively. (B) Similar traces obtained with rK430 dimers at 62.1 μM ATP (kobs=7.7 s−1) and 65.6 μM ADP (kobs=4.8 s−1). (C, D) Light-scattering traces obtained by stopped flow. (C) rK340 monomers (0.8 μM) were dissociated from MT (0.4 μM) by 750 μM ATP (kobs=51.6 s−1) or ADP (kobs=26.7 s−1). (D) rK430 dimers (0.8 μM) were dissociated from MT (0.4 μM) by 750 μM ATP (kobs=33.4 s−1) or ADP (kobs=29.3 s−1). (E, F) Combined flash-photolysis data and stopped-flow data. (E) Unbinding rate constants for ATP- (filled symbols) or ADP (empty symbols)-induced unbinding of rK340 monomers (0.8 μM) from MT (0.4 μM). (F) Unbinding rate constants for ATP- (filled symbols) or ADP (empty symbols)-induced unbinding of rK430 dimers (0.8 μM) from MT (0.4 μM). The flash-photolysis data (on the left of the vertical dashed line in (E) and (F)) were corrected for inhibition by caged nucleotide as described in Materials and methods. Hyperbolic fits to the corrected rate constants are shown as solid lines. Values obtained for the parameters of the hyperbolic fits are shown in Table I. In all cases, the concentrations given are mixing cell concentrations.
Table 1.
Kinetic parameters obtained for unbinding from MT of monomeric rat kinesin (rK340) and dimeric rat kinesin (rK430) in the presence of ATP or ADP
| Parameter | rK340 |
rK430 |
||
|---|---|---|---|---|
| ATP | ADP | ATP | ADP | |
| kmax (s−1) | 73.6±1.7 | 40.5±1.1 | 42.4±2.7 | 18.6±0.6 |
| Khalf (μM) | 63.1±7.9 | 109.0±15.7 | 138.4±35.4 | 133.6±15.5 |
| kon (s−1) | 3.8±1.2 | 1.5±0.6 | 1.7±0.8 | 1.3±0.2 |
| K1k+2 (μM−1 s−1) | 1.17 | 0.37 | 0.31 | 0.14 |
| KI (μM) | 432±30 | 383±40 | 477±33 | 394±43 |
To monitor nucleotide-induced unbinding at high nucleotide concentrations, we used stopped flow (Figure 1C and D). After compensation for the inhibitory effect of caged nucleotides in the flash-photolysis experiments, the flash photolysis and stopped-flow results were complementary and were combined (Figure 1E and F). The flash-photolysis technique allowed us to obtain data at low nucleotide concentrations and low occupancy, and thereby better to define Y-axis intercepts. Fitting these combined data to a hyperbola gave well-defined values for kmax (Table I).
Nucleotide-induced kinesin monomer (rK340) unbinding
The rK340 construct decorates MT at a stoichiometry of one kinesin monomer to one tubulin heterodimer (Hirose et al, 1995). The construct contains a full-length neck linker. Jiang et al (1997) have shown that intact neck linkers are important in triggering the rapid detachment of monomers. We measured the rates of nucleotide-induced unbinding of 0.8 μM rK340 monomers from 0.4 μM MT. kmax was approximately two-fold faster for ATP-induced unbinding at 73.6 s−1 than for ADP-induced unbinding at 40.5 s−1. In the ATP case the initial slope (kmax/Khalf) was steeper with a smaller Khalf and a faster kmax. The fitted hyperbolae showed nonzero Y-axis intercepts (kon in Table I), suggesting that a binding reaction of kinesin with MTs was significant. In this case kon should vary with the total protein concentration. We checked this, and found that the value of the intercept indeed varied linearly with the sum of the kinesin and MT concentrations, confirming that the binding reaction was significant under our conditions. A similar effect was described for the interaction of human nucleoside-diphosphate kinase with nucleotides (Schaertl et al, 1998). As expected, the deviation from single exponentials was more pronounced at lower nucleotide concentrations in the flash photolysis (Figure 1A) than at the greater nucleotide concentrations used in stopped-flow experiments (Figure 1C). This point is discussed in Supplementary data.
Nucleotide-induced kinesin dimer (rK430) unbinding
Analogous nucleotide-induced unbinding experiments were performed with rat kinesin dimers (rK430 0.8 μM head/0.4 μM MT sites) (Figure 1B, D and F). For ADP-induced dimer unbinding, fitting the combined flash photolysis and stopped-flow results to Equation 2 gave a kmax value of 18.6 s−1 (Table I). Accordingly, ADP-induced unbinding of dimers is at least two-fold slower than ADP-induced unbinding of monomers (40.5 s−1). Rates of ADP-induced dimer unbinding were insensitive to molecular crowding. By contrast, ATP-induced dimer unbinding was accelerated by molecular crowding.
Effects of molecular crowding
Under maximally crowded conditions (one rK430 molecule to one MT heterodimer), 750 μM ATP induced rK430 to unbind at 42 s−1 (Figure 1F). At one rK430 molecule to four MT heterodimers, rK430 unbound at 20.9 s−1, and at one rK430 molecule to eight MT heterodimers, rK430 unbound at 12.9 s−1 (not shown). ATP-induced unbinding of monomers was insensitive to molecular crowding (Figure 3D). We again observed that the values for Y-axis intercepts depended on MT and kinesin concentrations, consistent with the presence of a binding reaction.
Figure 3.

Effect of T93N on ATP-induced kinesin–MT unbinding. In total, 0.2 μM kinesin dimer was dissociated from 0.4 μM MT by flash photolytic release of ATP or by stopped-flow mixing with ATP. MTs were prereacted with T93N at the indicated occupancy. (A) Example of light-scattering traces obtained by microflash photolysis at 0, 0.2, 0.4 and 0.6 μM T93N. The superimposed solid lines are single exponential fits to the data (see Materials and methods). (B) Example of light-scattering traces obtained by stopped flow at 0.75 mM ATP to define kmax. The data are fitted to single exponentials. (C) Plots of the inverse of the observed rate constants (kobs−1, the residence time) against T93N concentration (data from stopped flow). (D) ATP dependence of the unbinding rate of 0.2 μM rK340 monomers in the absence of T93N (open circles) or in the presence of 0.4 μM T93N (filled circles). Crowding of rK340 in the absence of T93N also has no effect: filled squares show an unbinding of 0.8 μM rK340.
T93N construct
T93N is a kinesin monomer point mutant that binds essentially irreversibly to MTs. We reasoned that we could use this construct as a roadblock to prevent processive stepping completely, and then determine the rate of ATP-induced trail head detachment. The detailed properties of this construct are as follows.
Pelleting assay
We used this assay to check for the irreversibility of the binding of T93N to MT. After binding to MT and recovery of the complex by centrifugation, treatment with 5 mM ATP did not detectably dissociate T93N from MT (Figure 2A).
Figure 2.
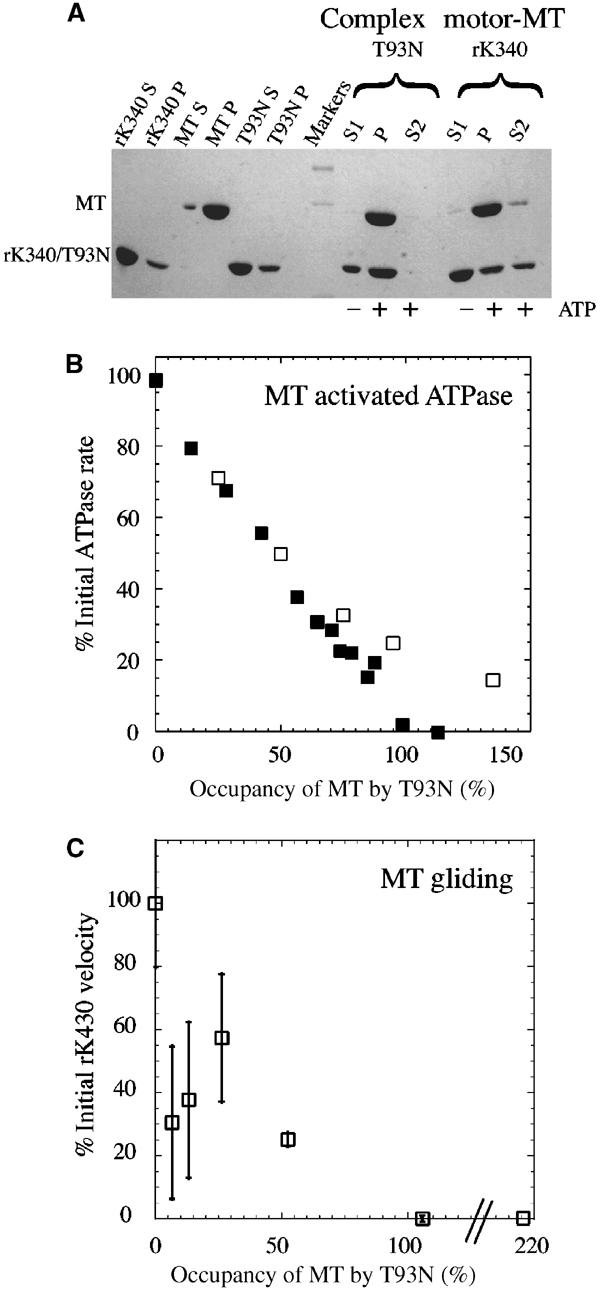
Characterisation of T93N. (A) MT pelleting experiment showing the irreversibility of the binding of T93N to MT. Left of markers: controls. Supernatants and pellets following centrifugation of wild-type kinesin alone, MT alone and T93N alone in the absence of ATP. Right of markers: ATP-induced unbinding. The first three lanes show the T93N experiment. A complex of MT and T93N was formed and centrifuged to produce a pellet P and a supernatant S1. The pellet P was resuspended in an ATP-containing solution and recentrifuged. The resulting supernatant S2 contained no detectable T93N, indicating that its binding to MT is effectively irreversible. The last three lanes show an rK340 positive control. In this case the ATP releases a large fraction of the rK340 into the supernatant S2. (B) Influence of the concentration of T93N on the rate of MT-activated ATPase of rK340 (open squares) and rK430 (filled squares). (C) Inhibition by T93N of MT gliding motility on a surface of rK430 GST. MTs were preloaded with T93N at the indicated occupancy and then introduced into the assay chamber. The maximum velocity (no T93N) under these conditions was 382 nm s−1. Both ATPase and motility assay data were obtained in the same buffer and at the same temperature as the transient data.
MT-activated ATPase
The effects of T93N on the MT-activated ATPase activity of the kinesin monomers and dimers were measured with a linked assay. Kinetically saturating concentrations of MT (3 μM for rK430 dimers, 20 μM for rK340 monomers) were incubated with T93N concentrations ranging from 0 to 4 μM for dimers, or from 0 to 30 μM for monomers, for at least 2 min. The wild-type motor was then added and the ATPase activity was measured. For both rK340 monomers and rK430 dimers, maximum inhibition corresponded to one T93N molecule per MT heterodimer (Figure 2B), assumed to correspond to 100% occupancy. For the monomer case, 30 μM MT was required to achieve full MT activation, and comparably high concentrations of T93N inhibitor were therefore used. Inhibition is accordingly partially masked at high T93N concentration by a small basal ATPase activity of T93N (0.029 s−1).
Motility
In MT sliding assays (Figure 2C), 6% decoration of MTs with T93N was sufficient to reduce MT velocity by a factor of two. At 52% T93N decoration, a wide variety of motility behaviours was observed: some MT moved with only their rear half attached, some moved slower than the rest, some stopped, some skated sideways before changing course, and some detached prematurely from the surface. For the 52% data point only, the velocity data were measured from that fraction of MT that moved smoothly (∼75%). Sporadically moving MT was not analysed.
Effects of T93N roadblock on ATP-induced kinesin unbinding
Control experiments (Figure 2) established that T93N binds stably to MT even in the presence of ATP. Control stopped-flow and flash-photolysis experiments showed no detectable nucleotide-induced unbinding of T93N from MT. A range of T93N concentrations was added to MT (0.4 μM) that had been partially decorated with wild-type rK430 (0.2 μM) (Figure 3). Observed rates of ATP-induced unbinding of RK430 increased with increasing T93N concentration, as determined by flash photolytic release of ATP (Figure 3A) or by stopped-flow mixing with ATP (Figure 3B). With all the free sites on the MT lattice blocked by T93N, the rate of rK430 unbinding reached a maximum of 42 s−1. The inverse of the unbinding rate constant kmax defines the average time (tav) that a kinesin molecule spends bound to the MT. Unbinding at 42 s−1 corresponds to a residence time of 23.8 ms (Figure 3C). The value obtained is similar to the unbinding rate obtained with rK430 alone when the MT lattice is entirely saturated with rK430 (Figure 1F). In contrast to the marked dependence on T93N of rK430 dimer unbinding, the rate for ATP-induced rK340 monomer (0.2 μM) unbinding from MT (0.4 μM) was not affected by 0.4 μM T93N (Figure 3D). The residence time shows an approximately linear dependence on T93N concentration (Figure 3C). Using the velocity of unimpeded stepping along the MT determined in motility assays (Figure 2) v=382 nm s−1 and a stepsize s=8 nm, the number of steps (n) taken can be estimated as n=(vtav)/s. The value of n decreased from ∼2.5 steps in moderately crowded conditions in the absence of T93N to ∼1.1 steps in 0.4 μM T93N. Note that the ∼50 steps per run were observed for rK430 under single-molecule conditions (Yajima et al, 2002), suggesting that even in our T93N-free control (0.2 μM rK430, 0.4 μM MT) run length is already considerably reduced by molecular crowding.
Effects of Pi and ionic strength on nucleotide-induced unbinding
The effect of 20 mM KPi on the nucleotide-induced unbinding of kinesin–MT was measured in the stopped-flow system in separate experiments under our standard conditions (0.4 μM MT site concentration, 0.8 μM kinesin head concentration) (Table II). Rates for nucleotide-induced unbinding in the presence of either 20 mM KPi or 40 mM KCl were measured at 200, 400 and 750 μM nucleotide concentration. Since 40 mM KCl provides the same ionic strength as 20 mM KPi the KCl experiments served as references. In the presence of 20 mM KPi, the kmax for ATP-induced monomer unbinding was reduced by 20% from 50.6 to 40.3 s−1, while that for ADP-induced monomer unbinding was reduced by 30% from 25.8 to 18.0 s−1. In ionic strength controls at 40 mM KCl, kmax was 49.4 and 23.2 s−1 for ATP and ADP, respectively, indicating that the effect of phosphate on nucleotide-induced monomer unbinding was not simply due to ionic strength-dependent charge masking, but was phosphate specific. For processive dimers in crowded conditions, added 20 mM KPi slowed the kmax for ATP-induced unbinding by 60% from 33.3 to 13.3 s−1, and the kmax for ADP-induced unbinding by 72% from 11.3 to 3.2 s−1. Again, ionic strength controls at 40 mM KCl (19.0 s−1 for ATP and 9.6 s−1 for ADP) indicated an effect of phosphate over and above that of ionic strength. Finally, we note that increased ionic strength not only slows down unbinding, but also weakens the affinity of kinesin for MT (Table II), implying that it must substantially slow down the binding of kinesin to MT. This is consistent with the two-step binding and unbinding scheme in Figure 4 (see below), wherein kinesin equilibrates between two bound states, a weak state that is destabilised by increasing ionic strength, and a strong state that is stabilised by increasing ionic strength.
Table 2.
Influence of phosphate on MT-activated ATPase and on the unbinding rates of monomeric rat kinesin (rK340) and dimeric rat kinesin (rK430)
| Effect of phosphate on MT-activated ATPase | rK340 |
rK430 |
||
|---|---|---|---|---|
| kcat (s−1) | K(0.5)MT (μM) | kcat (s−1) | K(0.5)MT (μM) | |
| Control | 28.83±2.09 | 3.87±0.86 | 47.85±3.07 | 0.71±0.20 |
| +40 mM KCl | 11.09±1.22 | 11.10±2.59 | 47.11±3.83 | 13.67±1.99 |
| +20 mM KPi | 16.01±3.81 | 9.75±6.23 | 42.84±2.82 | 3.43±0.72 |
| | ||||
| Effect of phosphate on unbinding rates | rK340 |
rK430 |
||
| |
ATP kmax (s−1) |
ADP kmax (s−1) |
ATP kmax (s−1) |
ADP kmax (s−1) |
| Control | 50.58±1.21 | 25.78±1.51 | 33.32±1.51 | 11.30±0.17 |
| +40 mM KCl | 49.36±0.77 | 23.19±0.65 | 19.04±0.48 | 9.56±0.21 |
| +20 mM KPi | 40.34±0.41 | 17.96±1.20 | 13.31±2.58 | 3.19±0.37 |
Figure 4.

Proposed kinetic scheme for monomers. Nucleotide binding is followed by an OPEN-to-CLOSED conformational change (see text). MT binding and unbinding are via an M.KTRAPPED.ADP intermediate. Rate constants for nucleotide binding and dissociation are consistent with the literature values (e.g. Ma and Taylor, 1997a, 1997b; Gilbert et al, 1998). The rate constant for complex formation of KTRAPPED.ADP and M is close to the diffusion limit (Hackney, 1995). Values for the other rate constants are estimates as indicated by the tildes (∼), and were made by simulating the scheme in Berkeley Madonna 8.1b6 and in KSIM (Neil Millar) using the current data (ADP-induced dissociation of M.K∼40 s−1; ATP-induced dissociation of M.K∼70 s−1; Km for ATP∼75 μM; Kd for ADP binding to M.K∼120 μM) as constraints, and the following further constraints: Kd for K.ADP-MT∼20–30 μM (Crevel et al, 1996); MT-activated transient release of ADP from K.ADP∼100 s−1 (Rogers et al, 2001).
Discussion
Kinetic scheme for monomers
To interpret the data, we adopt the generalised kinetic scheme for ATP turnover by single kinesin heads shown in Figure 4. The scheme has similarities to the scheme for myosin (Geeves et al, 1984) and has two notable features. First, nucleotide binding is followed by an isomerisation (a conformational change), corresponding to the well-known OPEN-to-CLOSED transition of the switch regions in myosin, kinesin and small G-proteins (Holmes and Geeves, 2000). Direct evidence for active site closure following binding of kinesin to MTs was recently described (Naber et al, 2003). In the scheme, this transition can be driven by either ATP binding or ADP binding. Second, the scheme postulates two-step MT binding and unbinding via a weakly bound M.K.TRAPPEDADP state (dotted box in Figure 4). A weak binding intermediate is functionally important for kif1a, a single-headed kinesin family member (Okada and Hirokawa, 2000). Evidence that a weak binding K.ADP intermediate is kinetically important for kinesin (kif5b) is given in Rogers et al (2001). Briefly, the Rogers data indicated that increasing the electrostatic attraction between the negative C-terminus of tubulin and the positive K-loop of kinesin both accelerated the recruitment of K.ADP on to the MT via an M.K.TRAPPEDADP state (dotted box in Figure 4) and inhibited a subsequent transition to a strongly bound state. It is well established that an ADP state of kinesin with a very low rate constant for ADP release is the default state for kinesin in solution (Hackney, 1994b). In our proposed scheme, this K.TRAPPEDADP state can attach weakly to MT to form M.K.TRAPPEDADP without the activation of ADP release. Flux through this state is the dominant pathway for MT binding and release.
Several lines of evidence suggest that detachment via K.ADP.Pi is a minor pathway. Kinesin in the presence of the transition state analogues ADP.AlF4 and ADP.BeFx binds tightly to MT, suggesting that a transient K.ADP.Pi intermediate also binds tightly to MT (Crevel et al, 1996; Rosenfeld et al, 1996; Ma and Taylor, 1997b). It is possible however that these analogues do not accurately mimic the K.ADP.Pi state. In direct measurements of Pi release (Gilbert et al, 1995), the transient rate of Pi release was found to be essentially identical to the unbinding rate, consistent with either Pi release limiting unbinding, or vice versa. Accordingly, kinesin models have been proposed in which K.ADP.Pi is an unbinding intermediate (Schief and Howard, 2001). Here we observed a substantial reduction in monomer detachment rates with added 20 mM Pi (Table II), consistent with a weak affinity for Pi and with the K.ADP.Pi state being tightly bound to MT. Based on these observations, we limit our attention to the case where detachment occurs only from M.K.TRAPPEDADP.
For rK340 monomers, we find that ATP-induced unbinding is faster than ADP-induced unbinding. For the scheme to be valid therefore, the longer pathway to detachment via ATP binding, hydrolysis and Pi release must be faster than the shorter pathway to detachment via ADP binding (Figure 4). Is this plausible? Ma and Taylor (1997b) found, for human K332 monomers, that kmax for ADP-induced unbinding at 65–90 s−1 was faster than kmax for ATP-induced unbinding at 40–50 s−1. Jiang and Hackney (1997) found, for Drosophila K357 monomers, that at low nucleotide concentration, ADP-induced unbinding was slower than ATP-induced unbinding, but that at saturating nucleotide the rates were equal (kmax 45 s−1). We found for rK340 that kmax for ATP-induced unbinding was 73.6 s−1 and that kmax of ADP-induced unbinding was 40.5 s−1. The apparent second-order rate constants for nucleotide-induced dissociation of the motor–MT complex (the bimolecular rates (kmax/Khalf)) that we obtained were similar to those obtained by Jiang and Hackney: we found 1.17 and 0.37 μM−1 s−1 for ATP and ADP, respectively (Table I), compared to Hackney's values for Drosophila kinesin of 1.60 and 0.41 μM−1 s−1. The scheme in Figure 4 has an isomerisation (a conformational change, corresponding to the OPEN-to-CLOSED transition) following binding of either ATP or ADP. To account for the data, we suggest that the OPEN-to-CLOSED transition that follows ADP binding is slower than that following ATP binding. A slight variation (approximately two-fold) in the rate constant for the ADP-driven OPEN-to-CLOSED transition would be enough to account for the differences between monomeric constructs from different species, while retaining the scheme in Figure 4.
ADP-induced unbinding of dimers
For ADP-induced unbinding of rK430 dimers, we found 18.6 s−1, which is close to the consensus figure of 12–13 s−1 for Drosophila and human kinesins given by Hackney (2002). ADP-induced dimer unbinding is thus about half as fast as ADP-induced monomer unbinding, indicating altered MT interaction in the dimer case. Hackney noted that the ADP-induced detachment of dimers must be close to 13 s−1 even in conditions where the MT lattice was sparsely populated. We measured ADP-induced detachment directly at both crowded and sparse stoichiometries, and confirmed that there was no effect of molecular crowding on the rate of ADP-induced detachment of dimers (data not shown).
The roadblock experiment and the mechanism of ATP-induced trail head detachment
The T93N reagent binds irreversibly to MT and its inhibition of sliding motility and of MT-activated ATPase is maximal at a 1:1 stoichiometry of T93N heads to MT heterodimers. The T93N roadblock allows us to measure the rate of ATP-induced trail head detachment in conditions where lead head attachment is prevented. We found that with all other sites on the MT blocked by the T93N, rK430 dimers detached at 42 s−1. Since available binding sites for the lead head are all blocked under these conditions, the value measured is the default rate constant for ATP-induced detachment of the trail head. The result obtained distinguishes between models in which trail head detachment occurs always with a fixed rate constant from those in which the rate constant depends on lead head attachment: during processive stepping at low load, trail head detachment must occur at least as fast as the overall stepping rate of ∼50–70 s−1. Our data indicate that at each step during a processive run under single-molecule conditions, trail head detachment as K.TRAPPEDADP is indeed accelerated by lead head attachment, but only by a maximum of less than two-fold. Presumably, the acceleration occurs because the plus-end-directed strain generated by lead head attachment accelerates trail head detachment.
Implications for the coordination of molecular walking
All candidate coordination mechanisms so far proposed incorporate the feature discovered by Hackney (1994a), that an ATP-dependent conformational change of the trail head works as a trigger for ADP release from the lead head. In the absence of ATP, the trail head attaches tightly to the MT in a rigor conformation, and the lead head is parked close to it, in a configuration (the tethered intermediate) that prevents the lead head undergoing MT-activated ADP release. ATP binding to the trail head then unparks the lead head and allows it to access its next site along the MT, with an associated shift in the dynamics of the neck linkers that join each head to the tail. The result is a bridge structure, a transient intermediate with both heads attached to the MT, an empty (apo or rigor) lead head and a trail head containing either ADP.Pi or ADP. The various candidate coordination schemes differ in their predictions for the pathway by which this bridge intermediate breaks down. The Rice et al (1999) scheme has ATP binding to the lead head causing docking of its neck linker, thereby applying strain to K.ADP trail heads and accelerating their detachment. By contrast, the scheme recently proposed by Rosenfeld et al (2003) has ATP binding to the lead head inhibited until trail head detachment has occurred, thereby avoiding a potential risk at high ATP concentration of futile cycling on the lead head. The scheme proposed by Gilbert and colleagues (Farrell et al, 2002) has MT-activated ADP release from the lead head occurring after binding but before “locking down” of the lead head, with locking down requiring ATP hydrolysis on the trail head. Howard and colleagues (Schief and Howard, 2001) have argued, based on the measured behaviour of a one-headed kinesin dimer, that the ATP-induced detachment of trail heads in intact dimers must be <1 s−1, requiring therefore a processivity scheme with substantial acceleration of trail head detachment by lead head attachment. Hackney (2002) has also argued for acceleration of trail head detachment by lead head attachment. Our own proposed scheme (Cross et al, 2000) has Pi release from the trail head occurring before the trail head detaches from the MT, and trail head detachment as K.TRAPPEDADP accelerated by plus-end-directed strain from the newly rigorised lead head. One point of agreement is that ATP binding is strain dependent. This emerges very clearly from single-molecule optical trapping, when each step corresponds to the binding of one ATP molecule, and the stepping frequency decreases with increasing load, indicating that ATP binding is inhibited by load (Svoboda and Block, 1994). Nishiyama et al (2002) reported that back steps also obey this relationship. Schnitzer and Block argue for a load-dependent isomerisation (a conformational change) of a K.ATP state (Schnitzer et al, 2000).
Our roadblock experiment using T93N indicates approximately two-fold acceleration of trail head detachment by lead head attachment. This modest value has general implications for the coordination mechanism. Specifically, it implies that lead head attachment during molecular walking must occur very rapidly. To see this, consider that at each step during a processive run, a kind of race develops between lead head attachment and trail head detachment. A processivity of ∼50, as for our construct under single-molecule conditions (Yajima et al, 2002), implies that lead head attachment wins this race 49 times out of 50. Our value of ∼42 s−1 for the default rate of trail head detachment therefore implies an effective rate constant for lead head attachment of ∼50 × 42 or ∼2000 s−1.
Rates approaching this were measured in a recent probe study wherein Rosenfeld et al (2002) found >800 s−1 for the ATP-induced separation of the two rhodamine-labelled neck linkers of a kinesin dimer. Hitherto, we and others have assumed that tight attachment of the lead head was essentially synchronous with mantADP release from the lead head (Crevel et al, 1999), so that ATP-induced mantADP release from the lead head at ∼100 s−1 was effectively a direct reporter of attachment. But in fact, tight attachment of the lead head might precede MT-activated ADP release by a considerable interval. Figure 5 illustrates this idea. In this scheme, ADP release is strain sensitive. Starting from the tethered intermediate with trail head empty and the lead head in the K.TRAPPEDADP state (top of Figure 5), we envisage the following sequence: ATP binds to the trail head, causing a conformational change that renders the active site hydrolysis-competent, and also releases the lead head from its parked position, allowing it to access its next site along the MT. Stable attachment to this next site occurs very fast; we estimate at ∼2000 s−1. As the lead head finds and binds strongly to its next site, it experiences negative (minus-end-directed) strain, which tends to stabilise its attachment and inhibit the release of ADP. Meanwhile the trail head experiences positive strain, and this tends to accelerate its detachment in the K.TRAPPEDADP state. Following trail head detachment, the strain on the lead head is relieved and ADP release from the lead head follows rapidly. The effect of this will be to make ADP release from the lead head act as a mechanochemical gating device (a one-way valve) in the walking mechanism, locking in the progress gained by diffusion-to-capture of the lead head. Coupling between ADP release from the lead head and detachment of the trail head would account for the observation that ADP release can be rate limiting on the overall cycle (Lockhart et al, 1995b). The general principle that minus end-directed strain tends to inhibit nucleotide exchange is reasonable, based on the known strain dependence of ATP binding (Nishiyama et al, 2002). Recent single-molecule studies by Ishiwata and colleagues have pointed to strain-dependent ADP release (Kawaguchi et al, 2003). Our own studies have shown that added ADP can efficiently inhibit single-molecule walking (Yajima et al, 2002).
Figure 5.

Logical gating scheme for dimers. The scheme shown refers to molecular walking at low external load, and corresponds to the chemical kinetic release of ADP from the lead head induced by the binding of ATP to the trail head. Following ATP binding to the trail head, tight attachment of the lead head to its next site occurs rapidly (>2000 s−1), but subsequent MT-activated ADP release from the lead head is inhibited by distortions due to minus endwards tension (black chevron). Relief of this strain by release of the trail head from the MT then allows ADP release from the lead head. The mechanochemical interdependencies of the two heads are like logic (IF...THEN DO) gates, as shown.
Control logic of kinesin
It is possible to regard kinesin as a robotic stepping machine and to propose a specific control scheme for its walking action. Such a scheme comprises a set of rules that make the actions of the two heads interdependent: at multiple points in the stepping cycle, each head pauses, interrogates its partner head and requires that something be true before it proceeds. For kinesin there is evidence for several such rules, as indicated in Figure 5. Starting from the tethered intermediate, the first such mutual dependency requires ATP binding to the trail head before allowing attachment of the lead head (Hackney, 1994a). As argued above, our data imply a second such dependency at the point of ADP release from the lead head, where strain due to the attached trail head must be relieved to sanction ADP release from the lead head. A third control point operates for trail head detachment, which as we have shown is accelerated, albeit modestly, by lead head attachment. These head–head dependencies are akin to logic gates in a control circuit, except that what is controlled is the probability of a particular outcome.
Consequences for the cell
Our data imply that the lead head must find and bind to its next site along the MT within a narrow kinetic window of opportunity. Following ATP binding to the trail head, its rate of detachment is enhanced by plus-end-directed strain, but in the absence of strain is simply on a timer. If the lead head fails to attach within this time window (as when roadblocked), then the trail head will in any case detach at a default rate of ∼42 s−1. Assuming a cycle rate (for each head) of 50–70 s−1, a default detachment rate of ∼42 s−1 will mean that on colliding with an obstacle, the molecule will wait for slightly less than one additional ATPase cycle before unbinding, meanwhile continuously probing for its next site. If the lead head ultimately fails to attach, the trail head will then detach. In the cell, this behaviour will maintain processive stepping until an obstacle is encountered, but in the event the obstacle is permanent it will allow the motor to detach. Tight ATPase coupling will thereby be maintained in the face of an immovable obstacle. If, as seems likely, this behaviour extends to collaborating teams of kinesins, either individually processive or individually nonprocessive, then one head encountering an obstacle will not block the progress of the entire team.
Materials and methods
Construction, expression and purification of proteins
rK340 was described in Lockhart et al (1995a). rK340-T93N (named T93N in this paper) was described in Krylyshkina et al (2002). rK430GST was described in Crevel et al (1997). RK430 was purified using the construct pETK430 described in Crevel et al (1997). RK430 cells were lysed into buffer A (20 mM PIPES (pH 6.9) containing 1 mM DTT, 5 mM MgCl2) and the clarified supernatant was applied to a 5 ml Hitrap S equilibrated in buffer A. After extensive washing of the column with buffer A, partially purified rK430 was eluted with buffer A supplemented with 100 mM NaCl. Pooled peak fractions were diluted two-fold in buffer A and applied to a 1 ml High trap Q column. After extensive washing with buffer A, rK430 was eluted from the Q column using buffer A supplemented with 200 mM NaCl. The concentrations of the proteins were determined using calculated coefficients of 15 300 M−1 cm−1 for rK340, 14 000 M−1 cm−1 for rK340-T93N (we assumed no nucleotide in the active site), 34 300 M−1 cm−1 for rK430 and 110 000 M−1 cm−1 for MT. Concentrations of motor are expressed throughout as concentrations of heads: there is one active site per head. Concentrations of MT are expressed as heterodimer concentrations: there is one kinesin head binding site per MT heterodimer.
Solution molecular weights of expressed proteins
The solution association state of the expressed and purified proteins was checked using glycerol gradient centrifugation and analytical gel filtration on Superose 12. Both techniques confirmed that rK340 behaves like a monomer in solution and that rK430 is dimeric. Gel filtration confirmed that T93N is a monomer.
Flash-photolysis experiments
The apparatus was recently described (Weiss et al, 2000). Briefly, a 20 μl cuvette containing the sample was illuminated with a 5 ns pulse of high-intensity UV laser light (355 nm) triggering release of ATP or ADP (up to ∼150 μM) from caged ATP or caged ADP, respectively (Walker et al, 1988). The sample was also illuminated with white light (>380 nm), and the absorbance at 405 nm and 90° light scattering through a 455 nm high-pass filter were recorded following each flash. The UV absorption at 405 nm was used to monitor the concentration of the aci-nitro breakdown product of the 1(2-nitrophenyl) ethyl ester photoreleased caging group, which tracks ATP release as described in Weiss et al (2000).
For the present experiments, MT and kinesin were mixed in the presence of apyrase, producing an apo (rigor) complex. Caged-ATP (adenosine 5′-triphosphate (1-(2-nitrophenyl) ethyl ester)) and caged-ADP (adenosine 5′-diphosphate (1-(2-nitrophenyl) ethyl ester) stocks were made at 10 mM in 20 mM MOPS (pH 7.0), 5 mM MgCl2, 100 mM KCl. MT:motor complex was formed in 20 mM MOPS buffer (pH 7.0) containing 10 mM DTT, 5 mM MgCl2, 20 μM paclitaxel and 500 μM caged nucleotide, except for the experiment shown in Figure 3A, for which 1000 μM was used. In our standard conditions, stock MTs were diluted to 0.4 μM heterodimer concentration and stock motor was added to yield the final concentration of 0.8 μM. The sample was spun 2 min at 3000 rpm to eliminate air bubbles and the sample (20 μl) was then loaded into the apparatus. The binding of uncaged nucleotide to kinesin caused dissociation of the MT–kinesin complex, and this was monitored using the associated decrease in light scattering. The excess photoreleased nucleotide was then hydrolysed by the kinesin and apyrase, so that the light-scattering signal recovered. A further laser flash could then be delivered. Typically four to six flashes were performed per sample, varying the dose of laser energy to release various amounts of nucleotide.
Stopped-flow experiments
Stopped-flow experiments were performed with a standard Hi-Tech SF-61DX2 stopped-flow spectrophotometer. The MT:motor complex was formed in 20 mM MOPS buffer (pH 7.0) containing 10 mM DTT, 5 mM MgCl2 and 20 μM paclitaxel to which 10 mM KCl was added (to provide the same conditions as in the flash-photolysis experiments). Apyrase was not added routinely. Control experiments showed that the presence of apyrase did not affect the kinetic parameters. The complex of kinesin (0.8 μM) or MT (0.4 μM) was mixed with ATP or ADP at 200, 400 and 750 μM. The concentrations given are those established after the stopped-flow mixing. The samples were illuminated at 320 nm and the 90° light scattering was recorded. We used the 90° scattering signal in preference to the turbidity signal in order to improve the signal to noise ratio.
Measurement of steady-state ATPase rates
The assay was performed as previously described (Crevel et al, 1997), using a pyruvate kinase/lactate dehydrogenase linked assay. The assay was performed in 20 mM MOPS buffer containing 10 mM DTT, 1 mM MgCl2 and 20 μM paclitaxel. To measure the effect of T93N, sufficient MTs were added to obtain maximal ATPase: 3 μM MT for rK430 and 20 μM MT for rK340.
MT motility
MT motility assays were performed according to Crevel et al (1997) using a buffer corresponding to that used in kinetic experiments. rK430GST was used to coat the surface of a coverslip, and MTs preincubated for 2 min with various amounts of T93N were loaded into the chamber. Motility was observed in 20 mM MOPS buffer (pH 7.2) containing 10 mM DTT, 2 mM MgCl2, 20 μM paclitaxel and 1 mM ATP, and quantitated using RETRAC 2 (http://mc11.mcri.ac.uk/retrac.html).
Pelleting assay
In all, 8.4 μM T93N or rK340 (wild type) and 2.8 μM MT heterodimer were incubated in 20 mM Pipes (pH 6.9), 10 mM DTT, 2 mM MgCl2 and 20 μM paclitaxel, and centrifuged in a Beckman TL100 centrifuge at 100 000 g for 10 min at 20°C. The supernatants were saved and the pellets were resuspended in the same buffer, with the addition of 5 mM ATP. A second centrifugation was performed, the supernatant was saved, and the pellet was resuspended in the same volume (40 μl). Both supernatants (+ and − ATP) and pellets (+ATP) were loaded on an SDS gel and stained with Coomassie brilliant blue. Controls confirmed negligible pelleting of motor in the absence of MT.
Reagents
Caged ATP was from Molecular Probes. Caged ADP was a kind gift from John Corrie (MRC Mill Hill); some later experiments were carried out using caged ADP from Molecular Probes.
Kinetic models and fitting procedures
For nucleotide-induced kinesin dissociation, the kinetic scheme presented in Scheme I was used. The scheme describes the ADP-induced dissociation of kinesin from MT. M, K and D represent MT, kinesin and ADP, respectively.
Figure 6.

Scheme I
In the model, step 1 and step 3 is a fast equilibrium and kobs then shows a hyperbolic dependence on nucleotide concentration:
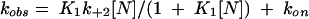 |
1 |
where K1=k−1/k+1 and K1k+2 is a second-order dissociation constant (the initial slope). kon is the rate of kinesin binding to MT and its value is a function of the concentration of kinesin and MT. Control experiments provided evidence for the inhibition of ATP or ADP binding to kinesin–MT by cATP or cADP. The inhibitory effects of these caged nucleotides were characterised in separate experiments to determine the value of the inhibition constants (KI) for each case. The method for this determination is described in Supplement A, and the KI values obtained from the analysis are presented in Table I. Using these KI values, the measured rate constants (kobs) were corrected at each caged nucleotide concentration ([cN]) with the following equation:
 |
2 |
Supplementary Material
Supplementary Material
Acknowledgments
This work was supported by a grant from the Human Frontiers Science Program, by a European Union Grant HPRN-CT-2000-00091 (MAG) and by a Wellcome Trust Programme Grant 055841 (MAG). MNy is an EMBO/HHMI Scientist.
References
- Case RB, Rice S, Hart CL, Ly B, Vale RD (2000) Role of the kinesin neck linker and catalytic core in microtubule-based motility [see comments]. Curr Biol 10: 157–160 [DOI] [PubMed] [Google Scholar]
- Coy DL, Wagenbach M, Howard J (1999) Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem 274: 3667–3671 [DOI] [PubMed] [Google Scholar]
- Crevel I, Carter N, Schliwa M, Cross R (1999) Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin. EMBO J 18: 5863–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel IM, Lockhart A, Cross RA (1996) Weak and strong states of kinesin and ncd. J Mol Biol 257: 66–76 [DOI] [PubMed] [Google Scholar]
- Crevel IM, Lockhart A, Cross RA (1997) Kinetic evidence for low chemical processivity in ncd and Eg5. J Mol Biol 273: 160–170 [DOI] [PubMed] [Google Scholar]
- Cross RA, Crevel I, Carter NJ, Alonso MC, Hirose K, Amos LA (2000) The conformational cycle of kinesin. Philos Trans R Soc Lond B 355: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell CM, Mackey AT, Klumpp LM, Gilbert SP (2002) The role of ATP hydrolysis for kinesin processivity. J Biol Chem 277: 17079–17087 [DOI] [PubMed] [Google Scholar]
- Geeves MA, Goody RS, Gutfreund H (1984) Kinetics of acto-S1 interaction as a guide to a model for the crossbridge cycle. J Muscle Res Cell Motil 5: 351–361 [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Moyer ML, Johnson KA (1998) Alternating site mechanism of the kinesin ATPase. Biochemistry 37: 792–799 [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Webb MR, Brune M, Johnson KA (1995) Pathway of processive ATP hydrolysis by kinesin. Nature 373: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1994a) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA 91: 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1994b) The rate-limiting step in microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains occurs while bound to the microtubule. J Biol Chem 269: 16508–16511 [PubMed] [Google Scholar]
- Hackney DD (1995) Implications of diffusion-controlled limit for processivity of dimeric kinesin head domains. Biophys J 68: 267s–269s [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (2002) Pathway of ADP-stimulated ADP release and dissociation of tethered kinesin from microtubules. Implications for the extent of processivity. Biochemistry 41: 4437–4446 [DOI] [PubMed] [Google Scholar]
- Hancock WO, Howard J (1998) Processivity of the motor protein kinesin requires two heads. J Cell Biol 140: 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi H, Muto E, Inoue Y, Yanagida T (1997) Kinetics of force generation by single kinesin molecules activated by laser photolysis of caged ATP. Proc Natl Acad Sci USA 94: 4395–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA (1995) Nucleotide-dependent angular change in kinesin motor domain bound to tubulin. Nature 376: 277–279 [DOI] [PubMed] [Google Scholar]
- Holmes KC, Geeves MA (2000) The structural basis of muscle contraction. Philos Trans R Soc Lond B 355: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua W, Young EC, Fleming ML, Gelles J (1997) Coupling of kinesin steps to ATP hydrolysis. Nature 388: 390–393 [DOI] [PubMed] [Google Scholar]
- Jiang W, Hackney DD (1997) Monomeric kinesin head domains hydrolyze multiple ATP molecules before release from a microtubule. J Biol Chem 272: 5616–5621 [DOI] [PubMed] [Google Scholar]
- Jiang W, Stock MF, Li X, Hackney DD (1997) Influence of the kinesin neck domain on dimerization and ATPase kinetics. J Biol Chem 272: 7626–7632 [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Uemura S, Ishiwata S (2003) Equilibrium and transition between single- and double-headed binding of kinesin as revealed by single-molecule mechanics. Biophys J 84: 1103–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylyshkina O, Kaverina I, Kranewitter W, Steffen W, Alonso MC, Cross RA, Small JV (2002) Modulation of substrate adhesion dynamics via microtubule targeting requires kinesin-1. J Cell Biol 156: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A, Crevel IM, Cross RA (1995a) Kinesin and ncd bind through a single head to microtubules and compete for a shared MT binding site. J Mol Biol 249: 763–771 [DOI] [PubMed] [Google Scholar]
- Lockhart A, Cross RA, McKillop DF (1995b) ADP release is the rate-limiting step of the MT activated ATPase of non-claret disjunctional and kinesin. FEBS Lett 368: 531–535 [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW (1995) Mechanism of microtubule kinesin ATPase. Biochemistry 34: 13242–13251 [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW (1997a) Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem 272: 724–730 [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW (1997b) Kinetic mechanism of a monomeric kinesin construct. J Biol Chem 272: 717–723 [DOI] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA (1998) Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry 37: 800–813 [DOI] [PubMed] [Google Scholar]
- Naber N, Minehardt TJ, Rice S, Chen X, Grammer J, Matuska M, Vale RD, Kollman PA, Car R, Yount RG, Cooke R, Pate E (2003) Closing of the nucleotide pocket of kinesin-family motors upon binding to microtubules. Science 300: 798–801 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Higuchi H, Yanagida T (2002) Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat Cell Biol 4: 790–797 [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N (2000) Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA 97: 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD (1999) A structural change in the kinesin motor protein that drives motility. Nature 402: 778–784 [DOI] [PubMed] [Google Scholar]
- Rogers KR, Weiss S, Crevel I, Brophy PJ, Geeves M, Cross R (2001) KIF1D is a fast non-processive kinesin that demonstrates novel K-loop-dependent mechanochemistry. EMBO J 20: 5101–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM (2003) Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem 278: 18550–18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Rener B, Correia JJ, Mayo MS, Cheung HC (1996) Equilibrium studies of kinesin-nucleotide intermediates. J Biol Chem 271: 9473–9482 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Xing J, Jefferson GM, Cheung HC, King PH (2002) Measuring kinesin's first step. J Biol Chem 16: 16. [DOI] [PubMed] [Google Scholar]
- Schaertl S, Konrad M, Geeves MA (1998) Substrate specificity of human nucleoside-diphosphate kinase revealed by transient kinetic analysis. J Biol Chem 273: 5662–5669 [DOI] [PubMed] [Google Scholar]
- Schief WR, Howard J (2001) Conformational changes during kinesin motility. Curr Opin Cell Biol 13: 19–28 [DOI] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388: 386–390 [DOI] [PubMed] [Google Scholar]
- Schnitzer MJ, Visscher K, Block SM (2000) Force production by single kinesin motors. Nat Cell Biol 2: 718–723 [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM (1994) Force and velocity measured for single kinesin molecules. Cell 77: 773–784 [DOI] [PubMed] [Google Scholar]
- Vugmeyster Y, Berliner E, Gelles J (1998) Release of isolated single kinesin molecules from microtubules. Biochemistry 37: 747–757 [DOI] [PubMed] [Google Scholar]
- Walker JW, Reid GP, McCray JA, Trentham DR (1988) Photolabile 1-(2-nitrophenyl)ethyl phosphate esters of adenine nucleotide analogues. Synthesis and mechanism of photolysis. J Am Chem Soc 110: 7170–7177 [Google Scholar]
- Weiss S, Chizhov I, Geeves MA (2000) A flash photolysis fluorescence/light scattering apparatus for use with sub microgram quantities of muscle proteins. J Muscle Res Cell Motil 21: 423–432 [DOI] [PubMed] [Google Scholar]
- Yajima J, Alonso MC, Cross RA, Toyoshima YY (2002) Direct long-term observation of kinesin processivity at low load. Curr Biol 12: 301–306 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


