Abstract
We encountered three clinical isolates of methicillin-resistant Staphylococcus aureus which were susceptible to netilmicin and arbekacin in the absence of β-lactam antibiotics but which were resistant to them in the presence of β-lactam antibiotics. One of these strains, KU5801, was used to further investigate the antagonism between aminoglycosides and β-lactam antibiotics. β-Lactam antibiotics induced bacterial synthesis of aminoglycoside-6′-N-acetyltransferase and 2"-O-phosphotransferase [AAC(6′)-APH(2")] in association with decreased antimicrobial activities of aminoglycosides. A 14.4-kb EcoRI fragment that included the genes that control for β-lactam-inducible aminoglycoside resistance was cloned from a 31-kb conjugative plasmid present in KU5801. Restriction fragment mapping and PCR analysis suggested that a Tn4001-like element containing a gene encoding AAC(6′)-APH(2") was located downstream from a truncated blaZ gene. The DNA sequence between blaR1 and a Tn4001-like element was determined. The Tn4001-IS257 hybrid structure was cointegrated into the blaZ gene, and the typical sequences for the termination of transcription were not found between these regions. We deduced that antagonism of aminoglycosides by β-lactam antibiotics in isolate KU5801 involved transcription of the aac(6′)-Ie-aph(2")-Ia gene under the influence of the system regulating penicillinase production.
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of nosocomial infections, and these bacteria have acquired resistance to multiple antibiotics among the wide range of antibiotics used to treat infections caused by these organisms, including aminoglycosides (9, 10, 11). The frequencies of isolation of gentamicin- and tobramycin-resistant (MICs > 8 μg/ml) MRSA strains in Japan are 61.7 and 95.3%, respectively (9). The activity of netilmicin against gentamicin- and tobramycin-resistant MRSA isolates was found to be more potent than those of gentamicin and tobramycin (20, 24). Arbekacin, a new aminoglycoside antibiotic, showed strong activity against these MRSA isolates because it was less modified by aminoglycoside-6′-N-acetyltransferase and 2"-O-phosphotransferase [AAC(6′)-APH(2")] or aminoglycoside-4′-O-phosphoryltransferase I [ANT(4′)-I] than gentamicin or tobramycin (1, 9, 10, 29). This antibiotic was approved for clinical use against MRSA infections by the Japanese Ministry of Health and Welfare in 1990. Since the effects of aminoglycosides used in combination with β-lactam antibiotics are frequently shown to be synergistic (2, 12), arbekacin is often used in combination with a β-lactam antibiotic against MRSA infections. The effects of combinations of arbekacin with β-lactam antibiotics such as ampicillin and sulbactam have been shown to be synergistic and have successfully been used to treat serious infections caused by MRSA (8).
Recently, we encountered three clinical isolates of MRSA which were susceptible to netilmicin and arbekacin in routine testing but which were resistant to them in the presence of β-lactam antibiotics. These MRSA isolates were simultaneously isolated from different patients in a neurosurgery ward. Their genomic DNA fingerprinting patterns obtained by pulsed-field gel electrophoresis were identical (data not shown). We presumed that these MRSA isolates originated from one clone and spread to patients in the same ward. The present report is a description and characterization of the antagonism between aminoglycosides such as netilmicin and β-lactam antibiotics in this strain. To better understand the mechanism of antagonism, we studied the influences of β-lactam antibiotics on bacterial aminoglycoside-modifying enzyme (AME) production and analyzed the genetic sequence located between penicillinase regulatory genes and the gene encoding an AME.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus KU5801 was isolated from a patient who developed a respiratory tract infection following subarachnoid hemorrhage. Strain KU5801 was serendipitously discovered by routine susceptibility testing in a hospital's clinical laboratory, in which truncation of the aminoglycoside zone of inhibition on the side of the adjacent disk containing β-lactam antibiotics was observed by disk diffusion susceptibility testing. For reference, KU5801 was resistant to methicillin, erythromycin, tobramycin, gentamicin, tetracycline, minocycline, and ofloxacin, while it was susceptible to streptomycin, netilmicin, arbekacin, chloramphenicol, and rifampin. No production of β-lactamase could be detected in this strain by an iodometric assay (15). We used S. aureus RN4220 and RN4220RIF, a rifampin-resistant mutant arising from RN4220, as recipients in transformation and conjugation studies.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| KU5801 | Clinical isolate | This study |
| RN4220 | Recipient strain for transformation | 13 |
| RN4220RIF | Rifampin-resistant mutant of RN4220 used as a recipient strain for conjugation | This study |
| E. coli JM109 | Cloning host strain | Takara Shuzo Co., Ltd. |
| Plasmids | ||
| pND50 | Shuttle cloning vector | 30 |
| pT7blueT | PCR-amplified fragment cloning vector | Novagen Inc. |
| pI258 | Reference plasmid | 25 |
| pKU111 | A 31-kb plasmid isolated from KU5801 | This study |
| pKU112 | Recombinant plasmid containing a 14.4-kb EcoRI fragment from pKU111 cloned into pND50 | This study |
| pKU113 | Derivative plasmid of pKU112 with reversed 2.5-kb ClaI fragment (see Fig. 4) | This study |
| pKU114 | Recombinant plasmid containing a 2.2-kb PCR-amplified fragment by P1 and A3 primers from pKU111 in pT7blueT | This study |
A 31-kb plasmid isolated from KU5801 was designated pKU111. pKU112 is a recombinant plasmid containing a 14.4-kb EcoRI fragment encoding a β-lactam-inducible aminoglycoside resistance gene from pKU111 that was inserted into pND50, a shuttle cloning vector for Escherichia coli and S. aureus (30). In pKU113, a plasmid derived from pKU112, a 2.5-kb ClaI fragment was recloned in the opposite orientation within the 14.4-kb EcoRI fragment (see below).
Conjugation and transformation.
Mating (16) and transformation (19) were carried out as described previously. Transconjugants were selected by using agar plates containing 25 μg of kanamycin (Meiji Seika Kaisha, Ltd., Tokyo, Japan) per ml and 50 μg of rifampin (Sigma Chemical Co., St. Louis, Mo.) per ml. Transformants were selected by using agar plates containing 25 μg of kanamycin per ml.
Antibiotics and susceptibility testing.
Powders of the different antibiotics of known potency were obtained from the indicated sources: arbekacin and streptomycin, Meiji Seika Kaisha, Tokyo, Japan; netilmicin and gentamicin, Schering-Plough Co., Ltd., Osaka, Japan; imipenem, Banyu Pharmaceutical, Tokyo, Japan; aztreonam, Bristol-Myers Co., Tokyo, Japan; and methicillin, cefazolin, cefotaxime, and lividomycin, Sigma Chemical Co. All antibiotics were dissolved in distilled water. Susceptibility disks containing 50 μg of antibiotic per disk were prepared from 8-mm paper circles (Advantec, Tokyo, Japan).
MIC determinations and disk diffusion tests were performed in Mueller-Hinton medium (Difco Laboratories, Detroit, Mich.) by the protocol of the National Committee for Clinical Laboratory Standards (17, 18). Antagonism between aminoglycosides and β-lactam antibiotics was confirmed by the double-disk diffusion test. The MICs of the aminoglycosides were determined with or without the addition of aztreonam (final concentration, 25 μg/ml) to the agar plates.
AME activity.
Cultures of RN4220RIF(pKU111) were grown to the early logarithmic phase with shaking at 37°C in Luria-Bertani (LB) broth (containing tryptone, 10 g; yeast extract, 5 g; and NaCl, 5 g per liter). Induction was then carried out by adding methicillin (Sigma Chemical Co.) at various concentrations and allowing growth to continue. After 2 h of induction, the bacterial cells were centrifuged, washed in 200 ml of 50 mM Tris-HCl buffer (pH 7.5), and resuspended in 5 ml of lysing solution (50 mM Tris-HCl [pH 7.5], 25 μg of lysostaphin per ml). After 30 min of incubation at 37°C, the cell extracts were separated from the precipitate by centrifugation. AME activity was measured in cell extracts by a bioassay with netilmicin as the substrate, as described previously (7). The assays for determination of AME activity were conducted in triplicate for each sample. The AME activity calculated from the remaining activity of netilmicin in the bioassay is indicated as the number of micrograms of netilmicin inactivated per hour per milligram of protein.
Quantitative analysis of mRNA.
The expression of mRNA coding the aac(6′)-Ie-aph(2")-Ia gene was analyzed by a real-time (TaqMan) PCR assay. Cultures of RN4220RIF(pKU111) were grown to the early logarithmic phase with shaking at 37°C in 200 ml of LB broth. Then, induction was carried out by adding aztreonam (Bristol-Myers Co.) at 25 μg/ml and allowing growth to continue. After 2 h of induction, the bacterial cells were centrifuged and washed in distilled water. Total cellular RNA was extracted from the bacterial pellet described above by using the TRIzol reagent by the protocol provided by the manufacturer (Life Technologies, Inc., Rockville, Md.). The RNA solution was treated with DNase I (Roche Molecular Biochemicals, Mannheim, Germany) and was purified by phenol-chloroform extraction and ethanol precipitation. Total RNA was reverse transcribed for single-strand cDNA synthesis with random hexadeoxynucleotide primers (Promega Co., Madison, Wis.) and Ready-to-Go You-Prime First-Strand Beads (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). The primers and internal probes for amplification of cDNA reverse transcribed from AAC(6′)-APH(2") mRNA were determined with the Primer Express computer program (Applied Biosystems Japan Ltd., Tokyo, Japan) and were prepared by Takara Shuzo Co., Ltd. (Kyoto, Japan), and Applied Biosystems Japan Ltd. The oligonucleotide sequences of the forward and reverse primers were 5′-CCAAGAGCAATAAGGGCATACC-3′ and 5′-CATTGCCTTAACATTTGTGGCA-3′, respectively. The sequence of the internal probe with a 6-carboxyfluorescein fluorescent label was 5′-CCAGAACATGAATTACACGAGGGCAAAAAA-3′. The PCR mixture was prepared with the TaqMan Universal PCR Master Mixture (Applied Biosystems) according to the instructions of the manufacturer, except that the final PCR mixture volume was 25 μl instead of 50 μl. PCR amplification was performed on an ABI Prism 7700 Sequence Detection System (Applied Biosystems). To quantify the gene transcripts precisely, 16S rRNA was monitored as an internal control, and each sample was normalized on the basis of its 16S rRNA transcript content. The primers and internal probe for 16S rRNA were determined and prepared as described above, and their oligonucleotide sequences were as follows: forward primer, 5′-GTGAAATGCGCAGAGATATGGA-3′; reverse primer, 5′-TCGCACATCAGCGTCAGTTAC-3′; and probe, 5′-ACACCAGTGGCGAAGGCGACTTTCT-3′. Standard curves for AAC(6′)-APH(2") mRNA and 16S rRNA were generated by using a serially diluted solution of total DNA extracted from RN4220RIF(pKU111) as the template. The amount of the aac(6′)-Ie-aph(2")-Ia gene expression was calculated from these standard curves, and quantitative normalization of the cDNA in each sample was performed by use of expression of the genes encoding 16S rRNA as an internal control. Real-time PCR assays were conducted in triplicate for each sample, and a mean value was indicated as the number of micrograms converted into the value of 16S rRNA.
PCR.
PCR primers chosen on the basis of previous reports were synthesized by using published DNA sequences (5, 19, 21, 22). These primers, obtained from Takara Shuzo Co., Ltd., are shown in Table 2. Total DNA from KU5801 was purified as described previously (19). PCR amplification was performed with a DNA thermal cycler (Perkin-Elmer Cetus, Emeryville, Calif.). The cycling program was repeated for 25 cycles and included a denaturing step at 94°C for 1 min, an annealing step at 55°C for 2 min, and an extension step at 72°C for 3 min. Samples of the reaction products were analyzed by electrophoresis on 1 or 2% agarose gels in TBE buffer (89 mM Tris-HCl, 89 mM borate, 2 mM EDTA [pH 8.0]). PCR products were detected by ethidium bromide staining followed by UV illumination.
TABLE 2.
Primers used for PCR
| Primer | Sense or antisense | Sequence | Positionsa |
|---|---|---|---|
| P1 | Sense | 5′-AATCCTGCAAGAAGAGTTAG-3′ | 5153-5172 |
| P2 | Antisense | 5′-TTCCTTCATTACACTCTTGG-3′ | 6238-6219 |
| A1 | Sense | 5′-GGACTTGACTGAGTTTATGG-3′ | 290-309 |
| A2 | Antisense | 5′-ATGGCAAGCTCTAGGATTAC-3′ | 618-599 |
DNA sequence.
The DNA fragment amplified by PCR with primers P1 and A2 (Table 2) was cloned into the pT7blueT vector (Novagen, Inc., Madison, Wis.), and a recombinant plasmid designated pKU114 was constructed. The nucleotide sequence of this fragment was determined with universal M-13 RV and M4 primers and with synthetic primers (Takara Shuzo Co.) by the dideoxynucleotide termination method of Sanger et al. (23).
Aminoglycoside and β-lactam antagonism in aac(6′)-Ie-aph(2")-Ia reverse recombinant.
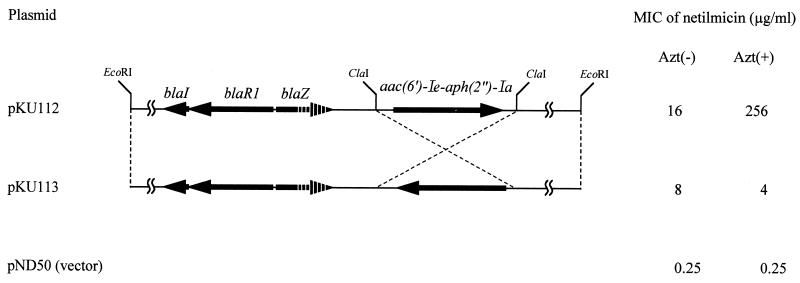
To prove that AAC(6′)-APH(2") expression is influenced by blaZ transcriptional regulation, pKU113 containing aac(6′)-Ie-aph(2")-Ia in the reverse orientation was constructed from pKU112 by self-ligation after ClaI digestion (see Fig. 4). pKU113 was transformed into S. aureus strain RN4220, and its recombinant strain was used in the study of antagonism between aminoglycosides and β-lactam antibiotics.
FIG. 4.
Influence of transcriptional orientation of aac(6′)-Ie-aph(2")-Ia on netilmicin resistance induced by aztreonam. Arrows indicate the relative transcriptional orientations of blaI, blaR1, blaZ, and aac(6′)-Ie-aph(2")-Ia. The 3′ end of the blaZ gene was deleted. The MICs of netilmicin were determined with or without aztreonam for S. aureus RN4220 transformed by each of the plasmids. Azt(−), without aztreonam; Azt(+), with 25 μg of aztreonam per ml.
Nucleotide sequence accession number.
The DNA sequence of the 2,158-bp DNA fragment amplified from RN4220RIF(pKU111) by PCR with primers P1 and A2 cloned into vector pT7blueT was determined and appears in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB074882.
RESULTS
Aminoglycoside and β-lactam antagonism.
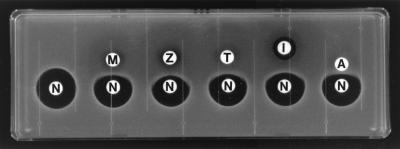
In the double-disk diffusion test, antagonism between arbekacin and some β-lactam antibiotics was evident as truncation of the arbekacin zone of inhibition on the side of the adjacent disk. By the double-disk diffusion test with KU5801, all β-lactam antibiotics studied produced such a distortion of the zone of inhibition (Fig. 1). When the MICs of aminoglycosides such as arbekacin, gentamicin, netilmicin, streptomycin, and lividomycin with or without aztreonam were determined for KU5801 (Table 3), aztreonam decreased the susceptibilities to arbekacin, gentamicin, and netilmicin; the MICs obtained when aztreonam was present were four to eight times higher than those when aztreonam was absent. The MICs of streptomycin and lividomycin were not changed by aztreonam.
FIG. 1.
Antagonism between netilmicin and β-lactam antibiotics for S. aureus KU5801 in the double-disk diffusion test. M, methicillin; Z, cefazolin; T, cefotaxime; I, imipenem; A, aztreonam; N, netilmicin. Each of the susceptibility test disks contains 50 μg for the β-lactam antibiotics and 125 μg for netilmicin.
TABLE 3.
Susceptibilities of S. aureus KU5801 and S. aureus RN4220RIF(pKU111) to aminoglycosides with or without aztreonam
| Strain | MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Netilmicin
|
Gentamicin
|
Arbekacin
|
Streptomycin
|
Lividomycin
|
||||||
| Azt(−) | Azt(+) | Azt(−) | Azt(+) | Azt(−) | Azt(+) | Azt(−) | Azt(+) | Azt(−) | Azt(+) | |
| KU5801 | 4 | 32 | 128 | 1,024 | 1 | 4 | 8 | 8 | 32 | 32 |
| RN4220RIF | 0.25 | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 | 8 | 8 | 4 | 4 |
| RN4220RIF(pKU111) | 4 | 64 | 64 | >1,024 | 1 | 16 | 8 | 8 | 4 | 4 |
Azt(−), without aztreonam; Azt(+), with aztreonam at 25 μg/ml.
Conjugal transfer and cloning.
Transconjugants with β-lactam-inducible aminoglycoside resistance were obtained from KU5801 at frequency of 6.4 × 10−9. A large 31-kb plasmid identified in all transconjugants (data not shown) was designated pKU111. The 14.4-kb EcoRI fragment was cloned from pKU111 into pND50, and the resulting recombinant plasmid was designated pKU112. Both the transconjugant [RN4220RIF(pKU111)] and the transformant [RN4220(pKU112)] showed β-lactam-inducible aminoglycoside resistance identical to that observed for KU5801(Table 3).
AME activity.
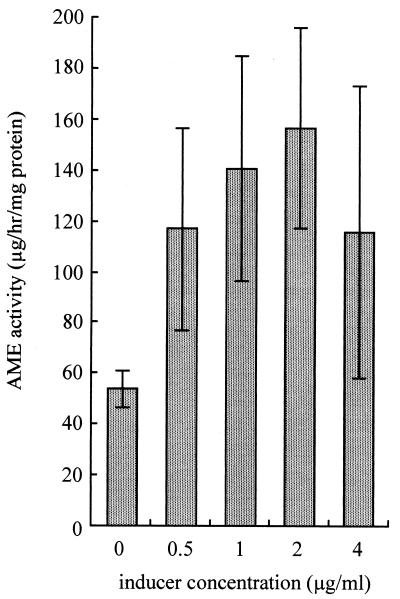
AME activity in RN4220RIF(pKU111) was measured with different concentrations of methicillin (Fig. 2). When RN4220RIF(pKU111) was grown in the absence of methicillin, AME activity was 53.9 μg/h/mg of protein. AME activity was induced by the addition of 2 μg of methicillin per ml, resulting in AME activity of 157 μg/h/mg of protein. Despite induction by methicillin, the cell extract could not inactivate streptomycin or lividomycin (data not shown).
FIG. 2.
AME activity of S. aureus RN4220RIF(pKU111) with methicillin present as an inducer. Data are expressed as means ± standard deviations for triplicate tests.
Gene expression.
Expression of the aac(6′)-Ie-aph(2")-Ia gene in RN4220RIF(pKU111) was analyzed under conditions with or without induction with 25 μg of aztreonam per ml. When RN4220RIF(pKU111) was grown in the absence of aztreonam, the amount of AAC(6′)-APH(2") mRNA was 0.0032 ± 0.00035 μg/μg of 16S rRNA (mean ± standard deviation). On the other hand, the amount of AAC(6′)-APH(2") mRNA detected when aztreonam was present was 0.10 ± 0.0074 μg/μg of 16S rRNA. This amount was 32 times higher than the amount of AAC(6′)-APH(2") mRNA detected when aztreonam was absent.
Cloning and mapping.
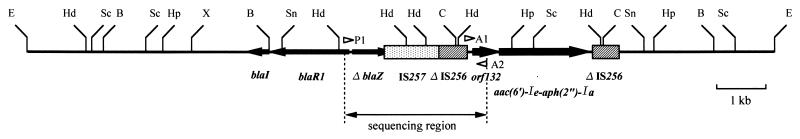
The restriction map of the 14.4-kb EcoRI fragment of pKU112 was compared with those of Tn552 (22) and Tn4001 (5, 21) (Fig. 3). In the central region of the 14.4-kb EcoRI fragment, some restriction sites corresponded to a portion of Tn552 or Tn4001. The Tn4001-like element appeared to be just downstream of the region corresponding to the bla operon carried on Tn552.
FIG. 3.
Restriction map and deduced structure of the 14.4-kb EcoRI fragment of pKU112. Arrows indicate the relative transcriptional orientations of blaI, blaR1, the 3′ end of truncated blaZ, orf132, and aac(6′)-Ie-aph(2")-Ia. The shaded boxes indicate IS257 and the truncated IS256 elements. The open arrowheads indicate the location and the direction of extension (3′ end) of each primer used in this study (cf. Table 2). E, EcoRI; Hd, HindIII; Sc, ScaI; B, BalI; Hp, HpaI; X, XbaI; Sn, SnaI; C, ClaI.
PCR analysis.
Two sets of primers (Table 2), one for detection of the promoter region of the aac(6′)-Ie-aph(2")-Ia gene and another for detection of the blaZ gene, were used. Amplification of DNA fragments from total DNA isolated from RN4220RIF(pKU111) and also RN4220(pI258) (a control strain carrying the complete Tn552 sequence) was confirmed by agarose gel electrophoresis (data not shown). PCR with primers A1 and A2, specific for the promoter region of aac(6′)-Ie-aph(2")-Ia, yielded a fragment of 329 bp. This DNA fragment was amplified only from total DNA isolated from RN4220RIF(pKU111). PCR with primers P1 and P2, specific for the blaZ gene, yielded a fragment of 1,086 bp. This DNA fragment was amplified only from total DNA isolated from RN4220(pI258) and not from total DNA isolated from RN4220(pKU111). PCR analysis was performed with primers P1 and A2 (Fig. 3), and a 2.2-kb DNA fragment was amplified from total DNA isolated from RN4220RIF(pKU111).
DNA sequence.
The 2,158-bp DNA fragment amplified from RN4220RIF(pKU111) by PCR with primers P1 and A2 was cloned into vector pT7blueT (Fig. 3). The sequence of the upstream region (nucleotide [nt] positions 1 to 246) corresponded to portions of blaR1 and blaZ carried on Tn552; the level of identity was 97.4%. The region downstream of the blaZ gene carried on pKU111 was deleted beginning at nt 625. A nonsense mutation causing defective blaZ translation was found at nt 650, 17 bp downstream from the blaZ deletion position. The DNA sequence of the downstream region (nt positions 245 to 2157) corresponded to the sequences of IS257, truncated IS256, and orf132; these genes are located upstream from aac(6′)-Ie-aph(2")-Ia carried on pSK41 (3), a staphylococcal conjugative multidrug resistance plasmid. The level of identity between the downstream region of our sequence and the pSK41 sequence was 99.7%. These results show that aac(6′)-Ie-aph(2")-Ia is located approximately 2 kb downstream from the blaZ promoter region and that both the aac(6′)-Ie-aph(2")-Ia gene and the blaZ gene are on the sense DNA strand.
Aminoglycoside and β-lactam antagonism in an aac(6′)-Ie-aph(2")-Ia reverse recombinant.
The MICs of netilmicin with or without aztreonam for RN4220(pKU113) were compared with those for RN4220(pKU112) (Fig. 4). The MICs of netilmicin without aztreonam for RN4220(pKU113) and RN4220(pKU112) were 16 and 8 μg/ml, respectively. The addition of aztreonam increased the MIC of netilmicin for RN4220(pKU112) 16 times and decreased the MIC for RN4220(pKU113) by half. As mentioned above, antagonism between aminoglycosides and β-lactam antibiotics was not observed in RN4220(pKU113).
DISCUSSION
Generally, transcription of genes encoding AME activities is believed to be constitutive; although it is costly in terms of cellular energy, such expression provides constant protection against aminoglycosides (26). Two known exceptions to this generalization are expression of the aac(6′)-Ic gene of Serratia marcescens (27) and the aac(2′)-Ia gene of Providencia stuartii (P. N. Rather, E. Orosz, K. J. Shaw, R. Hare, and G. H. Miller, Program Abstr. 32nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 436, 1992). Transcriptional regulation of these genes depends on unusual promoter regions. However, in KU5801, a clinical MRSA isolate, we found a novel aminoglycoside resistance phenotype that was induced by β-lactam antibiotics. In the present study of antagonism between aminoglycosides and β-lactam antibiotics, all β-lactam agents tested induced resistance to gentamicin, netilmicin, and arbekacin but not resistance to streptomycin or lividomycin (Fig. 1; Table 3). Gentamicin, netilmicin, and arbekacin all are modified by AAC(6′)-APH(2"), while streptomycin and lividomycin are not (26). Moreover, the AME activity in RN4220RIF(pKU111) under our experimental conditions with methicillin as the inducer was increased 2.9-fold by 2 μg of methicillin per ml. These results demonstrate that resistance to aminoglycosides in KU5801 involves enzymatic modification of these classes by AAC(6′)-APH(2") induced by β-lactam antibiotics.
We also showed that the genes encoding aminoglycoside resistance induced by β-lactam antibiotics are located on the 14.4-kb EcoRI fragment of pKU111. Sequence analysis showed that IS257 and a truncated IS256 element were integrated into the blaZ gene and that both genes were coded on the same DNA strand. The sequences of IS257 and the truncated IS256 element were identical to that of the upstream region of the aac(6′)-Ie-aph(2")-Ia gene carried on the Tn4001-IS257 hybrid structure, which is commonly found in conjugative plasmids that carry gentamicin resistance, e.g., pSK1 (3). Although the central 1.9-kb region of the Tn4001-IS257 hybrid structure including the orf132 and aac(6′)-Ie-aph(2")-Ia genes is identical to this region of Tn4001, the 899-bp region at the 5′ end of IS256 is truncated by insertion of IS257 on both sides of the central region (4). A 105-bp inverted repeat, proposed to be involved in prevention of overexpression of tnp in response to a foreign upstream promoter (5), is present in the 5′-end region of IS256 carried on Tn4001; however, in the Tn4001-IS257 hybrid structure, this region is truncated by the IS257 insertion and the inverted repeat corresponding to it is not found (3, 4). The observation that the 105-bp inverted repeat at the 5′ end of IS256 is deleted is very important for understanding how β-lactam antibiotics induce production of AAC(6′)-APH(2") in KU5801, because transcription of the blaZ gene regulated by the BlaI-BlaR1 system can produce an mRNA that includes the aac(6′)-Ie-aph(2")-Ia gene. To confirm this hypothesis, we measured the level of aac(6′)-Ie-aph(2")-Ia gene expression in RN4220RIF(pKU111) under conditions in which aztreonam was used as the inducer. It was found that induction by aztreonam led to an apparent increase in AAC(6′)-APH(2") mRNA levels. Furthermore, we constructed pKU113 from pKU112; this is a derivative of pKU112 with a reversed 2.5-kb ClaI fragment containing aac(6′)-Ie-aph(2")-Ia (Fig. 4). When we determined the susceptibility of RN4220(pKU113) to netilmicin with or without aztreonam, aztreonam did not increase the level of resistance to netilmicin. This result shows that antagonism between aminoglycosides and β-lactam antibiotics is caused by transcription of DNA to mRNA encoding the blaZ gene operating region through the action of the aac(6′)-Ie-aph(2")-Ia gene.
We believe that the sequence of pKU111 reflects one with genetic rearrangement of the bla gene and a Tn4001-IS257 hybrid structure from a conjugative staphylococcal plasmid such as pUW3626, which was among the plasmids isolated from an outbreak of multiple-drug-resistant S. aureus infections at a Kentucky hospital (6); this plasmid carried both the bla gene and the Tn4001-IS257 hybrid structure (4). Rearrangements caused by IS257-mediated cointegration into the staphylococcal chromosome or plasmid affect expression of resistance to antibiotics. For example, Leelaporn et al. (14) and Simpson et al. (28) reported that near its ends, IS257 carries a potentially outward-directed −35 promoter sequence that influences expression of downstream genes encoding trimethoprim resistance or tetracycline resistance by formation of hybrid promoters. Although the IS257-derived hybrid promoter did not play a direct role in the induction of AAC(6′)-APH(2") production by β-lactam antibiotics in KU5801, IS257 played a role in the increased level of expression of downstream drug resistance genes in both of these instances (14, 28).
We found a difference between increases in AME activity and increases in aac(6′)-Ie-aph(2")-Ia gene expression after induction by a β-lactam antibiotic in RN4220RIF(pKU111). The increase in aac(6′)-Ie-aph(2")-Ia gene expression after induction by aztreonam was almost consistent with the increase in β-lactamase activity after induction by methicillin in S. aureus RN4220(pI258), which harbored complete blaI, blaR1, and blaZ genes (19). However, in enzyme activity assays, RN4220RIF(pKU111) showed a 2.9-fold increase in AME activity after induction by methicillin. The reason for the inconsistency between these two analyses is not clear. Further studies will be required to explore the relationship between gene expression and enzyme activity.
In conclusion, we demonstrated a mechanism for β-lactam-induced expression of AAC(6′)-APH(2") in KU5801, a clinical MRSA isolate. As a result of genetic rearrangement, IS257 had an influence on the expression of adjacent genes, and this influence potentially enhances the ability of the organism to adapt to an environment containing antibiotics.
Acknowledgments
We thank Tsuyoshi Kojima and Jun-ichi Yamagishi, Dainippon Pharmaceutical Co., Ltd., for supplying pND50. We also thank Erumi Murase, Meiji Seika Kaisha, Ltd., for technical assistance.
This study was supported in part by the All Kitasato Project Study, Kitasato University.
REFERENCES
- 1.Akins, R. L., and M. J. Rybak. 2000. In vitro activities of daptomycin, arbekacin, vancomycin, and gentamicin alone and/or in combination against glycopeptide intermediate-resistant Staphylococcus aureus in an infection model. Antimicrob. Agents Chemother. 44:1925-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asada, K., Y. Inaba, E. S. Tateda, K. A. Kuwahara, T. Ito, and K. Hiramatsu. 1995. Evolution and resistance expression of MRSA. Evaluation of β-lactam antibiotics against a set of isogenic strains with different types of phenotypic expression. Acta Biochim. Pol. 42:517-524. [PubMed] [Google Scholar]
- 3.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmid. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne, M. E., T. G. Littlejoho, and R. A. Skurray. 1990. Transposons and insertion sequences in the evolution of multiresistant Staphylococcus aureus, p. 165-174. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, Inc., New York, N.Y.
- 5.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis for IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, M. L., E. S. Wong, and S. Falkow. 1982. Common R-plasmid in Staphylococcus aureus and Staphylococcus epidermidis during a nosocomial Staphylococcus aureus outbreak. Antimicrob. Agents Chemother. 21:210-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas, M. J., and J. E. Dowding. 1975. Aminoglycoside-modifying enzymes. Methods Enzymol. 42:611-628. [DOI] [PubMed] [Google Scholar]
- 8.Hiramatsu, K. 1998. Vancomycin resistance in staphylococci. Drug Resist. Updates 1:135-150. [DOI] [PubMed] [Google Scholar]
- 9.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue, M., M. Nonoyama, R. Okamoto, and T. Ida. 1994. Antimicrobial activity of arbekacin, a new aminoglycoside antibiotic, against methicillin-resistant Staphylococcus aureus. Drugs Exp. Clin. Res. 20:233-240. [PubMed] [Google Scholar]
- 11.Konno, M. 1995. Nosocomial infections caused by methicillin-resistant Staphylococcus aureus in Japan. J. Infect. Chemother. 1:30-39. [Google Scholar]
- 12.Kono, K., S. Takeda, I. Tatara, and K. Arakawa. 1994. In vitro activities of arbekacin, alone and in combination, against methicillin-resistant Staphylococcus aureus. Jpn. J. Antibiot. 47:710-719. [PubMed] [Google Scholar]
- 13.Kreiwirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by prophage. Nature (London) 305:709-712. [DOI] [PubMed] [Google Scholar]
- 14.Leelaporn, A., N. Firth, M. E. Byrne, E. Roper, and R. A. Skurray. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob. Agents Chemother. 38:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitch, C., and S. Boonlayangoor. 1992. β-Lactamase tests, p. 5.3.1-5.3.8. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 16.Murray, B. E., D. A. Church, A. Wanger, K. K. Zscheck, M. E. Levison, M. J. Ingerman, E. Abrutyn, and B. Mederski-Samoraj. 1986. Comparison of two β-lactamase-producing strains of Streptococcus faecalis. Antimicrob. Agents Chemother. 30:861-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility test, 7th ed. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Okamoto, R., T. Okubo, and M. Inoue. 1996. Detection of genes regulating β-lactamase production in Enterococcus faecalis and Staphylococcus aureus. Antimicrob. Agents Chemother. 40:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooishi, M., and M. Miyao. 1997. Antibiotic sensitivity of recent clinical isolates from patients with ocular infections. Ophthalmologica 211:15-24. [DOI] [PubMed] [Google Scholar]
- 21.Rouch, D. A., M. E. Byrne, Y. C. Kong, and R. A. Skurray. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J. Gen. Microbiol. 133:3039-3052. [DOI] [PubMed] [Google Scholar]
- 22.Rowland, S.-J., and K. G. H. Dyke. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 4:961-975. [DOI] [PubMed] [Google Scholar]
- 23.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheel, O., D. J. Lyon, V. T. Rosdahl, F. A. B. Adeyemi-Doro, T. K. W. Ling, and A. F. B. Cheng. 1996. In-vitro susceptibility of isolates of methicillin-resistant Staphylococcus aureus 1988-1993. J. Antimicrob. Chemother. 37:243-251. [DOI] [PubMed] [Google Scholar]
- 25.Shalita, Z., E. Murphy, and R. P. Novick. 1980. Penicillinase plasmids of Staphylococcus aureus: structural and evolutionary relationships. Plasmid 3:291-311. [DOI] [PubMed] [Google Scholar]
- 26.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw, K. J., P. N. Rather, F. J. Sabatelli, P. Mann, H. Munayyer, R. Mierzwa, G. L. Petrikkos, R. S. Hare, and G. H. Miller. 1992. Characterization of the chromosomal aac(6′)-Ic gene from Serratia marcescens. Antimicrob. Agents Chemother. 36:1447-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson, A., R. A. Skurray, and N. Firth. 2000. An IS257-derived hybrid promoter directs transcription of a tetA(K) tetracycline resistance gene in the Staphylococcus aureus chromosomal mec region. J. Bacteriol. 182:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubukata, K., N. Yamashita, A. Gotoh, and M. Konno. 1984. Purification and characterization of aminoglycoside-modifying enzymes from Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 25:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamagishi, J., T. Kojima, Y. Oyamada, K. Fujimoto, H. Hattori, S. Nakamura, and M. Inoue. 1996. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]