Abstract
The yeast high osmolarity glycerol (HOG) pathway signals via the Pbs2 MEK and the Hog1 MAPK, whose activity requires phosphorylation of Thr and Tyr in the activation loop. The Ptc1-type 2C Ser/Thr phosphatase (PP2C) inactivates Hog1 by dephosphorylating phospho-Thr, while the Ptp2 and Ptp3 protein tyrosine phosphatases dephosphorylate phospho-Tyr. In this work, we show that the SH3 domain-containing protein Nbp2 negatively regulates Hog1 by recruiting Ptc1 to the Pbs2-Hog1 complex. Consistent with this role, NBP2 acted as a negative regulator similar to PTC1 in phenotypic assays. Biochemical analysis showed that Nbp2, like Ptc1, was required to inactivate Hog1 during adaptation. As predicted for an adapter, deletion of NBP2 disrupted Ptc1-Pbs2 complex formation. Furthermore, Nbp2 contained separate binding sites for Ptc1 and Pbs2: the novel N-terminal domain bound Ptc1, while the SH3 domain bound Pbs2. In addition, the Pbs2 scaffold bound the Nbp2 SH3 via a Pro-rich motif distinct from that which binds the SH3 domain of the positive regulator Sho1. Thus, Nbp2 recruits Ptc1 to Pbs2, a scaffold for both negative and positive regulators.
Keywords: adapter, Hog1, MAP kinase, PP2C
Introduction
Mitogen-activated protein kinase (MAPK) pathways regulate growth, development and the response to stress in all eucaryotes (Gustin et al, 1998; Pearson et al, 2001; Hohmann, 2002; Johnson and Lapadat, 2002). The three-kinase cascade common to these pathways contains a MAP/ERK kinase kinase (MEKK) or Raf, which, when activated, phosphorylates and activates MAP/ERK kinase (MEK), which in turn activates MAPK via dual phosphorylation of a Thr and Tyr residue in the activation loop. The Saccharomyces cerevisiae high-osmolarity glycerol (HOG) MAPK pathway is activated primarily by osmotic stress (reviewed in Gustin et al, 1998; Hohmann, 2002; de Nadal et al, 2002) and also by heat stress (Winkler et al, 2002). Upstream of the three-kinase cascade are two putative membrane-bound stress sensors, Sln1, a two-component signaling protein, and Sho1, a novel protein containing an SH3 domain (Figure 1A). The Sln1 branch contains Ypd1 and Ssk1, which forms a three-component signaling system orthologous to bacterial signaling proteins. Ssk1 negatively regulates the downstream MEKKs, Ssk2 and Ssk22. The Sho1 branch signals via the Cdc42 GTPase, Ste50 a novel protein, the Ste20 p21-activated protein kinase (PAK) and the Ste11 MEKK. Both the Sln1 and Sho1 branches converge on the Pbs2 MEK-Hog1 MAPK module. In addition to activating Hog1, Pbs2 has also been shown to act as a scaffold in this pathway, binding Sho1, Ste11, Ssk2/22 and Hog1 (Posas and Saito, 1997; Tatebayashi et al, 2003).
Figure 1.
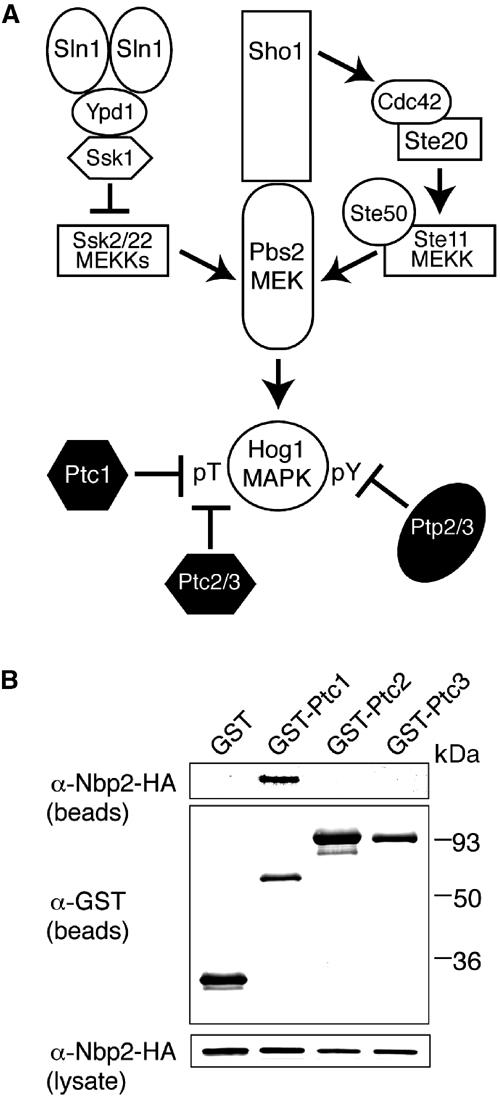
Activation of the HOG MAPK pathway and its negative regulation by protein phosphatases. (A) The yeast stress-activated HOG MAPK pathway contains a three-kinase cascade, which comprises the Ssk2, Ssk22 and Ste11 MEKKs, the Pbs2 MEK and the Hog1 MAPK. The Sln1-Ypd1-Ssk1 branch negatively regulates the Ssk2/22 MEKKs, while Sho1 positively regulates the Ste11 MEKK. Two classes of phosphatases regulate Hog1, which requires phosphorylation of Thr and Tyr in the sequence TGY for activation. Ptp2 and Ptp3 dephosphorylate pY, while Ptc1, Ptc2 and Ptc3 dephosphorylate pT. (B) Ptc1 but neither Ptc2 nor Ptc3 co-precipitates with Nbp2. The ability of Ptc1, Ptc2 and Ptc3 to associate with Nbp2 was examined in the nbp2Δ strain JHM17 expressing GST-PTCs and Nbp2-HA. Yeast were cultured, lysed, and co-precipitation assays performed as described in Materials and methods. The bound material was examined by SDS–PAGE and immunoblotting with anti-GST and anti-HA antibodies. The amount of Nbp2-HA in the lysate is also shown.
As important as the kinases that activate these pathways are the protein phosphatases that inactivate them. Mutation of phosphatases leads to aberrant signaling in yeast and Drosophila melanogaster MAPK pathways (Maeda et al, 1993; Jacoby et al, 1997; Martin-Blanco et al, 1998; Mattison et al, 1999; Zeitlinger and Bohmann, 1999; Winkler et al, 2002). Two groups of protein phosphatases inactivate the HOG pathway (Figure 1A). One group is the protein tyrosine phosphatases (PTPs) Ptp2 and Ptp3, which inactivate Hog1 by dephosphorylating the phosphotyrosine residue in the activation loop (Jacoby et al, 1997; Wurgler-Murphy et al, 1997). Ptp2 has been shown to reside in the nucleus, and is the stronger negative regulator in this pathway (Jacoby et al, 1997; Mattison and Ota, 2000).
The second group of phosphatases that inactivate the HOG pathway is the type 2C Ser/Thr phosphatases (PP2C), which are called PTCs in yeast. Three PTCs, Ptc1, Ptc2 and Ptc3, inactivate this pathway (Maeda et al, 1994; Warmka et al, 2001; Young et al, 2002). Ptc1 maintains low basal Hog1 activity and inactivates Hog1 during adaptation to osmotic stress (Warmka et al, 2001), while Ptc2 and Ptc3 play a complementary role by setting the maximal limit to which stress can activate Hog1 (Young et al, 2002). None of these three PTCs has an obvious effect on the upstream Pbs2 (Warmka et al, 2001; Young et al, 2002). Similarly, the Schizosaccharomyces pombe PP2C phosphatases inactivate the Spc1/Sty1 stress-activated MAPK, but do not appear to act on the upstream MEK (Nguyen and Shiozaki, 1999). However, PP2C in vertebrate signaling pathways can inactivate MEKK, MEK and MAPK in stress-activated pathways. PP2Cβ-1 inactivates the TAK1 MEKK (Hanada et al, 2001), while PP2Cα inactivates the MKK4 and MKK6 MEKs, and the p38 MAPK (Hanada et al, 1998; Takekawa et al, 1998).
Specificity is crucial to MAPK signaling pathways. In yeast and in vertebrates, multiple MAPK cascades coexist, regulating responses to a variety of signals. In each pathway are MEKKs, MEKs and MAPKs, which share significant sequence similarity within each group. In the absence of specificity conferring mechanisms, inappropriate crosstalk could occur between these pathways. One means to insulate these pathways is via scaffold proteins such as Pbs2 and Ste5 in the yeast HOG and pheromone response pathways (Posas and Saito, 1997; Elion, 2001), respectively, and JIP and MP-1 in vertebrate JNK and ERK1 pathways (Schaeffer et al, 1998; Whitmarsh et al, 1998), respectively. Another mechanism is via binding motifs contained within MEKs, MAPKs, its regulators and its substrates (Bardwell et al, 2001; Tanoue and Nishida, 2002).
Although protein phosphatases are often considered to be nonspecific, there are several examples of MAPK phosphatases that counter this notion. For example, the vertebrate dual-specificity phosphatase (DSP) MKP-3 is highly specific for ERK. Within its N-terminal noncatalytic domains lies the MAPK docking site known as the D domain or kinase interaction motif (KIM), which interacts with the common docking (CD) motif found in ERK (Tanoue and Nishida, 2002). In yeast, the PTPs also attain specificity for MAPK family members via their N-terminal noncatalytic domains. Ptp3 contains a D domain/KIM and a Cdc25 homology (CH) domain that bind to the Fus3 CD motif (Zhan and Guan, 1999), and Ptp2 binds Hog1 via the CD motif (Mattison and Ota, 2000) (D Heidysch, C Mattison and IM Ota, unpublished).
The means by which PP2Cs target the three-kinase MAPK cascade are not understood. Previous studies showed that PP2C proteins co-precipitate with MAPK cascade members, suggesting that they specifically bind them. For example, the S. pombe Ptc1 co-precipitates with Spc1/Sty1 MAPK (Nguyen and Shiozaki, 1999), and vertebrate PP2Cβ-1 binds TAK1 specifically, as it did not bind MEKK2, MKK4, MKK6, JNK or p38 (Hanada et al, 2001). Other studies suggested that PP2C can discriminate between phosphorylated and unphosphorylated substrates. PP2Cα bound phosphorylated, but not unphosphorylated p38 (Takekawa et al, 1998). However, PP2Cβ-1 binding to TAK1 did not require phosphorylation (Hanada et al, 2001). For each of these PP2C-MAPK pathway substrate interactions, it is not clear whether they are direct or occur via a third protein, since co-precipitations were performed using cell lysates. However, an Arabidopsis PP2C, MPC2, can bind the stress-activated SIMK MAPK directly (Meskiene et al, 2003).
We sought to identify how the yeast Ptc1 might target Hog1. A proteome-wide analysis of protein–protein interactions in S. cerevisiae by two groups using the yeast two-hybrid method found that Ptc1 interacted with Nbp2 (Ito et al, 2000; Uetz et al, 2000). Not much is known about Nbp2, a 237-residue protein containing an SH3 domain. Nbp2 was initially identified as a Nap1-binding protein (Shimizu et al, 2000), where Nap1 plays a role in bud morphogenesis and cell cycle progression (Kellogg et al, 1995; Altman and Kellogg, 1997). Beyond Nbp2 interaction with Ptc1, additional evidence suggested a possible role for Nbp2 in the HOG pathway. An analysis of protein complexes in yeast showed that Pbs2 co-precipitated with Nbp2 and Ptc1 (Ho et al, 2002). Since previous studies showed that Pbs2 acted as a scaffold that binds Hog1, Sho1, Ste11 Ssk2/22 (Posas and Saito, 1997; Tatebayashi et al, 2003), and Ptc1 was shown to inactivate Hog1 (Warmka et al, 2001), we predicted that Nbp2 might act as an adapter, recruiting Ptc1 to the MAPK cascade via the Pbs2 scaffold. Indeed, we find that Nbp2 mediates Ptc1 binding to the Pbs2-Hog1 MEK-MAPK module. In this way, Nbp2 enables Ptc1 inactivation of Hog1.
Results
Nbp2 binds Ptc1 but not other PTCs in the HOG pathway
In the HOG pathway, three PTCs have been shown to inactivate Hog1. Ptc1 maintains low Hog1 activity in the absence of stress, and inactivates Hog1 during adaptation to stress (Warmka et al, 2001), while Ptc2 and Ptc3 set maximal Hog1 activity (Young et al, 2002). Ptc2 and Ptc3 are closely related to each other, but differ structurally from Ptc1 in containing a noncatalytic C-terminal domain (Young et al, 2002). Since Nbp2 and Ptc1 were shown to interact by the yeast two-hybrid method (Ito et al, 2000; Uetz et al, 2000; Ho et al, 2002), but no evidence was found for Ptc2 and Ptc3, we tested whether Nbp2 was specific for Ptc1. GST-PTCs and Nbp2-HA were coexpressed in yeast from multicopy plasmids, and their ability to associate was examined in lysates. GST-Ptc1 co-precipitated with Nbp2-HA, but GST-Ptc2 and GST-Ptc3 could not bind Nbp2-HA (Figure 1B). GST-Ptc1 also bound Nbp2-HA when they were each expressed at lower levels from their endogenous promoters in low-copy CEN-based plasmids (data not shown). Thus, Nbp2 is specific for Ptc1.
NBP2 negatively regulates the HOG pathway
Since Ptc1 inactivates the HOG pathway by dephosphorylating Hog1 (Warmka et al, 2001), and Nbp2 binds Ptc1, we tested whether NBP2 might also act as a negative regulator of the HOG pathway similar to PTC1. Previously, it was shown that deletion of the phosphatases that regulate Hog1 dual phosphorylation on the activation loop Thr and Tyr exhibited a severe growth defect (Maeda et al, 1993), and that this was due to Hog1 hyperactivation (Jacoby et al, 1997). That is, ptc1Δ ptp2Δ had a severe growth defect while ptc1Δ ptp2Δ hog1Δ grew nearly as well as wild type (Jacoby et al, 1997). We tested whether deletion of NBP2 together with PTP2 might also exhibit growth defects. Indeed, the nbp2Δ ptp2Δ spore clones from a cross between nbp2Δ and ptp2Δ grew significantly worse than either strain alone (Figure 2A). To test whether this defect was due to HOG1, ptp2Δ hog1Δ was crossed to the nbp2Δ strain. The resulting nbp2Δ ptp2Δ hog1Δ spore clones grew significantly better than the nbp2Δ ptp2Δ double mutant (Figure 2B), demonstrating that HOG1 was responsible for these defects. It was further tested whether the nbp2Δ ptp2Δ growth defect was simply due to delayed germination, or whether defects could be seen in vegetatively growing cells. When grown in liquid culture and plated onto rich media, both the nbp2Δ ptp2Δ and ptc1Δ ptp2Δ mutants had significant growth defects relative to the wild type and single mutants (Figure 2C). However, the triple mutants nbp2Δ ptp2Δ hog1Δ and ptc1Δ ptp2Δ hog1Δ grew better than the double mutants (Figure 2C). Thus, the synthetic growth defect of the nbp2Δ ptp2Δ double mutant is due to HOG1, similar to the ptc1Δ ptp2Δ mutant.
Figure 2.

Strains lacking NBP2 exhibit growth defects due to HOG1. (A) A strain lacking NBP2 and PTP2 has growth defects due to HOG1. A tetrad dissection of JHM20 (MATα nbp2Δ∷kanMX) crossed to IMY21a (MATα ptp2Δ∷HIS3) is shown. Dissectants were grown at 30°C for 72h on standard rich media, YPD. (B) The nbp2Δ ptp2Δ growth defect is due to HOG1. Tetrad dissection of JHM20 (MATα nbp2Δ∷kanMX) crossed to CMY10 (MATα ptp2Δ∷HIS3 hog1Δ∷TRP1) is shown. Dissectants were grown at 30°C for 72h on YPD. (C) The growth defect of nbp2Δ ptp2Δ, which is suppressed in nbp2Δ ptp2Δ hog1Δ, can be seen in vegetatively growing cells. The growth of isogenic strains lacking HOG1, PTP2, PTC1 and NBP2 alone or in combination with each other in the BBY background as described in Table 1 was examined. The strains were grown in liquid YPD media, diluted to 1 unit (A600nm), and six-fold serial dilutions were spotted onto solid YPD and incubated at 30°C for 3 days. (D) Deletion of NBP2 or PTC1 is lethal when the hyperactive MEKK allele SSK2ΔN is expressed. Wild-type, ptc1Δ and nbp2Δ strains in the JD52 background (Table 1) were transformed with pSSK2ΔN (Warmka et al, 2001), a multicopy plasmid expressing SSK2ΔN under regulation of the GAL1 promoter, or the empty vector pYES2 (Invitrogen). Strains were grown on selective medium containing galactose or glucose at 30°C for 3 days. (E) Lethality resulting from SSK2ΔN overexpression in the nbp2Δ and ptc1Δ strains is due to HOG1. The ptc1Δ, nbp2Δ and hog1Δ single mutants, and the ptc1Δ hog1Δ and nbp2Δ hog1Δ double mutants in the JD52 background (Table 1) were transformed with pSSK2ΔN, and grown on selective medium containing galactose or glucose at 30°C for 3 days.
Another means to hyperactivate the HOG pathway is to overexpress the hyperactive MEKK allele SSK2ΔN, which leads to growth defects due to hyperactivation of the downstream Pbs2 MEK and Hog1 MAPK (Figure 1A) (Maeda et al, 1995). As predicted for a negative regulator, deletion of PTC1 in the background of the SSK2ΔN overexpressor results in further activation of Hog1, which is lethal (Figure 2D and E; Warmka et al, 2001). That ptc1Δ SSK2ΔN lethality was due to HOG1 was evident since ptc1Δ hog1Δ SSK2ΔN is viable (Figure 2E; Warmka et al, 2001). To test whether deletion of NBP2 would have a similar effect, SSK2ΔN was overexpressed in the nbp2Δ and nbp2Δ hog1Δ strains. SSK2ΔN overexpression was lethal in the nbp2Δ strain (Figure 2D and E), while it was not lethal in the nbp2Δ hog1Δ strain (Figure 2E). These results confirm that NBP2 is a negative regulator of the HOG pathway and suggest that Nbp2 likely acts downstream of the Ssk2 MEKK.
Nbp2 negatively regulates Hog1 kinase activity
Since genetic analysis showed that NBP2 is a negative regulator of the HOG pathway, we tested whether Nbp2 could affect Hog1 MAPK activity as does Ptc1. Consistent with its role as a negative regulator, we showed that deletion of PTC1 led to increased basal Hog1 kinase activity as well as an inability to inactivate Hog1 during adaptation (Warmka et al, 2001). Here, we find that deletion of NBP2 has similar effects. In the nbp2Δ mutant, Hog1 basal activity was elevated ∼3.5-fold (Figure 3) similar to ptc1Δ. In response to osmotic stress, Hog1 was activated, but like ptc1Δ, the nbp2Δ mutant showed an obvious defect in inactivating Hog1 during adaptation (Figure 3). As the level of Hog1 protein did not change, elevated Hog1 activity was most likely due to its increased phosphorylation. Thus, deletion of NBP2 led to defects in Hog1 inactivation as does deletion of PTC1, suggesting that NBP2 plays a role in Hog1 dephosphorylation in vivo.
Figure 3.

A strain lacking NBP2 has increased basal Hog1 kinase activity and a defect in adaptation. (A) Hog1 kinase activity was examined prior to and following stress in the NBP2 wild-type (IMY100) and in the nbp2Δ (JHM29) strains each carrying the Hog1-HA expression plasmid pHOG1-ha2 (Warmka et al, 2001). Before (time 0) and after exposure to osmotic stress (0.4M NaCl) for various times, Hog1-HA was immunoprecipitated and incubated with MBP and [γ-32P]ATP. Radiolabel incorporated into MBP was examined by SDS–PAGE and visualized using the PhosphorImager. Hog1-HA expression was the same in the two strains and unaltered by stress as indicated by the anti-HA blots of the lysates. (B) Hog1 kinase assays performed as in (A), for wild type (triangles) and nbp2Δ (circles) from three independent experiments. The plot shows the mean±s.d.
Nbp2 mediates binding between the Ptc1 phosphatase and the Pbs2 MEK
Since Nbp2 has no sequence similarity to PP2C or other protein phosphatases, it must downregulate Hog1 by a novel mechanism. One possibility is that Nbp2 acts as an adapter, recruiting Ptc1 to Hog1 via the Pbs2-Hog1 MEK-MAPK module. This seems possible, since Nbp2 was found in a complex with the scaffold protein Pbs2 and Ptc1 (Ho et al, 2002). Furthermore, the Pbs2 scaffold has been shown to bind Hog1 (Posas and Saito, 1997), which would provide Ptc1 with access to Hog1. If the role of Nbp2 is to recruit Ptc1 to Pbs2, then deletion of NBP2 should affect the ability of Ptc1 and Pbs2 to associate. Therefore, Ptc1-Pbs2 complex formation was examined in yeast lysates in the presence and absence of NBP2. Indeed, GST-Pbs2 co-precipitated with HA-Ptc1 in the wild type, but not in the nbp2Δ mutant (Figure 4). The GST-Pbs2 and HA-Ptc1 fusion proteins were expressed similarly in wild type and in nbp2Δ, indicating that their lack of interaction in the latter was due to the absence of Nbp2. Therefore, Nbp2 is required for Ptc1 to associate with Pbs2.
Figure 4.
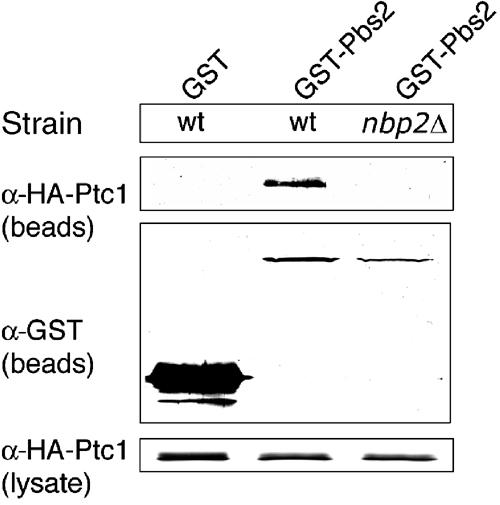
Ptc1 and Pbs2 associate in an Nbp2-dependent manner. Binding of HA-Ptc1 to GST-Pbs2 and GST was examined in the wild-type (JD52) and nbp2Δ (JHM17) strains. The strains carried pHA-PTC1 and pGST-PBS2 or the empty vector p(EG)KT, which expressed GST alone. GST-Pbs2 was isolated from yeast extracts using glutathione-sepharose, and the precipitated material was examined by SDS–PAGE and immunoblotting with anti-GST and anti-HA antibodies as described in Materials and methods. The amount of HA-Ptc1 in the lysates is also shown.
The Nbp2 SH3 domain binds Pbs2 and the novel N-terminal domain binds Ptc1
The above results showed that Nbp2 is needed to form a complex between Pbs2 and Ptc1. If Nbp2 recruits Ptc1 to Pbs2, then Nbp2 should be able to bind Ptc1 and Pbs2 simultaneously. If so, separate binding sites should exist on Nbp2 for Pbs2 and Ptc1. We predicted that the Nbp2 SH3 domain might bind Pbs2, as Pbs2 contains five Pro-rich or SH3 interaction motifs (SIMs) having the sequence PXXP, and one of these mediates binding to the upstream activator Sho1, which contains an SH3 domain (Posas and Saito, 1997). To map Nbp2 binding sites for Pbs2, GST-Nbp2 and -Nbp2 fragments containing or lacking the SH3 domain (Figure 5A) were coexpressed in yeast with Pbs2-HA. Full-length Nbp2 and Nbp2 fragments containing the SH3 domain bound Pbs2-HA, while Nbp2 fragments that lacked the SH3 domain could not bind Pbs2-HA (Figure 5B). Furthermore, the Nbp2 SH3 domain alone, residues 113–170, bound Pbs2-HA (Figure 5B). Thus the Nbp2 SH3 domain is necessary for binding Pbs2. Next, the Nbp2 binding site for Ptc1 was mapped. GST-Nbp2 associated with HA-Ptc1, as well as GST-Nbp2 fragments, which contained the N-terminal domain of this protein (Figure 5C). A fragment that contained the N-terminal domain alone, residues 1–115, also bound Ptc1, while neither the SH3 domain alone nor the C-terminal domain alone bound Ptc1 (Figure 5C). Therefore, the N-terminal region is necessary for binding Ptc1.
Figure 5.

Identification of Nbp2 domains required for association with Ptc1 and Pbs2. (A) Schematic representation of Nbp2 and its fragments used in the Pbs2 and Ptc1 binding experiments. Nbp2 is a 237-residue protein with a central SH3 domain corresponding to residues 116–166. The N- and C-terminal domains are novel. (B) The Nbp2 SH3 domain (residues 113–170) binds Pbs2. The yeast strain AWY3 (pbs2Δ) expressed GST fused to full-length Nbp2, Nbp2 fragments or GST alone from the CUP1 promoter, together with Pbs2-HA expressed from the CUP1 promoter. GST-containing proteins were isolated using glutathione-sepharose, and the bound material was examined by SDS–PAGE and immunoblotting with anti-GST and anti-HA antibodies. The level of Pbs2-HA in the lysates is also shown. (C) The Nbp2 N-terminal domain (residues 1–115) binds Ptc1. The yeast strain ASY1 (ptc1Δ) expressed GST fusions to NBP2 and its fragments or GST alone and HA-Ptc1 from the plasmid pHA-PTC1. Association between GST-containing proteins and HA-Ptc1 was examined as described in (B). (D) Recombinant Nbp2 SH3 domain and Nbp2 N-terminal domain bind Pbs2 and Ptc1, respectively. The ability of GST-Nbp2 N-terminal, GST-Nbp2 SH3 and GST-Nbp2 C-terminal domains, or GST alone to bind 6xHis-Pbs2 and 6xHis-Ptc1 was assessed as described in Materials and methods. The bound material was examined using anti-GST and anti-6xHis antibodies. The rightmost lane shows one-tenth the amount of 6xHis-Pbs2 and 6xHis-Ptc1 that was incubated with the GST-Nbp2 fusion proteins.
The above studies do not show whether Nbp2 bound Ptc1 and Pbs2 directly, as other proteins might have mediated their interaction in yeast lysates. Therefore, Nbp2 fragments, full-length Pbs2 and Ptc1, were expressed individually in Escherichia coli, purified and their ability to associate was examined in vitro. Corroborating the results in yeast lysates, the GST-Nbp2 N-terminal domain alone bound 6xHis-Ptc1, while neither the GST-Nbp2 SH3 domain nor the GST-Nbp2 C-terminal domain could bind 6xHis-Ptc1 (Figure 5D). In addition, the Nbp2 SH3 domain bound Pbs2, while neither the Nbp2 N-terminal domain nor the Nbp2 SH3 domain could bind Pbs2 (Figure 5D). Taken together with the analysis in yeast lysates, these studies show that the Nbp2 N-terminal and SH3 domains are necessary and sufficient for binding Ptc1 and Pbs2, respectively.
We then examined how the Pbs2 scaffold might be bound to the SH3 domain of the positive regulator Sho1, and the SH3 domain of the negative regulator Nbp2. One possibility is that they might be bound via the same Pro-rich motif in Pbs2, since binding of the positive and negative regulators to Pbs2 could be mutually exclusive. Previously, the Sho1 SH3 domain was shown to bind Pbs2 via the sequence PLPPLP, residues 94–99 (SH3 interaction motif 1 (SIM1); Figure 6A), where the mutation, P96S, blocked signaling via this branch of the HOG pathway and inhibited binding to Sho1 (Maeda et al, 1995). We tested whether Nbp2 might also be bound at this site. However, Nbp2 bound the Pbs2-P96S mutant protein as well as the wild-type Pbs2 (Figure 6B). There are two other SIMs in the N-terminal noncatalytic domain of Pbs2 (Figure 6A). Therefore, we tested whether they might be involved in binding Nbp2 SH3. Mutation of one of these sites SIM3, PTRP residues 196–199 to AAAA, did not alter its ability to bind to Nbp2. However, mutation of SIM2, PRRP residues 187–190 to AAAA, blocked binding to Nbp2 (Figure 6B). Since Pbs2-SIM2(AAAA) was still able to complement the osmosensitivity of the pbs2Δ mutant (data not shown), this mutant protein was not misfolded, and argues that the SIM2 site is essential for Nbp2 binding. Therefore, two different SIMs within Pbs2 provide binding sites for Sho1 and Nbp2. Since these sites are more than 90 residues apart, it is possible that the positive regulator Sho1 and the negative regulator Nbp2 are bound simultaneously to Pbs2.
Figure 6.
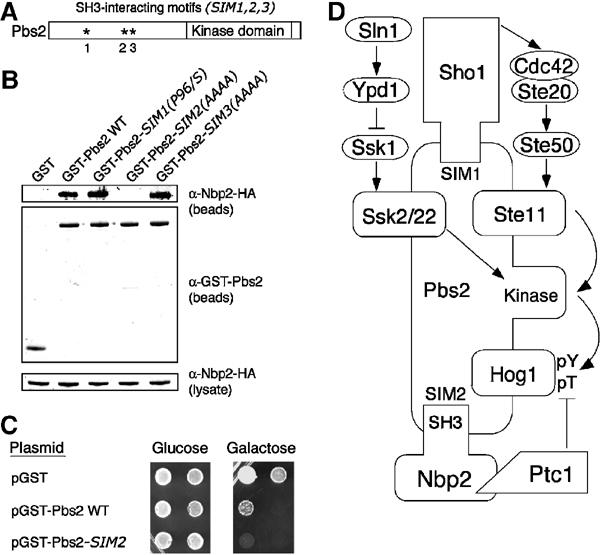
Nbp2 and Sho1 recognize different SH3 interaction motifs in Pbs2. (A) Schematic representation of the SH3 interaction motifs found in Pbs2. SH3 interaction motif 1 (SIM1, PLPPLP residues 94–99) is required for the Sho1-SH3 to bind. SIM2 (PRRP residues 187–190) and SIM3 (PTRP residues 196–199) have not been implicated previously in SH3 domain binding. (B) Pbs2-SIM2 is required to bind Nbp2. GST fusions to Pbs2 and Pbs2 with mutations in SIM1, 2 or 3 were coexpressed with Nbp2-HA in the pbs2Δ strain JHM61. GST fusion proteins were isolated using glutathione-sepharose, and the co-precipitated material was examined by SDS–PAGE and immunoblotting with anti-GST and anti-HA antibodies. The amount of Nbp2-HA in lysates was similar in all experiments as shown in the anti-HA blot. (C) Pbs2-SIM2 mediates Nbp2-Ptc1 negative regulation of the HOG pathway. Growth of the pbs2Δ ptp2Δ strain IMY108 containing empty vector (pGST), GST-Pbs2 and GST-Pbs2-SIM2(AAAA) was compared. GST and GST-Pbs2 fusions were under control of the GAL1 promoter and overexpressed on galactose-containing media. Cells at 30 units (A600nm) were diluted 10-fold (left to right) and were plated as described in Figure 2C. (D) Model for Nbp2-Ptc1 regulation of the HOG pathway via the scaffold protein Pbs2. We show here that Nbp2 negatively regulates the HOG pathway by recruiting Ptc1 to the Pbs2 scaffold, facilitating Hog1 inactivation. Previously, we showed that Ptc1 inactivates this pathway by dephosphorylating Hog1-pT (Warmka et al, 2001). Thus, the Pbs2 scaffold binds the negative regulators in this pathway (Nbp2-Ptc1) and the positive regulators, including Sho1, the Ste11, Ssk2 and Ssk22 MEKKs and the Hog1 MAPK. Since Pbs2 bound Nbp2 and Sho1 via two different Pro-rich motifs (SIMs), Pbs2 could bind the negative and positive regulators simultaneously.
Last, we tested whether Nbp2-Ptc1 exerted its negative regulatory effect via binding Pbs2-SIM2 in vivo. If so, a strain overexpressing the PBS2-SIM2(AAAA) mutant should be more susceptible to HOG pathway hyperactivation than a strain overexpressing wild-type PBS2, as it lacks the ability to dephosphorylate Hog1-pThr. Since strong growth defects due to Hog1 hyperactivation require increased phosphorylation on both Thr and Tyr in its activation loop, we examined the effects of PBS2-SIM2(AAAA) and PBS2 overexpression in a strain that lacked PTP2, which is defective in Hog1-pY dephosphorylation. Overexpression of PBS2 was detrimental to growth, but overexpression of the mutant PBS2-SIM2(AAAA) was lethal (Figure 6C). The control strain expressing empty vector was viable under these conditions. Thus, by binding SIM2 within Pbs2, Nbp2-Ptc1 exerts its negative regulatory effect on this pathway.
Discussion
The mechanisms that target PP2C Ser/Thr phosphatases to their substrates in MAPK pathways are not clear. The significance of this work is the identification and characterization of a novel PP2C targeting subunit important for inactivating the HOG pathway. We showed that Nbp2 acts as an adapter in this pathway, mediating Ptc1 interaction with the scaffold protein Pbs2 MEK, as shown in the model in Figure 6D. As predicted for an adapter, deletion of NBP2 blocked the ability of Ptc1 to bind Pbs2 (Figure 4), and Nbp2 contained separate binding sites for Ptc1 and Pbs2 (Figure 5). Furthermore, the Pbs2 scaffold, previously shown to bind Sho1 and other positive regulators (Posas and Saito, 1997), bound Sho1 and Nbp2 via distinct Pro-rich motifs (Figure 6). Consistent with the role of Nbp2 in recruiting Ptc1, Nbp2 acted as a negative regulator of Hog1 in genetic and biochemical studies. Deletion of the negative regulators NBP2 and PTP2 led to growth defects due to HOG1 (Figure 2), and a strain lacking NBP2 showed defects in downregulating Hog1 kinase activity (Figure 3). Thus, these defects are due to an inability to target Ptc1 to the Pbs2-Hog1 complex to inactivate Hog1 normally. Since NBP2 influenced Hog1 activity, we tested the possibility that Nbp2 bound Hog1, but we could not reproducibly detect an interaction between Nbp2 and Hog1. However, as Pbs2 is a scaffold that binds Hog1 (Posas and Saito, 1997), Nbp2-Ptc1 binding to Pbs2 would provide access to Hog1.
We also tested whether Nbp2 interaction with Ptc1, or its interaction with Pbs2, was altered by osmotic stress. However, no difference could be detected in co-precipitation assays. Therefore, the Nbp2-Ptc1 complex does not appear to require Pbs2 phosphorylation for interaction. As a result of this, it is possible that the Nbp2-Ptc1 complex has constitutive activity against this pathway. This seems possible, as even in the absence of stress, deletion of the Hog1 negative regulators NBP2 or PTC1 together with PTP2 led to pathway hyperactivation as evidenced by HOG1-dependent growth defects (Figure 2B and C), and deletion of NBP2 or PTC1 was sufficient to increase basal Hog1 kinase activity (Figure 3; Warmka et al, 2001).
Another important consideration for the proposed role for Nbp2 as an adapter, which mediates Ptc1 inactivation of Hog1 via Pbs2, is the subcellular localization of these components. We found that Nbp2, similar to Ptc1 (Warmka et al, 2001), is evenly distributed between the cytoplasm and the nucleus and that its localization does not change in response to stress (J Mapes and IM Ota, unpublished). The target of Nbp2-Ptc1, Hog1, accumulates in the nucleus in response to activation similar to other MAPKs (Ferrigno et al, 1998; Reiser et al, 1999), while the Pbs2 scaffold can be cytoplasmic and nuclear. Although previous studies had shown that Pbs2 was cytoplasmic and was excluded from the nucleus (Ferrigno et al, 1998; Reiser et al, 1999), a recent study showed that it contains a nuclear exclusion signal, which, when masked, allows Pbs2 to accumulate in the nucleus (Tatebayashi et al, 2003). As a result of this, it is possible for Ptc1, bound to Pbs2 via Nbp2, to inactivate nuclear-localized Hog1. However, since phosphorylated Hog1 appears to exit the nucleus (Mattison and Ota, 2000), cytoplasmic Pbs2-Nbp2-Ptc1 could also inactivate Hog1.
Since Nbp2 acts as an adapter, recruiting Ptc1 to Pbs2 to inactivate Hog1, it is reasonable to propose that Ptc1 inactivates Pbs2, and this in turn leads to Hog1 inactivation. However, in previous studies where we examined the role of Ptc1 in this pathway, we could not detect Ptc1 inactivation of Pbs2, although it clearly inactivated Hog1. For example, overexpression of PTC1 from a multicopy plasmid blocked osmotic stress-induced Hog1 kinase activation, but did not block Hog1-Tyr phosphorylation (Warmka et al, 2001). The latter suggests that PTC1 overexpression does not downregulate Pbs2 activity. If it did, a decrease in the level of both Hog1-pThr and pTyr should have been detected. We also attempted to measure Pbs2 kinase activity, but we could not detect an increase in Pbs2 kinase activity upon osmotic stress. However, overexpression of PTC1 diminished Pbs2 kinase activity, while its deletion increased Pbs2 activity (J Mapes and IM Ota, unpublished). At this time, we cannot exclude the possibility that Nbp2-Ptc1 inactivates Pbs2; however, our current data suggest that its activity toward Hog1 may be more significant.
Nbp2 exhibited another feature characteristic of an adapter for Ptc1. Nbp2 was not as strong a negative regulator as the Ptc1 phosphatase itself. For example, deletion of SLN1, which encodes a putative osmosensor and is a negative regulator of this pathway, is lethal due to Hog1 hyperactivation (Maeda et al, 1994). Overexpression of PTC1 from a multicopy plasmid suppressed sln1Δ lethality (Warmka et al, 2001), while overexpression of NBP2 from a multicopy plasmid did not (J Mapes and IM Ota, unpublished). In addition, overexpression of NBP2 did not enhance the ability of Ptc1 to inactivate Hog1 kinase activity in vivo (data not shown). These outcomes seem reasonable, since overexpressing a gene encoding phosphatase catalytic activity should have a greater effect than overexpressing an adapter protein. Furthermore, these results show that Ptc1 does not become a better negative regulator of Hog1 when the level of Nbp2 protein is increased. Thus Nbp2 is unlikely to affect Ptc1 catalytic activity. Since the level of Pbs2 protein is ∼10-fold lower than Ptc1 or Nbp2 (J Mapes and IM Ota, unpublished), we believe that the limiting factor in Nbp2-Ptc1 inactivation of Hog1 is the amount of Pbs2.
A study predicting interactions between yeast proteins containing SH3 domains and those proteins containing Pro rich motifs suggested that Nbp2 would bind Pbs2 in addition to other signaling proteins (Tong et al, 2002). One of these is the Bck1 MEKK in the cell wall integrity MAPK pathway. It is possible that Nbp2-Ptc1 could act in this pathway, since genetic analysis has linked Ptc1 to the Pkc1 protein kinase C (Huang and Symington, 1995), which acts upstream of Bck1. However, a role for Ptc1 as a negative regulator of this pathway has not been established. Another group of proposed Nbp2 interacting proteins were the p21-activated kinases (PAKs) Ste20, Cla4 and Skm1. Ste20 has been shown to act upstream of the MAPK cascades in the pheromone response and HOG pathways (Wu et al, 1995; Raitt et al, 2000). Although CLA4 can substitute for STE20 in the HOG pathway, this may only occur in the ste20Δ strain. We tested whether Nbp2 might bind Ste20, but were not able to detect their interaction (data not shown). However, Ptc1 was found to co-precipitate with Skm1 (Ho et al, 2002). It will be interesting to ask whether Nbp2 mediates their interaction, and to examine the possibility that Skm1 has a role in MAPK pathways.
That PP2C and its targeting subunits are important in PAK regulation has recently been demonstrated in mammalian cells. A PP2C targeting subunit called PIX recruits PP2C phosphatases POPX1 and POPX2 to PAKs (Koh et al, 2002). PIX is a guanine nucleotide exchange factor, which also contains an SH3 domain that mediates the interaction with PAKs. We examined the possibility that there might be a common sequence motif that serves as a docking site for PP2C proteins on PIX and Nbp2. However, the PIX and Nbp2 PP2C binding domains share no obvious similarities. As more PP2C binding proteins are identified, it may be possible to identify a PP2C binding site consensus sequence.
A yeast synthetic lethal analysis conducted with NBP2 and ∼4700 viable yeast deletion mutants placed NBP2 at a hub in a multinode network of interactions (Tong et al, 2002). Deletion of NBP2 and genes involved in nuclear migration, spindle function, actin assembly, and actin and tubulin folding was synthetically lethal, suggesting that Nbp2 has a role in regulating the cytoskeleton. Furthermore, other nodes interacting with Nbp2 were occupied by Bni1 (which plays various roles in regulating the actin cytoskeleton), Bim1 (which binds tubulin and regulates spindle orientation), and the recently characterized Mti1/Bbc1 (which regulates the actin cytoskeleton by binding type I myosins) (Mochida et al, 2002). Since the HOG pathway has been implicated in cell morphogenesis in response to growth and stress signals (reviewed in Hohmann, 2002), and the cytoskeleton reorganizes in response to stress, Nbp2 might regulate these functions by acting as an adapter for Ptc1. It would be interesting to test whether deletion of PTC1 with the same subset of genes is synthetic lethal. Alternatively, Ptc1 may have functions independent of Nbp2. If so, ptc1Δ may be synthetic lethal with other genes. The combination of molecular genetic, genomic and proteomic approaches such as these will shed more light on the role of this phosphatase adapter protein in yeast.
Materials and methods
Yeast strains and genetic techniques
Yeast strains (Table 1) were derived from BBY45 and BBY48 and the galactose-inducible strains JD52 and JD53. The nbp2Δ strains were obtained by transformation with an nbp2Δ∷kanMX allele, which was produced by PCR using oligonucleotides complementary to NBP2 and kanMX contained in pRS400 (Brachmann et al, 1998). The pbs2Δ ptp2Δ strain IMY108 was produced by transforming JHM61 with the ptp2Δ∷HIS3 allele (Ota and Varshavsky, 1992). CMY1973 (MATα hog1Δ) was produced by transforming JD53 with hog1Δ∷hisG-URA3-hisG (O'Rourke and Herskowitz, 1998) and growing on media containing 5-fluoroorotic acid to isolate the hog1Δ∷hisG strain. Strains lacking NBP2 in combination with PTP2 and/or HOG1 deletions were produced using standard genetic techniques.
Table 1.
Yeast strains used in this study
| Strains derived from JD52 and JD53 | ||
| JD52 | MATa trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | Warmka et al (2001) |
| JD53 | MATα trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | Warmka et al (2001) |
| JHM17 | MATa nbp2Δ∷kanMX trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| CMY1973 | MATα hog1Δ∷hisG trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| JHM34 | MATα nbp2Δ∷kanMX hog1Δ∷hisG trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| CMYX | MATa ptp2Δ∷HIS3 hog1Δ∷hisG trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| IMY107 | MATα ptc1Δ∷LEU2 trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | Warmka et al (2001) |
| JHM38 | MATa ptc1Δ∷LEU2 hog1Δ∷hisG trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| JHM61 | MATa pbs2Δ∷kanMX trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| IMY108 | MATa pbs2Δ∷kanMX ptp2Δ∷HIS3 trp1-Δ63 ura3-52 his3Δ200 leu2-3,112 lys2-801 GAL+ | This study |
| Strains derived from BBY45 and BBY48 | ||
| BBY45 | MATa trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Warmka et al (2001) |
| BBY48 | MATα trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Warmka et al (2001) |
| JHM20 | MATα nbp2Δ∷kanMX trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This study |
| IMY21a | MATa ptp2Δ∷HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Ota and Varshavsky (1992) |
| CMY10 | MATa ptp2Δ∷HIS3 hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Mattison and Ota (2000) |
| JHM25 | MATanbp2Δ∷kanMX ptp2Δ∷HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This study |
| JHM29 | MATanbp2Δ∷kanMX hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This study |
| JHM21 | MATanbp2Δ∷kanMX ptp2Δ∷HIS3 hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This study |
| IMY100 | MATa hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Jacoby et al (1997) |
| ASY1 | MATα ptc1Δ∷LEU2 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Jacoby et al (1997) |
| ASY2 | MATα ptc1Δ∷LEU2 ptp2Δ∷HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Jacoby et al (1997) |
| ASY3 | MATα ptc1Δ∷LEU2 ptp2Δ∷HIS3 hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Jacoby et al (1997) |
| IMY103 | MATa∷ptc1Δ∷LEU2 hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Warmka et al (2001) |
| AWY3 | MATa pbs2∷LEU2 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | Winkler et al (2002) |
| JHM29 | MATanbp2Δ∷kanMX hog1Δ∷TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This study |
Plasmids
Nbp2 fused to the hemagglutinin epitope (Nbp2-HA) was expressed in yeast from its endogenous promoter in the plasmid pNBP2-HA (HIS3, 2 μm). NBP2 was isolated by PCR from genomic DNA producing a fragment containing 340 bp upstream of the start codon and a NotI site substituted for the stop codon. The fragment was cloned into pRS423 (Sikorski and Hieter, 1989) and a NotI fragment containing three repeats of the HA epitope was ligated to produce pNBP2-HA. Nbp2-HA was functional as it suppressed the temperature sensitivity of the nbp2Δ strain.
GST-Nbp2 and its fragments were expressed from the CUP1 promoter in pCUP1-GST (URA3, 2 μm). The vector was constructed by subcloning a fragment containing the CUP1 promoter–GST–thrombin cleavage site from the modified pYEX4T-1 plasmid (Exclone, Invitrogen) into pRS426 (Sikorski and Hieter, 1989). NBP2 was isolated from genomic DNA using PCR and cloned into pCUP1-GST to produce pGST-NBP2. GST-Nbp2 fragments produced by PCR were expressed in yeast from pCUP1-GST. The fragments contained EcoRI and SmaI sites at their 5′ and 3′ ends, respectively, which were used to clone into pCUP1-GST.
GST-PTC fusion proteins were expressed in yeast from pKT-PTC1 (Warmka et al, 2001), pKT-PTC2 (Young et al, 2002) and pKT-PTC3. PTC3 was isolated from genomic DNA by PCR and cloned into p(EG)KT (Mitchell et al, 1993). HA-Ptc1 was expressed from the multicopy plasmid pHA-PTC1. pGFP-PTC1 (Warmka et al, 2001) was digested with NotI and BamHI to remove GFP, and a fragment containing three repeats of the HA epitope was substituted. The HA-Ptc1 protein was functional as it complemented the ptc1Δ temperature-sensitive phenotype.
Plasmids expressing PBS2 in yeast were constructed as follows. GST-Pbs2 was expressed from the GAL promoter in pGST-PBS2, which contains a 2.4 kb BamHI–SacI fragment of PBS2 obtained using PCR. Pbs2-HA was expressed from the CUP1 promoter in YEplac181 (LEU2, 2 μm) (Gietz and Sugino, 1988). The PBS2 stop codon was replaced with a NotI site, and three repeats of the HA epitope were ligated to produce pCUP1-PBS2-HA. GST fused to Pbs2, or Pbs2 where SIMs were mutated, were expressed in pEG(KT). Pbs2-P96S was constructed by producing two PCR products that overlapped at the mutated Pro96, using the oligonucleotides 5′-GGGAAGAGGCGACAATGGCTTATTAACAATCTG-3′ (mutation underlined) and 5′-GCCATTGTCGCCTCTTCCCGTAGC-3′ (mutation underlined). Two other SIMs in Pbs2 were mutated using similar methods.
Plasmids for expression in E. coli were constructed as follows. GST fusions to Nbp2 fragments were expressed from pGEX-6P-1 (Novagen). EcoRI–SmaI fragments from the pCUP1-Nbp2 yeast plasmids were subcloned into pGEX-6P-1. The plasmid expressing 6xHis-Ptc1 was described previously (Warmka et al, 2001). 6xHis-Pbs2 was expressed from pRSETa-Pbs2, which was constructed by subcloning the 2.4 kb BamHI–SacI fragment from pGST-Pbs2 into pRSETa (Invitrogen).
Expression of Nbp2, Ptc1 and Pbs2 in E. coli and purification by affinity chromatography
NBP2 fragments were expressed from the pGEX-6P-1 plasmids in E. coli BL21(DE3) pLysS. The strains were grown in 2xYT medium at 37°C to 0.6 units at A600 nm, cooled to 23°C, and induced with 300 mM isopropyl-β-D-thiogalactoside at 23°C for 2 h. Cells were lysed in sonication buffer (Tris–HCl (pH 8.0), 100 mM NaCl, 0.05% Triton X-100 and 1 mM phenyl methylsulfonyl fluoride (PMSF)), then mixed with glutathione-sepharose (Pharmacia) for 1 h at 4°C. The resin was washed with the same buffer and eluted with buffer containing 20 mM glutathione. Fractions containing the GST fusion proteins were dialyzed against phosphate-buffered saline, and the GST-tagged protein was quantified using the Bradford assay. The GST, GST N-terminal and GST SH3 domains were greater than 99% pure and the GST C-terminal fragment was ∼95% pure as judged by SDS–PAGE and Coomassie staining. Expression and purification of 6xHis-Ptc1 and 6xHis-Pbs2 was performed as described previously (Warmka et al, 2001).
Co-precipitation assays
Binding between Nbp2-HA and GST-PTCs or GST alone was examined in yeast strain JHM17 (nbp2Δ) carrying pNBP2-HA and pKT-PTC1, pKT-PTC2, pKT-PTC3, or empty vector p(EG)KT. Yeast were grown in synthetic media lacking uracil and histidine and containing 2% galactose. Cells from 150 ml of culture grown to ∼1 unit (A600 nm) were harvested by centrifugation and disrupted using glass beads in 600 μl of lysis buffer (50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.1% Triton X-100, 5 mM MgCl2, 5 mM MnCl2 and 0.1% 2-mercaptoethanol) with protease inhibitors leupeptin, pepstatin A, antipain, aprotinin and chymostatin, each at 20 μg/ml, and 1 mM PMSF. The lysates were incubated with 30 μl of glutathione-sepharose and incubated for 1.5 h at 4°C. The resin was washed extensively, boiled in sample buffer, and the bound material was analyzed by SDS–PAGE and immunoblotting with anti-GST (Pharmacia) and anti-HA (HA.11, Covance) antibodies. All other binding experiments in yeast lysates were performed similarly.
Binding between GST-tagged Nbp2 fragments or GST, and 6xHis-Ptc1 or 6xHis-Pbs2 purified from E. coli was performed as follows. A measure of 1 μl of 100 μM GST-tagged protein was incubated with 5 μl of 20μM 6xHis-Ptc1 for 1h at 4°C with 50μl of glutathione-sepharose in 100μl of buffer (50mM Tris–HCl (pH 7.5), 50mM NaCl and 0.05% Triton X-100). The resin was washed four times with the same buffer, and then four times with buffer containing 150mM NaCl. Binding between GST-Nbp2 fragments or GST and 6xHis-Pbs2 was carried out as above, except that the wash buffer contained 250mM NaCl. The bound material was examined by SDS–PAGE and immunoblotting with anti-GST and anti-His6 antibodies (Covance).
Hog1 kinase assay
Hog1 kinase activity was examined in wild-type and nbp2Δ strains as described previously (Warmka et al, 2001). Strains were grown in synthetic media lacking histidine to exponential phase, and then left untreated or exposed to 0.4M NaCl for various times. Cells were harvested by centrifugation, lysed using glass beads in buffer A, and Hog1-ha was immunoprecipitated with anti-HA antibody (Covance). The immunoprecipitates were washed extensively, and incubated with [γ-32P]ATP (0.1mM, 8000cpm/pmol) and myelin basic protein (MBP) (1μM; Sigma) for 30min at 30°C. Incorporation of 32P into MBP was assessed by SDS–PAGE and PhosphorImager analysis (Molecular Dynamics).
Acknowledgments
We thank Ji Lee and Janel Warmka for technical assistance, and Chris Mattison, Christian Young and Mark Winey for their helpful comments. We also thank Mark Winey, Martin Horvath and Janet Shaw for allowing us to complete this work in their laboratories. This work was funded by a grant from the American Cancer Society.
References
- Altman R, Kellogg D (1997) Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol 138: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell AJ, Flatauer LJ, Matsukama K, Thorner J, Bardwell L (2001) A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J Biol Chem 276: 10374–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- de Nadal E, Alepuz PM, Posas F (2002) Dealing with osmostress through MAP kinase activation. EMBO Rep 3: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA (2001) The Ste5p scaffold. J Cell Sci 114: 3967–3978 [DOI] [PubMed] [Google Scholar]
- Ferrigno P, Posas F, Koepp D, Saito H, Silver PA (1998) Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J 17: 5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada M, Kobayashi T, Ohnishi M, Ikeda S, Wang H, Katsura K, Yanagawa Y, Hiraga A, Kanamaru R, Tamura S (1998) Selective suppression of stress-activated protein kinase pathway by protein phosphatase 2C in mammalian cells. FEBS Lett 437: 172–176 [DOI] [PubMed] [Google Scholar]
- Hanada M, Ninomiya-Tsuji J, Komaki K, Ohnishi M, Katsura K, Kanamaru R, Matsumoto K, Tamura S (2001) Regulation of the TAK1 signaling pathway by protein phosphatase 2C. J Biol Chem 276: 5753–5759 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Hellbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutiller K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Desar D, Lin Z, Michallckova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford JVP, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KN, Symington LS (1995) Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics 141: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y (2000) Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between yeast proteins. Proc Natl Acad Sci USA 97: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby T, Flanagan H, Faykin A, Seto AG, Mattison C, Ota I (1997) Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem 272: 17749–17755 [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298: 1911–1912 [DOI] [PubMed] [Google Scholar]
- Kellogg D, Kikuchi A, Fujii-Nakata T, Turck CW, Murray AW (1995) Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol 130: 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C-G, Tan E-J, Manser E, Lim L (2002) The p21-activated kinase PAK is negatively regulated by POPX1 and POPX2, a pair of serine/threonine phosphatases of the PP2C family. Curr Biol 12: 317–321 [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H (1995) Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554–558 [DOI] [PubMed] [Google Scholar]
- Maeda T, Tsai AY, Saito H (1993) Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol Cell Biol 13: 5408–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245 [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 12: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison CP, Ota IM (2000) Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcellular localization of the Hog1 MAP kinase in yeast. Genes Dev 14: 1229–1235 [PMC free article] [PubMed] [Google Scholar]
- Mattison CP, Spencer SS, Kresge KA, Lee J, Ota IM (1999) Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol Cell Biol 19: 7651–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelenik H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9: 715–722 [DOI] [PubMed] [Google Scholar]
- Mochida J, Yamamoto T, Fujimura-Kamada K, Tanaka K (2002) The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott–Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics 160: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AN, Shiozaki K (1999) Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev 13: 1653–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I (1998) The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12: 2874–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota IM, Varshavsky A (1992) A gene encoding a putative tyrosine phosphatase suppresses lethality of an N-end rule-dependent mutant. Proc Natl Acad Sci USA 89: 2355–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Gibson TB, Xu B-E, Karandikar M, Berman K, Cobb MH (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22: 153–183 [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H (1997) Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276: 1702–1705 [DOI] [PubMed] [Google Scholar]
- Raitt DC, Posas F, Saito H (2000) Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J 19: 4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V, Ruis H, Ammerer G (1999) Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell 10: 1147–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ (1998) MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281: 1668–1671 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Akashi T, Okuda A, Kikuchi A, Fukui K (2000) NBP1 (Nap1 binding protein 1), an essential gene for G2/M transition of Saccharomyces cerevisiae, encodes a protein of distinct sub-nuclear localization. Gene 246: 395–404 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa M, Maeda T, Saito H (1998) Protein phosphatase 2Calpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J 17: 4744–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Nishida E (2002) Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther 93: 193–202 [DOI] [PubMed] [Google Scholar]
- Tatebayashi K, Takekawa M, Saito H (2003) A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J 22: 3624–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CWV, Fields S, Boone C, Cesareni G (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295: 321–324 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Warmka J, Hanneman J, Lee J, Amin D, Ota I (2001) Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol Cell Biol 21: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ (1998) A mammalian scaffold complex that selectively mediates MAP kinase activation. Science 281: 1671–1674 [DOI] [PubMed] [Google Scholar]
- Winkler A, Arkind C, Mattison CP, Burkholder A, Knoche K, Ota I (2002) Heat stress activates the yeast high osmolarity glycerol MAPK pathway; protein tyrosine phosphatases are essential under heat stress. Eukaryot Cell 1: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Whiteway M, Thomas DY, Leberer E (1995) Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem 270: 15984–15992 [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H (1997) Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol 17: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C, Mapes J, Hanneman J, Al-Zarban S, Ota I (2002) Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot Cell 1: 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Bohmann D (1999) Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126: 3947–3956 [DOI] [PubMed] [Google Scholar]
- Zhan XL, Guan KL (1999) A specific protein–protein interaction accounts for the in vivo substrate selectivity of Ptp3 towards the Fus3 MAP kinase. Genes Dev 13: 2811–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]


