Abstract
Compounds that interact with DNA or RNA generally act as inhibitors of enzymes that unwind DNA or RNA. In the present study we describe the synthesis and properties of some nucleoside analogues that interact with double-stranded DNA but that, in contrast, facilitate the unwinding reaction mediated by West Nile (WN) virus nucleoside triphosphatase (NTPase)/helicase. The nucleoside analogues described, 1-(2′-O-methyl-β-d-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5H,6H)-dione (HMC-HO4), 1-(β-d-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5H,6H)-dione, and 1-(2′-deoxy-α-d-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5H,6H)dione, all contain the imidazo[4,5-d]pyridazine ring system. The extent of the enhancing effect on helicase activity was found to be dependent on the time of exposure of the DNA substrate to the compounds and their concentrations. The nucleoside analogues were nevertheless found to be capable of uncoupling the ATPase and helicase activities of the enzyme by a mechanism operating on the level of the enzyme. Thus, in the case of HMC-HO4, the direct interaction with the enzyme caused inhibition of its helicase activity, with a half-maximal inhibitory concentration of 30 μM. The similar potency of the compound against replication of WN virus in cell culture suggests that inhibition of the helicase activity of the viral enzyme is responsible for the observed antiviral activity of HMC-HO4 and may indeed represent an important mode of action of antiviral drugs in general. Comparative studies performed with the related NTPase/helicase from hepatitis C virus revealed that the extent of the effects mediated by imidazo[4,5-d]pyridazine nucleosides is enzyme specific. The substances described may represent a starting point for the development of a new class of helicase-specific antivirals.
The members of the family of Flaviviridae are small, enveloped RNA viruses with similar structures (31). The viral genome encodes a polyprotein of approximately 3,000 amino acids that is processed into three structural proteins (C, prM, and E) and at least seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (1, 6, 27). Among the nonstructural proteins, NS3 appears to be the most promising target for antiviral agents because of the multiple enzymatic activities associated with this protein. NS3 exhibits serine protease activity that resides in the NH2 terminus of the protein and nucleoside triphosphatase (NTPase) and RNA helicase activities located in the COOH terminus (10, 11, 12). NTPase/helicase, together with the NS5-associated RNA polymerase, is thought to be an essential component of the viral replicase complex (19).
The NTPases/helicases are capable of enzymatically unwinding duplex RNA or DNA structures by disrupting the hydrogen bonds that keep the two strands together (17, 21). This is accomplished by a reaction that is coupled to the hydrolysis of a nucleoside triphosphate (NTP). Nonhydrolyzable ATP analogues did not substitute for ATP in the RNA-unwinding reaction, suggesting that ATP hydrolysis is required for this reaction (10). Although the helicase activity is dependent on the energy produced in the course of NTP hydrolysis, numerous observations show that the number of events of NTP hydrolysis per unwinding cycle is not a constant value (2, 5). Thus, potential specific inhibitors of the NTPases/helicases of members of the family Flaviviridae could act by any one or more of the following mechanisms: (i) inhibition of NTPase activity by interference with NTP binding (3), (ii) inhibition of NTPase activity by an allosteric mechanism (3), and (iii) inhibition of the coupling of NTP hydrolysis to the unwinding reaction (5). Other inhibitory mechanisms are also conceivable. These may involve modulation of interaction of the enzyme with its RNA or DNA substrate, for example, (iv) competitive inhibition of RNA binding (28) and (v) inhibition of the unwinding by steric blockage of translocation of the helicase along the polynucleotide chain (25).
Because of the well-established antihelicase activities of numerous DNA-interacting agents, we were interested in developing NTPase/helicase inhibitors that act by interaction with a DNA substrate. In this report we describe the compounds 1-(2′-O-methyl-β-d-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5H,6H)-dione (HMC-HO4), 1-(β-d-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5H,6H)-dione (HMC-HO5), and 1-(2′-deoxy-α-d-ribofuranosyl)imidazo[4,5-d]pyridazine-4,7(5 H,6H)dione (HMC-HO1α). These compounds are analogues of purine nucleosides in which a pyridazine moiety replaces a pyrimidine fused to an imidazole ring. Our preliminary molecular modeling studies as well as the results of the experiments presented here suggest an interaction of these compounds with DNA. Surprisingly, the detailed kinetic analyses reported in this study revealed that this interaction results instead in an enhancement of the unwinding activities of the NTPases/helicases of the West Nile (WN) virus and of the related virus hepatitis C virus (HCV). On the other hand, the compounds were also discovered to interact directly with the enzymes investigated and uncouple their ATPase and helicase activities. In the case of HMC-HO4, this interaction resulted in a decrease in the level of the unwinding reaction mediated by the WN virus enzyme. These two very different effects occur at quite different concentrations of HMC-HO4. The HMC-HO4-mediated inhibition of the helicase activity correlated with the corresponding reduction in the level of WN virus replication in cell culture.
MATERIALS AND METHODS
Sources of enzymes.
The starting material for the purification of the WN virus NTPase-helicase was the medium harvested from cultures of WN virus-infected Vero E6 cells. The purification procedure has previously been described in detail (5). The NTPase/helicase domain of HCV NS3 was expressed in Escherichia coli and was purified as described previously (14, 20). Since the enzyme preparation contained some background proteins, the purification procedure was completed by gel exclusion chromatography on a Superdex-200 column (Amersham-Pharmacia) (4). The final preparation obtained contains homogeneous HCV NTPases/helicases when analyzed on a sodium dodecyl sulfate (SDS)-polyacrylamide gel stained with Coomassie blue.
ATPase and helicase assays.
A standard ATPase assay was performed by the charcoal adsorption method described previously (3, 4). The helicase activity of the enzyme was determined by using a DNA substrate that was obtained by annealing two partly complementary cDNA oligonucleotides that were synthesized with a sequence that corresponds to the deoxynucleotide versions of the RNA strands described previously (10). The helicase activity was tested with 2 pmol of enzyme incubated in reaction mixture (final volume, 25 μl) containing 20 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol, 0.01% Triton X-100, 0.1 mg of bovine serum albumin per ml, 9.5 μM ATP, and 4.7 pM DNA substrate. The reaction was allowed to proceed for 30 min at 30°C and was stopped by addition of 5 μl of termination buffer (100 μM Tris-HCl [pH 7.5], 20 mM EDTA, 0.5% SDS, 0.1% Triton X-100, 25% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol). The samples were separated on a Tris-borate-EDTA (TBE)-15% polyacrylamide gel containing 0.1% SDS (5). The gels were dried and exposed to Kodak X-ray films at −70°C. Subsequently, the parts of the gels corresponding to the released strand and to the not-unwound substrate were cut out, and the amount of 32P radioactivity was measured.
Assays for determination of effect of interaction of imidazo[4,5-d]-pyridazine nucleosides with DNA on unwinding efficacy of NTPase/helicase.
Assays for determination of the effect of the interaction of imidazo[4,5-d]-pyridazine nucleosides with DNA on the unwinding efficacy of NTPase/helicase were performed in two steps: (i) step 1, preincubation of the DNA substrate with the compound, and (ii) step 2, determination of the helicase activities of the enzymes investigated against the preincubated DNA substrate. Preincubation of the DNA substrate with imidazo[4,5-d]pyridazine nucleosides was performed at 30°C in 20 μl of TGT buffer (20 mM Tris-HCl [pH 7.5], 10% glycerol, 0.05% Triton X-100, 1 mM EDTA, 1 mM β-mercaptoethanol). The preincubation proceeded for various periods of time and with various concentrations of imidazo[4,5-d]pyridazine nucleosides, as indicated in the appropriate figure legends. In step 2, the unwinding reaction was started by addition of 2 pmol of WN virus or HCV NTPase/helicase in 10 μl of TGT buffer. In control experiments, the DNA substrate was preincubated under the same conditions, except that imidazo[4,5-d]pyridazine nucleosides were added together with the enzyme at the start of the reaction.
Synthesis of HMC-HO4, HMC-HO1α, and HMC-HO5. (i) Synthesis of HMC-HO4.
HMC-HO4 was synthesized in four steps starting from methyl 1-β-d-ribofuranosyl-4,5-imidazoledicarboxylate by the procedure described by Chen and Hosmane (7).
(ii) Synthesis of HMC-HO5.
HMC-HO5 was synthesized in two steps starting from methyl 1-β-d-ribofuranosyl-4,5-imidazoledicarboxylate by the procedure described in the literature (8, 29).
(iii) Synthesis of HMC-HO1α.
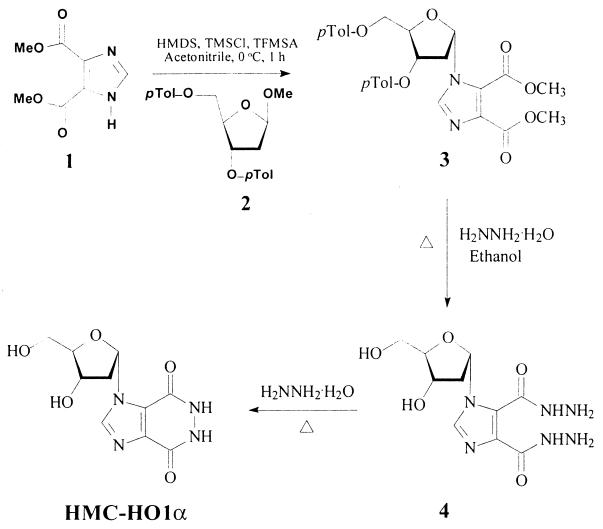
HMC-HO1α was synthesized in three steps starting from methyl 4,5-imidazoledicarboxylate (compound 1). The synthesis is outlined in Fig. 1; and the synthesis procedure for each step, along with the physical, spectral, and analytical data for the new compounds, are described below.
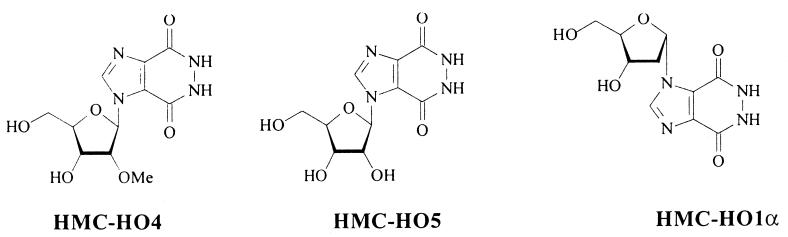
FIG. 2.
Chemical structures of the imidazo[4,5-d]pyridazine nucleosides used in this study. The procedures for synthesis and purification of the compounds are presented in Materials and Methods. Me, methyl.
Step 1: synthesis of compound 3.
A solution of compound 1 (1.84 g, 10 mmol) and methyl 2-deoxy-3,5-di-O-p-toluoyl-β-d-erythropentofuranoside (compound 2; 3.84 g, 10 mmol) in dry acetonitrile (50 ml) was placed into a flame-dried, 100-ml round-bottom flask. Note that compound 2 was prepared by the procedure described in the literature starting from 2-deoxy-d-ribose (16, 32), and separation of compound 2 from its β-anomer was done by silica gel column chromatography. The solution was stirred in an ice bath for 10 min. Then, 1,1,1,3,3,3-hexamethyldisilazane (HMDS; 7 ml, 33 mmol), chlorotrimethylsilane (TMSCl; 4.5 ml, 36 mmol), and trifluoromethanesulfonic acid (TFMSA; 3 ml, 36 mmol) were consecutively added to the solution of compounds 1 and 2 described above. The resulting solution was stirred in an ice bath for 1 h. The reaction was completed by thin-layer chromatography analysis (silica gel plates with chloroform-methanol [30:1]). The reaction mixture was evaporated to dryness in vacuo. The resulting residue was dissolved in chloroform and washed with saturated aqueous sodium bicarbonate and then water. After the chloroform solution was dried over anhydrous MgSO4 and filtering, it was evaporated to dryness in vacuo. The residue was purified by column chromatography and eluted with chloroform to give a pure colorless oily product, methyl 1-(3′,5′-di-O-p-toluoyl-2′-deoxy-α-d-ribofuranosyl)imidazole-4,5-dicarboxylate (compound 3; 4.6 g, 86%). Rf, 0.83 (chloroform-methanol [10:1]); 1H nuclear magnetic resonance (NMR) (CDCl3) δ 7.97 (s, 1H, imidazole), 7.95 (d, 2H, J = 8.1 Hz, o-phenyl [o-Ph]), 7.62 (d, 2H, J = 8.1 Hz, o-Ph), 7.28 (d, 2H, J = 8.1 Hz, m-Ph), 7.20 (d, 2H, J = 8.1 Hz, m-Ph), 6.65 (d, 1H, J = 6.6 Hz, 1′-H), 5.65 (d, 1H, J = 6.6 Hz, 3′-H), 4.92 (t, 1H, J = 3.6 Hz, 4′-H), 4.58 (d, 2H, J = 3.6 Hz, 5′-H), 3.94 (s, 3H, OCH3), 3.92 (s, 3H, OCH3), 3.07 (dt, 1H, J = 15.6 and 6.6 Hz, 2′-βH), 2.57 (d, 1H, J = 15.6 Hz, 2′-αH), 2.44 (s, 3H, PhCH3), 2.39 (s, 3H, PhCH3); 13C NMR (CDCl3) δ 21.58 (2PhCH3), 40.93 (C-2′), 52.23 (OCH3), 52.48 (OCH3), 63.99 (C-5′), 74.59 (C-3′), 85.52 (C-4′), 89.18 (C-1′), 129.28 (2m-Ph), 129.33 (2m-Ph), 129.59 (2o-Ph), 129.68 (2o-Ph), 136.47 (C-4 or C-5), 137.11 (C-2), 138.58 (C-5 or C-4), 144.16 (Ph-C1), 144.47 (Ph-C1), 160.48 (C=O), 162.87 (C=O), 165.76 (PhC=O), 166.02 (PhC=O). Analysis calculated for C28H28N2O9 · 0.5H2O: C, 61.65; H, 5.36; N, 5.13. Found: C, 62.03; H, 5.38; N, 4.79; high-resolution mass spectrum (fast atom bombardment): Calculated for C28H29N2O9: 537.1875. Found: 537.1869.
Step 2: synthesis of 1-(2′-deoxy-α-d-ribofuranosyl)imidazole-4,5-dicarboxhydrazide (compound 4).
A solution of compound 3 (0.55 g, 1 mmol), ethanol (15 ml), and hydrazine hydrate (99%, 0.8 ml) was refluxed for 6 h. After the solution was cooled, the precipitate was filtered, washed with ethanol, and recrystallized from methanol to provide 270 mg (90%) of white crystals. Melting point, 163 to 165°C; Rf, 0.63 (chloroform-methanol-30% ammonium hydroxide [2:2:1]); 1H NMR (dimethyl sulfoxide [DMSO]-d6) δ 12.23 (brs, 1H, NH, exchangeable with D2O), 9.81 (brs, 1H, NH, exchangeable with D2O), 8.02 (s, 1H, imidazole), 6.75 (d, 1H, J = 6.3 Hz, 1′-H), 5.03 (d, 1H, J = 2.7 Hz, 3′-OH, exchangeable with D2O), 4.88 (t, 1H, J = 5.4 Hz, 5′-OH, exchangeable with D2O), 4.60 (brs, 2H, NH2, exchangeable with D2O), 4.59 (brs, 2H, NH2, exchangeable with D2O), 4.30 (m, 1H, 4′-H), 4.20 (m, 1H, 3′-H), 3.43 (t, 2H, J = 5.1 Hz, 5′-H), 2.64 (dt, 1H, J = 14.4 and 6.3 Hz, 2′β-H), 2.00 (d, 1H, J = 14.4 Hz, 2′α-H); 13C NMR (DMSO-d6) δ 43.06 (C-2′), 61.61 (C-5′), 70.55 (C-3′), 88.65 (C-4′), 89.79 (C-1′), 124.32 (C-4 or C-5), 133.70 (C-5 or C-4), 137.05 (C-2), 158.09 (C=O), 161.57 (C=O). Analysis calculated for C10H16N6O5: C, 40.00; H, 5.37; N, 27.99. Found: C, 40.10; H, 5.40; N, 27.81.
Step 3: synthesis of HMC-HO1α.
A solution of compound 4 (0.15 g, 0.5 mmol) and 99% hydrazine was refluxed for 1 h. The excess hydrazine was removed by distillation in vacuo, and the residue was coevaporated several times with water. The crystalline residue was recrystallized from methanol to give white crystals (0.1 g, 75%). Melting point, >250°C; Rf, 0.36 (chloroform-methanol-30% ammonium hydroxide [2:2:1]); 1H NMR (DMSO-d6) δ 8.48 (s, 1H, imidazole), 6.80 (d, 1H, J = 6.3 Hz, 1′-H), 4.28 (m, 1H, 4′-H), 4.24 (m, 1H, 3′-H), 3.43 (t, 2H, J = 4.2 Hz, 5′-H), 2.69 (dt, 1H, J = 14.4 and 6.3 Hz, 2′β-H), 2.19 (d, 1H, J = 14.4 Hz, 2′α-H). Analysis calculated for C10H12N4O5 · 1.75H2O (299.7552): C, 40.07; H, 5.21; N, 18.69. Found: C, 39.85; H, 5.25; N, 18.47; high-resolution mass spectrometry (fast-atom bombardment) calculated for C10H13N4O5: 269.0886. Found: 269.0872.
Cell culture and infections.
Vero E6 cells were cultivated in RPMI 1640 medium containing 10% fetal calf serum (Gibco), 100 μg of ampicillin per ml, and 80 μg of gentamicin per ml. The cells in the logarithmic phase of growth were infected with WN virus (strain ATCC VR-82) as described previously (30). For determination of the antiviral activities of the compounds investigated, the cells (5 × 105 cells suspended in 1 ml of RPMI 1640 medium) were seeded in 24-well flat-bottom tissue culture plates. After incubation for 24 h, the compounds were added to the medium as described in the legend to Fig. 7. For cell infection, the virus inoculum was added in 50 μl of the complete medium and was allowed to be adsorbed for 3 h at 37°C. Thereafter, the inoculum was removed, and the cells were washed twice with the medium and further incubated for 2 days in the presence of the compounds under the conditions described above. It should be mentioned that at concentrations up to 0.5 mM the imidazo[4,5-d]pyridazine nucleoside did not influence the growth of the cells.
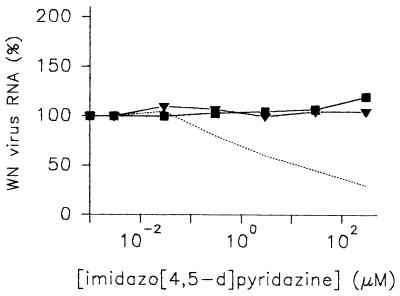
FIG. 7.
Effects of imidazo[4,5-d]pyridazine nucleosides on the replication of WN virus. Vero E6 cells were cultivated in 24-well tissue culture plates for 24 h and treated for 2 h with HMC-HO4 (dashed line), HMC-HO5 (▾), and HMC-HO1α (▪), which were applied at the indicated concentrations. Thereafter, the virus inoculum was added for 3 h. The cells were washed, fresh medium was added, and the compound concentrations were adjusted accordingly. After 2 days, the cells were harvested and WN virus RNA was quantitatively determined as described in Materials and Methods. The amounts of viral RNA extracted from the cells infected in the absence of imidazo[4,5-d]pyridazine nucleosides were referred to as 100%. The results shown are representative of those for three independent experiments.
Kinetics of viral replication and quantitative determination of WN virus RNA.
Viral replication was assessed by quantitative determination of WN virus RNA as follows. The medium from the wells was removed, the cell monolayers were washed with fresh medium, and the RNA was prepared with the RNeasy RNA preparation kit (Qiagen) by the procedure recommended by the manufacturer. The WN virus RNA was quantitatively determined by 5′-nuclease real-time reverse transcription (RT)-PCR. Briefly, primers WnS1 (5′-TGC TGA TAT GAT TGA CCC TTT TCA-3′) and WnAS1 (5′-AGT GTA AGT AAT GCC TCC AAA CAA-3′) were used to amplify a 151-bp fragment in the NS3 region of WN virus. The PCR products were detected with probe WnP1 (5′-TTC GCA AGA GGT GGA CAG CCA-3′). The 3′ end of the probe was labeled with 6-carboxy-N,N,N′,N-tetramethylrhodamine (TAMRA) and was phosphorylated to prevent elongation during PCR. The 5′ end of the probe was labeled with 6-carboxyfluorescein (FAM). RT-PCR was performed with the Platinum RT-PCR ThermoScript One-Step system (Life Technologies), as recommended by the manufacturer. The reaction mixture contained 280 nM primers WnS1 and WnAS1, 140 nM probe WnP1, and 40 μg of bovine serum albumin per ml in 20 μl of reaction buffer. A total of 0.8 μl of a ThermoScript reverse transcriptase-platinum Taq enzyme mixture and 2 μl of prepared RNA were added to the reaction mixture. Thermal cycling, carried out in a LightCycler thermal cycler (Roche Molecular Biochemicals), involved RT at 50°C for 20 min, denaturation at 95°C for 5 min, and 45 cycles of 95°C for 5 s and 60°C for 20 s (33). In the course of PCR, probe WnP1 was digested by the 5′ nuclease activity of the Taq DNA polymerase when it was specifically annealed to a PCR product generated from WN virus (18). Probe digestion liberated FAM from the TAMRA dye, causing an increase in the level of FAM-specific fluorescence during PCR (23). FAM-specific fluorescence was measured on detection channel F1 (530 nm) and was divided by the fluorescence measured on channel F2 (640 nm) for normalization. As a standard for real-time PCR, a strongly positive WN virus stock was diluted in negative cell lysate prior to RNA preparation. One unit of WN virus RNA detected by PCR was defined as the lowest possible dilution that could be amplified in five of five replicate reactions. The procedure for quantitative determination with the LightCycler thermal cycler has been described previously (9).
Other methods.
Kinetic parameters of the reactions were determined by nonlinear regression analysis with ENZFITTER (BioSoft) and SIGMA PLOT (Jandel Corp.) software. The protein concentration was measured by the method of Lowry et al. (24). The purity of proteins was determined by densitometric analysis of SDS-polyacrylamide gels stained with Coomassie blue (22).
RESULTS
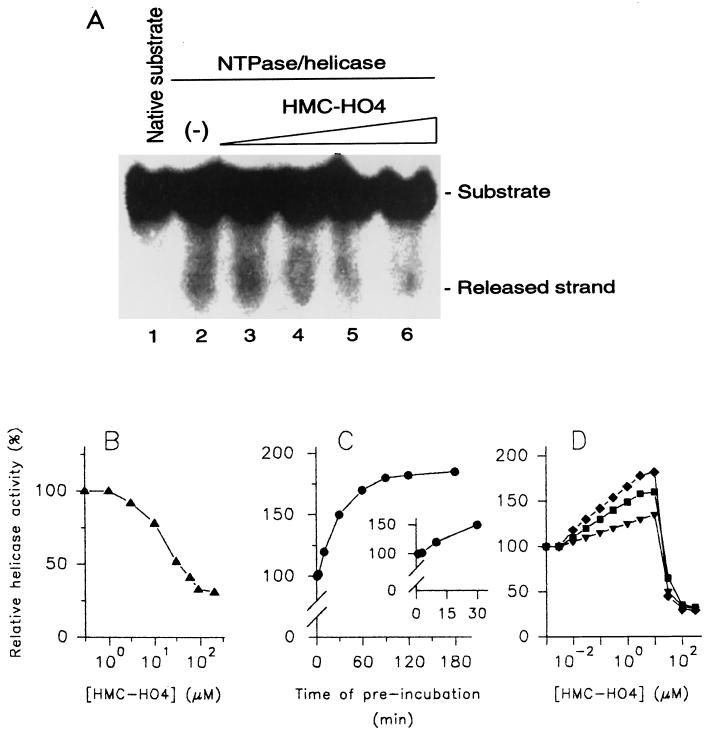
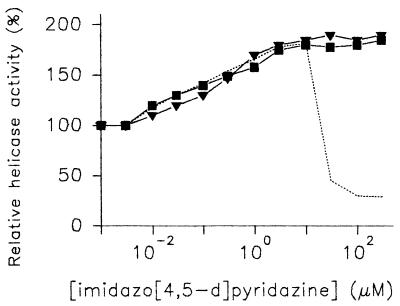
In our recent work we have described the purification and biochemical properties of the WN virus NTPase/helicase obtained from a native source (5). In the present report, the modulating effects of three imidazo[4,5-d]pyridazine nucleoside analogues, namely, HMC-HO4, HMC-HO5, and HMC-HO1α (Fig. 2), on the unwinding reaction mediated by this enzyme have been investigated. For the present studies we selected the reaction conditions and concentrations of NTPase/helicase and DNA substrate that were previously found to be most appropriate (5). When the helicase activity of the WN virus NTPase/helicase was tested as a function of the HMC-HO4 concentration, an inhibitory effect with a 50% inhibitory concentration (IC50) of 30 μM was obtained (Fig. 3A and B). The inhibition was not complete and reached 30 to 35% of the value for the control at HMC-HO4 concentrations that corresponded to three times the IC50 (approximately 100 μM). The experimental inhibitory curve was, however, markedly altered when the DNA substrate was preincubated in the presence of HMC-HO4 before the enzyme was added and the reaction was started. For a detailed investigation of the influence of the interaction of the DNA substrate with the compound on the helicase activity, the unwinding reaction was carried out in two steps. In the first step, aliquots of the DNA substrate were incubated with HMC-HO4 for various durations, and in the second step, the enzyme was added to the preincubated samples as described in the Materials and Methods section. Under the reaction conditions used, preincubation of the DNA substrate with the compound (20 μM) results in an enhancement of the unwinding activity of the enzyme (Fig. 3C). The activating effect was measurable after 5 to 10 min (Fig. 3C, inset) and approached a plateau after 90 min of preincubation. Next, we performed experiments in which the DNA substrate was preincubated for constant times in the presence of increasing concentrations of HMC-HO4. The activating effect increased linearly at HMC-HO4 concentrations ranging from 50 nM to 1.5 μM, with a measured 50% effective dose (ED50) equal to 120 nM. The experimental curve reached a maximum level at 10 μM (170 to 180% of the value for the control for preincubations longer than 90 min) and declined at concentrations higher than 20 μM, thus yielding nonsaturation kinetics of activation. Figure 3D presents the experimental activation curves obtained for durations of preincubation of 20, 40, and 90 min. To verify that HMC-HO4 induces this activating effect by direct interaction with the enzyme, we preincubated the WN virus NTPase/helicase with 15, 30, and 100 μM compound for different times prior to the start of the helicase reaction. However, we found that enzyme preincubated with HMC-HO4 was no more competent for the unwinding reaction than the control sample not preincubated with the compound.
FIG. 3.
Modulation of the helicase activity of WN virus NTPase/helicase by HMC-HO4. (A and B) Investigation of helicase activity as a function of increasing concentrations of HMC-HO4. The reaction took place in the absence (lanes 1 and 2) or in the presence (lanes 3 to 6) of the compound, which was added to the reaction mixture simultaneously with the enzyme. The concentration of HMC-HO4 was adjusted to 1 μM (lane 3), 10 μM (lane 4), 100 μM (lane 5), and 1 mM (lane 6). The substrate (lane 1) and the released strand were separated in a TBE-polyacrylamide gel and visualized by exposure of the dried gel to X-ray film for 20 h (A). Alternatively, the parts of the gels corresponding to the released strand were excised and the 32P radioactivity was quantified as described in Materials and Methods. The unwinding activity of the enzyme measured in the absence of HMC-HO4 was referred to as 100% (B). (C) Helicase activity of WN virus NTPase/helicase as a function of the duration of the interaction of HMC-HO4 with the DNA substrate. Aliquots of the DNA substrate were incubated in the presence of 20 μM HMC-HO4 or in the absence of the compound. At the indicated time points, the unwinding reaction was started by addition of the enzyme or the enzyme and HMC-HO4 as described above. The activity of the enzyme was determined as described above for panel B. The unwinding activity of the enzyme measured without the preincubated substrate was referred to as 100%. The inset demonstrates an expanded view of the region from 0 to 30 min of preincubation. (D) Effect of preincubation of the DNA substrate with HMC-HO4 on helicase activity as a function of increasing concentrations of the compound. Aliquots of the DNA substrate were incubated in the presence of increasing concentrations of HMC-HO4 or in the absence of the compound for 20 min (▾), 40 min (▪), and 90 min (⧫). Thereafter, the reaction was started by addition of the enzyme, and the unwinding activity was determined as described above for panel C. The results shown are representative of those for three independent experiments.
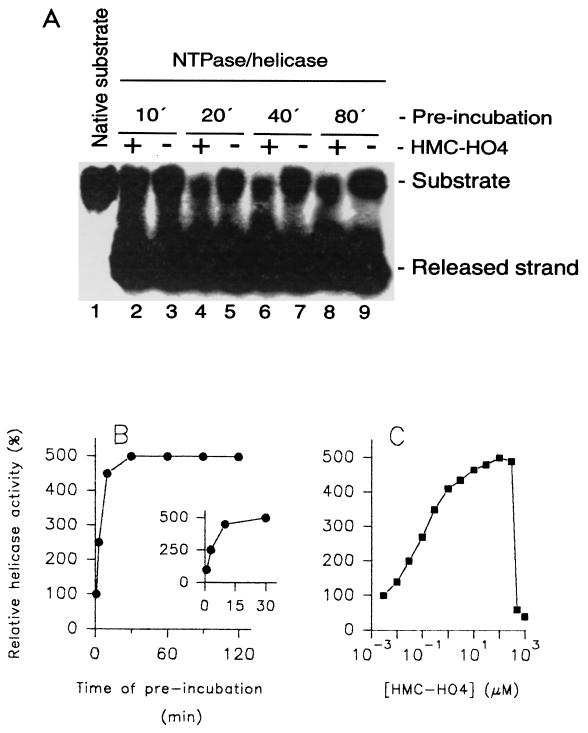
In order to investigate whether this kind of interaction is specific for the WN virus enzyme, we performed similar kinetic studies with the NTPase/helicase from a related virus, HCV, expressed in E. coli. Contrary to the results obtained with the WN virus enzymes, the observed inhibition of the helicase activity of the HCV NTPase/helicase by HMC-HO4 was relatively weak. The measured IC50 was 450 μM. Surprisingly, the activating effect of HMC-HO4 upon preincubation with the DNA substrate was more pronounced compared with that observed with the WN virus enzyme. The increase in helicase activity reached a maximum after 15 to 20 min of preincubation (Fig. 4A and B) and was measurable just after 2.5 min (Fig. 4B, inset). The extent of activation was dose dependent and reached a maximum of 450 to 500% of that for the control at HMC-HO4 concentrations between 100 to 200 μM and an ED50 equal to 150 nM (Fig. 4C). Furthermore, when the HCV enzyme was preincubated with 150 or 450 μM HMC-HO4 for durations longer than 30 min, weak inhibition of the enzyme was observed compared with the inhibition observed with the control sample not preincubated with the compound (data not shown).
FIG. 4.
Modulation of the helicase activity of HCV NTPase/helicase by HMC-HO4. (A and B) Investigation of the helicase activity of HCV NTPase-helicase as a function of the time of duration of the interaction of HMC-HO4 with the DNA substrate. Aliquots of the DNA substrate (lane 1) were incubated in the presence of 150 μM HMC-HO4 or in the absence of the compound. At the indicated time points, 10 min (lanes 2 and 3), 20 min (lanes 4 and 5), 40 min (lanes 6 and 7), and 80 min (lanes 8 and 9), the unwinding reaction was started by addition of the enzyme or the enzyme and HMC-HO4, as described in Materials and Methods. The samples were separated in a TBE-polyacrylamide gel and visualized by exposure of the dried gel to X-ray film for 20 h (A). For determination of the activity of the enzyme, the parts of the gels corresponding to the released strand were excised and the 32P radioactivity was quantified as described in Materials and Methods. The unwinding activity of the enzyme measured without the preincubated substrate was referred to as 100% (B). The inset demonstrates an expanded view of the region from 0 to 30 min of preincubation. (C) Effect of preincubation of the DNA substrate with HMC-HO4 on the activities of HCV NTPase/helicase as a function of increasing concentrations of the compound. Aliquots of the DNA substrate were incubated in the presence of increasing concentrations of HMC-HO4 or in the absence of the compound for 20 min. The reaction was started by addition of the enzyme, and the unwinding activity was determined as described above. The results shown are representative of those for three independent experiments.
In light of the results presented above, it is inferred that HMC-HO4 interacts with the DNA substrate and thereby facilitates the unwinding reaction. This notion was further corroborated by the following experiments. We synthesized two additional nucleoside analogues of HMC-HO4, namely, HMC-HO5 and HMC-HO1α (Fig. 2), each of which bore the same heterocyclic base (the imidazo[4,5-d]pyridazine ring system) as HMC-HO4 but which differed in the type and the mode of attachment of the sugar to the base. It was hoped that because the heterocyclic base is the principal site of recognition in DNA, the presence of the same heterocyclic moiety with the same functional groups in HMC-HO5 and HMC-HO1α would not considerably alter their characteristics of binding with the DNA substrate compared with the characteristics of binding for HMC-HO4. On the other hand, the modified sugar ring or the mode of attachment of the sugar ring to the heterocyclic base was anticipated to have a significant effect upon the enzyme binding capabilities of HMC-HO5 and HMC-HO1α. This prediction was indeed borne out in experiments in which neither of the two compounds displayed any inhibitory effect toward the helicase activity of either the WN virus NTPase/helicase or the HCV NTPase/helicase, even up to concentrations as high as 1 mM. By contrast, and as anticipated, when the DNA substrate was preincubated with either HMC-HO5 or HMC-HO1α, increases in the unwinding activities of the enzymes were observed. The extent of this activation was comparable to that measured with HMC-HO4. In the case of the WN virus enzyme, increases in unwinding activities of 190 and 185% (compared to that for the control) for HMC-HO5 and HMC-HO1α, respectively, were measured, with ED50s of 280 and 185 nM, respectively (Fig. 5). In addition, the extent of the modulating effects that the imidazo[4,5-d]pyridazine nucleosides exerted on the helicase activity was not dependent on the concentration of ATP at which the reaction was carried out.
FIG. 5.
Activating effect of preincubation of the DNA substrate with imidazo[4,5-d]pyridazine nucleosides on the helicase activity of WN virus NTPase/helicase. Aliquots of the DNA substrate were incubated in the presence of increasing concentrations of HMC-HO4 (dashed line), HMC-HO5 (▾), and HMC-HO1α (▪) or in the absence of the compounds. After 90 min of preincubation the unwinding reaction was started by addition of the enzyme or enzyme and the respective compound. The reaction proceeded, and the unwinding rate was determined as described in Materials and Methods. The unwinding activity of the enzyme measured with the preincubated substrate in the absence of the compounds was referred to as 100%. The results shown are representative of those for three independent experiments.
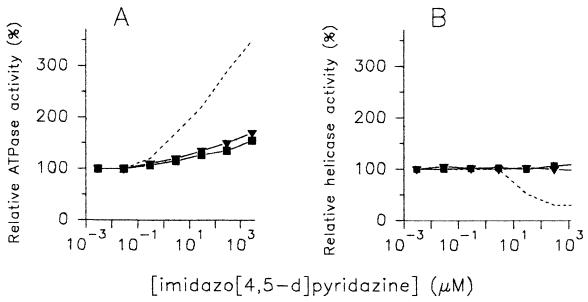
We further investigated the kinetic basis for the stimulatory and inhibitory effects of HMC-HO4 on the helicase activity in more detail. We tested whether the inhibitory effect was accompanied by corresponding changes in the NTPase activity of the WN virus enzyme. The assays, performed with an ATP concentration equal to the enzyme's Km value (9.5 μM), revealed that HMC-HO4 stimulated the ATPase activity of the enzyme. The activating effect corresponded closely to the increasing concentrations of the compound and did not decline up to a nucleoside concentration of 3 mM. When the influences of HMC-HO5 and HMC-HO1α on the ATPase activity of the WN virus enzyme were investigated, linear activation was again obtained. The extent of activation was, however, significantly lower compared with that measured with HMC-HO4 (increases in ATPase activity of 350, 170, and 155% were measured in the presence of 3 mM HMC-HO4, HMC-HO5, and HMC-HO1α, respectively) (Fig. 6A). Analogous to the helicase reaction, the activation of ATP hydrolysis was not dependent on the ATP concentration. Similar experimental curves of the activating effect were obtained when 100 μM (approximately 10 times the Km) or 1 μM (approximately 1/10 the Km) ATP was used. Also, the presence of poly(A) or poly(U), activators of the ATPase activities of the NTPases/helicases of WN virus and HCV (5, 28), respectively, did not alter the responses of the enzymes to imidazo[4,5-d]pyridazine nucleosides (data not shown).
FIG. 6.
Comparison of the modulating effects of imidazo[4,5-d]pyridazine nucleosides on the ATPase and helicase activities of WN virus NTPase/helicase. The ATPase (A) and helicase (B) reactions were performed in the presence of increasing concentrations of HMC-HO4 (dashed line), HMC-HO5 (▾), and HMC-HO1α (▪). In both assays, the compounds were added to the reaction mixture simultaneously with the enzyme. The reactions were performed and the enzymatic activities were determined by the procedures described in Materials and Methods. The activities of the enzyme measured in the absence of the compounds were referred to as 100%. The results shown are representative of those for three independent experiments.
It should be mentioned that qualitatively the same effects were observed when the ATPase activity of the HCV enzyme was measured as a function of increasing concentrations of the imidazo[4,5-d]pyridazine nucleosides (data not shown). Preincubation of the enzymes with the compounds (before the start of the ATPase reaction) did not influence the kinetics of ATP hydrolysis. Thus, the turnover rate of the ATPase reaction did not correspond to that of the helicase activities of the enzymes. Figures 6A and B show the juxtaposition of the modulating effects that the imidazo[4,5-d]pyridazine nucleosides exert on the helicase and ATPase activities of the WN virus NTPase/helicase. The fact that both activities of the enzymes could be modulated independently from each other suggests that the imidazo[4,5-d]pyridazine nucleosides interact directly not only with the DNA substrate but also with the enzyme.
Recently, it was demonstrated that the enzymatic activities of NTPase/helicase of bovine virus diarrhea virus are essential for viral replication (13). This led us to investigate the biological significance of the modulating activities of the imidazo[4,5-d] pyridazines on the NTPase and helicase activities of the WN virus NTPase/helicase. We therefore examined the ability of HMC-HO4 to enhance or inhibit the replication of the WN virus in Vero E6 cells. The cells were incubated with increasing concentrations of HMC-HO4 for 2 h, after which the virus inoculum was applied as described in the Materials and Methods section. Two days after the infection the RNA from the infected cells was extracted and viral replication was estimated by quantitative determination of the WN virus RNA, as described above. The amounts of viral RNA extracted from the HMC-HO4-treated cells were compared with the amounts of RNA obtained from untreated control cells. Figure 7 demonstrates the relative amounts of viral RNA as a function of increasing concentrations of HMC-HO4. The observed increase in the unwinding activity of the enzyme mediated by HMC-HO4 was not reflected in enhanced virus replication. In experiments in which the cells were pretreated with the compound for 1, 2, or 3 days before infection, no activating effect on virus replication was seen. On the other hand, the concentration of HMC-HO4 at which the amount of viral RNA was reduced by 50% compared with the amount of RNA for the untreated control cells was 25 to 30 μM. Thus, the IC50 measured in the in vivo assay corresponded closely to the IC50 measured in the helicase assay in vitro. Similar to the inhibition of the helicase activity of the enzyme, the reduction of virus replication was not complete and reached 35 to 40% of that for the untreated control at HMC-HO4 concentrations higher than 70 to 100 μM. The experiments did not reveal any apparent correlation between the effect of HMC-HO4 on the ATPase activities of the WN virus NTPase/helicase and on virus replication.
In further experiments we tested the effects of HMC-HO5 and HMC-HO1α on the replication of viral RNA. Similar to HMC-HO4, the stimulating effects of the compounds on the helicase activity, measured in vitro, were not reflected by any corresponding alterations in the levels of virus replication. As shown in Fig. 7, neither compound influenced the amounts of viral RNA extracted from infected cells up to concentrations as high as 300 μM.
DISCUSSION
The investigations described here were carried out with WN virus or HCV NTPase/helicase preparations that were free of other ATPase and helicase activities (4, 5). The results obtained with the imidazo[4,5-d]pyridazine nucleosides, presented here, show unequivocally that the ATPase and helicase activities of the NTPase/helicase of WN virus and HCV may be modulated independently from each other. Although HMC-HO5 and HMC-HO1α stimulated both the ATPase and the helicase activities of the enzymes, the experimental curves fitted for the ATPase and helicase activities as a function of the compound concentrations did not parallel each other. The concentrations of HMC-HO5 and HMC-HO1α required for the maximum stimulation of ATPase activity were higher by 1 or more orders of magnitude than those required for maximum stimulation of the unwinding reaction. Moreover, HMC-HO4, which was the imidazo[4,5-d]pyridazine nucleoside that was found to enhance enzyme-mediated ATP hydrolysis the most efficiently, acts as an inhibitor of helicase activity. The experimental curves reflecting the activities of the enzyme as a function of the HMC-HO4 concentration were partially reciprocal.
These observations led us to conclude that the activation of the helicase activity of the enzyme mediated by the imidazo[4,5-d]pyridazine nucleosides did not result from the enhancement of the rate of turnover of ATP hydrolysis. The uncoupling of the NTPase and helicase activities of the viral enzymes is not without precedent. Previous reports demonstrated that the stimulation of the ATPase activity of HCV NTPase/helicase by single-stranded nucleic acids is not directly related to procession of the helicase along the RNA or DNA substrate (15, 28). Furthermore, numerous chemically unrelated compounds like 5-fluoro-2-cytosine or O6-benzylguanine are able to inhibit or enhance the ATPase activity without affecting the helicase activity of the enzyme (5). On the other hand, some chloroethylguanine derivatives stimulate the helicase activity but have no apparent effect on the ATPase activity of the enzyme (5). Nevertheless, while the compounds mentioned above acted exclusively at the enzyme level, the imidazo[4,5-d]pyridazine nucleosides described here influenced the unwinding activity by interaction with the enzyme as well as its DNA substrate. To the best of our knowledge, little is known to date on how imidazo[4,5-d]pyridazine nucleosides interact with DNA. Chemically related compounds like 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and its derivatives are reported to be noncovalent DNA groove-binding agents with slight specificity for adenine-thymidine (AT) sequences (26). Whether this kind of interaction between the imidazo[4,5-d]-pyridazine nucleosides and DNA may lead to decreased stability of the DNA duplex and therefore to an enhanced level of unwinding remains to be verified. The DNA version of the helicase RNA substrate reported previously (10), which we used in the present study, contains some AT pairs. In this context, one could speculate that AT-poor DNA sequences or double-stranded RNA may be less susceptible to activation of the unwinding reaction mediated by the imidazo[4,5-d]pyridazine nucleosides. These investigations are under way.
This hypothesis may be corroborated by our in vivo experimental data. If the AT specificity of the interaction between DNA and imidazo[4,5-d]pyridazines mentioned above does indeed exist, the stability of the viral double-stranded RNA may not be influenced by the compounds. Consequently, the activating effects exerted by imidazo[4,5-d]pyridazines on the helicase activity observed in vitro with the DNA substrate could not occur in vivo. On the other hand, the interaction of HMC-HO4 with the enzyme is rather independent of the status of the substrate. Thus, it is conceivable that the inhibition of the unwinding activity of the enzyme that we determined with the DNA substrate also occurs under in vivo conditions.
As a final note, inhibition of the unwinding activity of the NTPase/helicase of members of the family Flaviviridae may represent a novel antiviral strategy. Thus, compounds like HMC-HO4 may be a starting point for the development of potent therapeutic compounds.
FIG. 1.
Synthesis of HMC-HO1α. Me, methyl; Tol, toluoyl.
Acknowledgments
This research was supported by a grant (grant BWB E/B31E/MO171/M5916-Rj.2000) to P. Borowski from the Bundesministerium der Verteidigung and by a grant (grant 1RO1 CA71079) to R. S. Hosmane from the U.S. National Institutes of Health.
REFERENCES
- 1.Bazan, J. F., and R. Fletterick. 1989. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology 171:637-639. [DOI] [PubMed] [Google Scholar]
- 2.Bianco, P., and S. C. Kowalczykowski. 2000. Translocation step size and mechanism of the RecBC DNA helicase. Nature 405:368-372. [DOI] [PubMed] [Google Scholar]
- 3.Borowski, P., R. Kuehl, O. Mueller, L.-H. Hwang, J. Schulze zur Wiesch, and H. Schmitz. 1999. Biochemical properties of a minimal functional domain with ATP-binding activity of the NTPase/helicase of hepatitis C virus. Eur. J. Biochem. 266:715-723. [DOI] [PubMed] [Google Scholar]
- 4.Borowski, P., O. Mueller, A. Niebuhr, M. Kalitzky, L.-H. Hwang, H. Schmitz, A. Siwecka, and T. Kulikowski. 2000. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim. Pol. 47:173-180. [PubMed] [Google Scholar]
- 5.Borowski, P., A. Niebuhr, O. Mueller, M. Bretner, K. Felczak, T. Kulikowski, and H. Schmitz. 2001. Purification and characterization of West Nile virus NTPase/helicase. Evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 75:3220-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castle, E., U. Leidner, T. Nowak, G. Wengler, and G. Wengler. 1986. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology 149:10-26. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H.-M., and R. S. Hosmane. 2001. Synthesis of 1-(2′-O-methyl-β-d-ribofuranosyl)-1 H-imidazo[4,5-d]pyridazine-4,7(5 H,6 H)-dione: an attractive building block for antisense and triple-helical applications. Molecules 6:203-207. [Google Scholar]
- 8.Cook, P. D., P. Dea, and R. K. Robins. 1978. Synthesis of imidazo[4,5-d]pyridazine nucleosides related to inosine. J. Heterocyclic Chem. 15:1-8. [Google Scholar]
- 9.De Silva, D., M. Herrmann, K. Tabiti, and C. Wittwer. 1998. Rapid genotyping and quantification on the LightCyclerTM with hybridization probes. Biochemica 2:12-15. [Google Scholar]
- 10.Gallinari, P., D. Brennan, C. Nardi, M. Brunetti, L. Tomei, C. Steinkühler, and R. De Francesco. 1998. Multiple enzymatic activities associated with recombinant NS3 of hepatitis C virus. J. Virol. 72:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 13.Gu, B., C. Liu, J. Lin-Goerke, D. R. Maley, L. L. Gutshall, C. A. Fletenberger, and A. Del Vecchio. 2000. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J. Virol. 74:1794-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gwack, Y., D. W. Kim, J. Han, and J. Choe. 1996. Characterization of RNA binding activity and RNA helicase activity of hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 215:654-659. [DOI] [PubMed] [Google Scholar]
- 15.Hesson, T., A. Mannarino, and M. Cable. 2000. Probing the relationship between RNA-stimulated ATPase and helicase activities of HCV NS3 using 2′-O-methyl RNA substrates. Biochemistry 39:2619-2625. [DOI] [PubMed] [Google Scholar]
- 16.Hodge, R. P., C. K. Brush, C. M. Harris, and T. M. Harris. 1991. Synthesis of 1- and 1,2,2′-deuteriated deoxyribose and incorporation into deoxyribonucleosides. J. Org. Chem. 56:1553-1564. [Google Scholar]
- 17.Hodgman, T. C. 1988. A new superfamily of replicative proteins. Nature 333:22-23. [DOI] [PubMed] [Google Scholar]
- 18.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′- to 3′-exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor, M., L. Zhang, M. Ramachandra, J. Kusukawa, K. E. Ebner, and R. Padmanabhan. 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270:19100-19106. [DOI] [PubMed] [Google Scholar]
- 20.Kim, D. W., Y. Gwack, H. J. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 225:160-166. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J., K. Morgenstern, J. Griffith, J. Dwyer, M. Thomson, M. Murcko, C. Lin, and P. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 24.Lowry, O. H., N. J. Rosebrough, A. J. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 25.Lun, L., P.-M. Sun, C. Trubey, and N. Bachur. 1998. Antihelicase action of Cl-958, a new drug for prostate cancer. Cancer Chemother. Pharmacol. 42:447-453. [DOI] [PubMed] [Google Scholar]
- 26.Marsch, G., R. L. Ward, M. Colvin, and K. W. Turteltaub. 1994. Non-covalent DNA groove-binding by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Nucleic Acids Res. 22:5408-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, H., and R. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tai, C.-L., W.-K. Chi, D.-S. Chen, and L.-H. Hwang. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 70:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tapiero, C., J. L. Imbach, R. P. Panzica, and L. B. Townsend. 1976. The synthesis of 1-(β-d-ribofuranosyl) imidazo[4,5-d]pyridazine-4,7-dione via ring closure of an imidazole nucleoside. J. Carbohydr. Nucleosides Nucleotides 3:191-195. [Google Scholar]
- 30.Wengler, G., T. Nowak, and E. Castle. 1990. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology 177:795-801. [DOI] [PubMed] [Google Scholar]
- 31.Westaway, E. G., M. A. Brinton, S. Gaidamovich, M. C. Horzinek, A. Igrashi, L. Kääriäinen, D. K. Lvov, J. S. Porterfield, P. K. Russell, and D. Trent. 1985. Flaviviridae. Intervirology 24:183-192. [DOI] [PubMed] [Google Scholar]
- 32.Wierenga, W., and H. I. Skulnick. 1981. Stereochemical control as a function of protecting-group participation in 2-deoxy-d-erythro-pentofuranosyl nucleosides. Carbohydr. Res. 90:41-52. [Google Scholar]
- 33.Wittner, C. T., M, G. Herrman, A. A. Mass, and R. P. Rasmussen. 1997. A microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]