Abstract
Here we describe 11 crystal structures of nucleosome core particles containing individual point mutations in the structured regions of histones H3 and H4. The mutated residues are located at the two protein–DNA interfaces flanking the nucleosomal dyad. Five of the mutations partially restore the in vivo effects of SWI/SNF inactivation in yeast. We find that even nonconservative mutations of these residues (which exhibit a distinct phenotype in vivo) have only moderate effects on global nucleosome structure. Rather, local protein–DNA interactions are disrupted and weakened in a subtle and complex manner. The number of lost protein–DNA interactions correlates directly with an increased propensity of the histone octamer to reposition with respect to the DNA, and with an overall destabilization of the nucleosome. Thus, the disruption of only two to six of the ∼120 direct histone–DNA interactions within the nucleosome has a pronounced effect on nucleosome mobility and stability. This has implications for our understanding of how these structures are made accessible to the transcription and replication machinery in vivo.
Keywords: chromatin remodeling, crystallography, nucleosome structure, protein–DNA interactions
Introduction
The nucleosome is the elemental repeating unit in all eucaryotic chromatin. One tetramer of histones (H3–H4) and two dimers of (H2A–H2B) form the histone octamer, around which 147 base pairs of DNA are wrapped in 1.65 turns of a tight superhelix (Luger et al, 1997a). The massive distortion of the DNA is brought about by tight interactions between the rigid framework of the histone proteins with the DNA at 14 independent DNA-binding locations, numbered superhelix location (SHL) 0.5–6.5 in Figure 1, either using L1L2 loops or α1α1 DNA-binding motifs see Luger, 2003; Muthurajan et al, 2003 for a detailed description of nucleosome structure. DNA binding at each site is accomplished mainly by three to six hydrogen bonds between protein main-chain amides and the DNA phosphate backbone (Davey et al, 2002). Side chains contribute to binding by contacting the DNA phosphate backbone, but avoid making base-specific contacts. The packaging of DNA into nucleosomes causes severe impediments to gene regulation. Therefore, the transcription of many eucaryotic genes in the context of chromatin depends on the action of ATP-dependent chromatin remodeling factors. ATP is used to alter nucleosomes in a way that facilitates transcription factor binding and access of the transcription machinery to the DNA that is normally constrained by tight interaction with the histone octamer (Workman and Kingston, 1998). This altered state of the nucleosome is structurally ill defined, and is characterized by a changed DNase I footprinting pattern (Utley et al, 1997; Phelan et al, 2000; Aoyagi et al, 2002 and references therein) and by increased access of restriction endonucleases to nucleosomal DNA (Aoyagi et al, 2002; Kassabov et al, 2002). The presence of a full complement of histone proteins has been established (reviewed in Imbalzano, 1998). In some cases, the remodeled state appears to persist after the passage of the remodeling complex (Lorch et al, 1998). Some chromatin remodeling factors (e.g. SWI/SNF) enhance sliding of the histone octamer along the DNA (Becker, 2002; Kassabov et al, 2002 and references therein) potentially leading to the exposure of promoters and regulatory sequences. Alternately, SWI/SNF can also cause ‘looping' out of DNA or transfer the octamer in trans (Fan et al, 2003). Nucleosome sliding is also observed in vitro, where the energy barrier is overcome by increasing the temperature. The two phenomena may be mechanistically related, since both require the permanent or transient disruption of an undetermined number of protein–DNA contacts.
Figure 1.
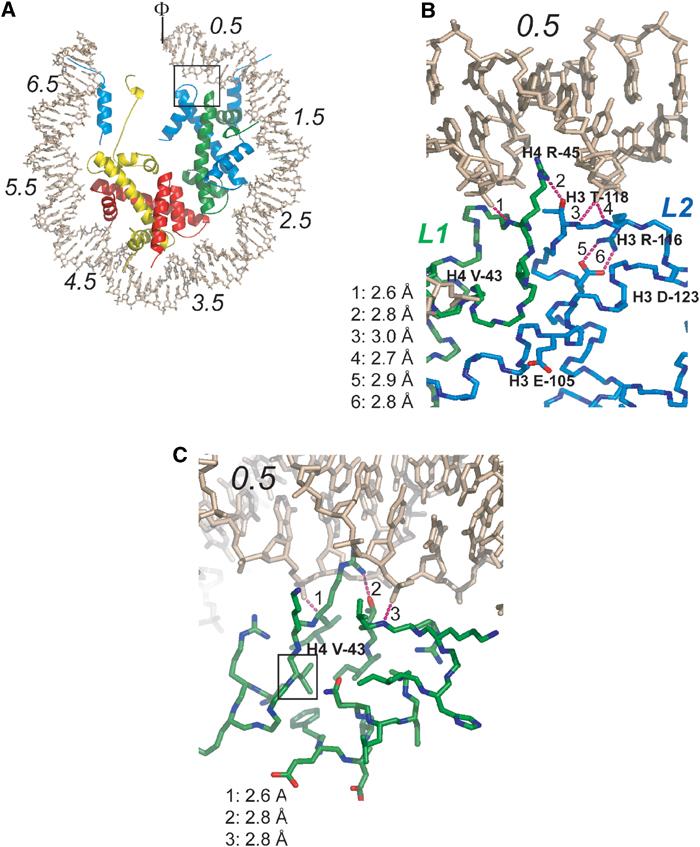
Location and structural context of histone Sin mutants. (A) Overview of the NCP structure, viewed down the superhelical axis. Only half of the DNA and associated proteins are shown. SHLs are indicated by italic numbers from 0.5 (flanking the nucleosomal dyad) to 6.5 (at the entry and exit point of the DNA). The locations of the H4 L1 loop and the H3 L2 loop are indicated in green and blue, respectively. Histone H3 is colored in blue, H4 in green, H2A in yellow, and H2B in red. The box indicates the location of Sin mutants. The nucleosomal dyad is indicated (Φ). (B) Close-up of the Sin region showing the five residues affected by Sin mutations. Dotted lines represent hydrogen bonds. (C) Location of H4 Val-43 in the hydrophobic core at the underside of the L1L2 loop arrangement. The view is similar as in (B), with a slight rotation around the y-axis.
Mutations in the genes for histones H3 and H4 partially alleviate the transcription defects caused by the inactivation of the yeast chromatin remodeling factor SWI/SNF (Kruger et al, 1995). These genetic screens provided further support to the hypothesis that SWI/SNF might function by affecting chromatin structure (Kruger and Herskowitz, 1991). The so-called Sin (Swi-INdependent) mutations are clustered in the L1L2 region of the (H3–H4)2 tetramer that organizes the central two turns of nucleosomal DNA (SHL±0.5, Figure 1). Surprisingly, only one of the affected residues is involved in making direct contacts with the DNA. Analysis of the effect of some of the Sin mutants on chromatin structure and transcription in vivo and in vitro demonstrated decreased ability to supercoil DNA, increased susceptibility to Dam methylase and DNase, and impaired nucleosome-mediated repression of PHO5 expression (Kurumizaka and Wolffe, 1997; Wechser et al, 1997). Previous crystallographic studies have shown that interactions between DNA and the histone octamer are strongest at the dyad region of the nucleosome (SHL±0.5), and hence disruption of contacts in this region may be the rate-limiting step in nucleosomal dissociation and sliding (Luger and Richmond, 1998). This led to the hypothesis that Sin mutations compensate for inactivation of SWI/SNF by destabilizing protein–DNA interactions in this region, either leading to facilitated sliding or to the formation of the ‘remodeled state' in the absence of ATP-dependent remodeling factor. Intriguingly, it was recently shown that nucleosomal arrays reconstituted with one particular Sin mutant (H4 R45C) completely lose the ability to undergo magnesium-dependent condensation, suggesting that the Sin mutations may affect chromatin structures beyond the nucleosome (Horn et al, 2002).
We tested the different hypotheses by which histone Sin mutations may overcome an SWI/SNF deficiency by determining the crystal structures of nucleosome core particles (NCPs) reconstituted with histones bearing the original Sin mutants, and by analyzing the stability and sliding properties of mutant NCPs in solution. To distinguish whether the observed in vivo and in vitro effects are the result of a loss-of-function from mutating a specific residue, or a result of the introduction of a more bulky and structurally more disruptive mutant, we also analyzed several control mutants. The 11 mutant structures show that the overall structure of the nucleosomes and the path of the DNA across the histone octamer surface are maintained. However, the dynamic behavior and stability of mutant nucleosomes is changed as a result of the local disruption of only very few selected histone–DNA interactions. This results in the increased mobility, or sliding rate, of the histone octamer on the DNA in vitro. Our data show that interactions at the dyad region are indeed rate limiting for histone octamer sliding, and that only very few need to be disrupted for increased sliding rates. Thus, histones have evolved to balance for seemingly contradictory tasks of binding and bending DNA, at the same time permitting DNA fluidity that appears to be necessary for all processes involving the DNA template.
Results
Histone Sin mutations fall into four different classes
The originally identified histone Sin mutants can be grouped into four classes based on the function of the affected residues in nucleosome structure. As a consequence of the partially symmetric architecture of the histone octamer, this classification can also be applied to other regions in the NCP, in particular SHL±2.5, 3.5, and 5.5 (Figure 1A, Muthurajan et al, 2003).
Class I mutations affect either H3 T118 or H4 R45. The side chain of H4 R45 is inserted into the minor groove of the DNA at SHL±0.5, but is kept from making site-specific contacts with the DNA base by a hydrogen bond with H3 T118 (Figure 1B). Both residues are conserved throughout evolution, and a similar arrangement is observed at 12 of the 14 SHLs (Luger and Richmond, 1998; Luger, 2003). The Sin mutations identified in Kruger et al (1995) change H4 R45 to cysteine or histidine, and H3 T118 to isoleucine. We also analyzed mutant nucleosomes where H4 R45 was mutated to alanine or glutamate, and H3 T118 was mutated to either alanine or histidine (Table I).
Table 1.
Summary of crystallographic data for all mutant nucleosome structures
| H3 mutants |
H4 mutants |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R116A | R116H* | T118A | T118H | T118I* | R45A | R45C* | R45E | R45H* | V43A | V43I* | |
| Data collection | |||||||||||
| Resolution (Å) | 50–2.7 | 50–3.0 | 50–2.9 | 50–2.4 | 35–2.9 | 35–3.0 | 40–2.9 | 50–2.7 | 100–2.3 | 50–2.75 | 35–2.7 |
| Redundancy | 4.3 | 4.3 | 7.1 | 3.7 | 1.67 | 4.98 | 3.4 | 2.95 | 4.7 | 5.0 | 3.32 |
| No. of unique hkl | 59 471 | 42 653 | 47 057 | 83 303 | 43 053 | 43 674 | 47 812 | 59 509 | 93 817 | 55 911 | 58 985 |
| Completion % (last shell) | 97.6 (97.9) | 83.8 (87.4) | 99.8 (99.5) | 98.7 (96.8) | 87.4 (84.8) | 99.9 (99.8) | 97.6 (94.7) | 95.5 (88.3) | 99.0 (98.0) | 98.7 (97.0) | 97.5 (89.0) |
| Rmerge (last shell) | 0.044 (0.246) | 0.1 (0.282) | 0.124 (0.385) | 0.066 (0.273) | 0.071 (0.377) | 0.059 (0.271) | 0.062 (0.324) | 0.051 (0.381) | 0.069 (0.236) | 0.083 (0.36) | 0.036 (0.174) |
| Unit cell | |||||||||||
| a (Å) | 105.9 | 105.4 | 105.0 | 105.9 | 105.7 | 106.2 | 105.7 | 106.0 | 106.6 | 105.9 | 105.8 |
| b (Å) | 110.0 | 109.7 | 109.7 | 109.6 | 109.6 | 109.5 | 109.5 | 110.0 | 109.7 | 109.8 | 109.6 |
| c (Å) | 182.7 | 180.9 | 180.7 | 181.4 | 181.5 | 182.4 | 182.8 | 182.8 | 182.3 | 181.6 | 181.4 |
| Refinement | |||||||||||
| Resolution (Å) | 50–2.7 | 50–3.0 | 50–2.9 | 50–2.4 | 35–2.9 | 35–3.0 | 40–2.9 | 50–2.7 | 70–2.3 | 50–2.75 | 35–2.7 |
| R-factor (last shell) | 0.22 (0.34) | 0.20 (0.32) | 0.21 (0.36) | 0.23 (0.35) | 0.21 (0.38) | 0.22 (0.34) | 0.21 (0.33) | 0.22 (0.39) | 0.24 (0.35) | 0.22 (0.37) | 0.21 (0.30) |
| Free R-factor (last shell) | 0.27 (0.43) | 0.26 (0.37) | 0.29 (0.44) | 0.26 (0.36) | 0.27 (0.42) | 0.29 (0.40) | 0.27 (0.43) | 0.26 (0.44) | 0.27 (0.39) | 0.28 (0.41) | 0.27 (0.36) |
| No. atoms in model (B-factor statistics) | |||||||||||
| Total (mean B-factor Å2) | 12 222 (60.7) | 12 037 (53.2) | 12 123 (54.4) | 12 245 (56.0) | 12 072 (51.2) | 12 134 (53.2) | 12 153 (45.1) | 12 217 (58.9) | 12 264 (56.4) | 12 184 (53.2) | 12 363 (48.4) |
| Protein (mean B-factor Å2) | 6004 (46.3) | 5990 (44.2) | 5981 (45.3) | 6047 (47.7) | 5975 (41.9) | 5990 (44.1) | 6002 (49.2) | 6023 (49.2) | 5994 (47.6) | 5966 (44.1) | 6097 (39.5) |
| DNA (mean B-factor Å2) | 5980 (96.7) | 5980 (86.9) | 5980 (94.8) | 5980 (94.2) | 5980 (93.1) | 5980 (86.9) | 5980 (77.3) | 5980 (101.5) | 5980 (98.7) | 5980 (95.9) | 5980 (88.9) |
| Solvent (mean B-factor Å2) | 238 (43.2) | 164 (58.7) | 162 (46.3) | 218 (45.8) | 117 (41.1) | 164 (58.7) | 172 (48.3) | 214 (51.5) | 290 (46.5) | 238 (39.8) | 286 (39.2) |
| RMSD from ideality | |||||||||||
| Bond lengths (Å) | 0.0107 | 0.013 | 0.015 | 0.0112 | 0.0092 | 0.0067 | 0.025 | 0.011 | 0.0102 | 0.0092 | 0.013 |
| Bond angles (deg) | 1.38 | 1.41 | 1.53 | 1.46 | 1.36 | 1.19 | 2.40 | 1.63 | 1.48 | 1.39 | 1.70 |
| PDB entry id |
1P34 |
1P3A |
1P3K |
1P3L |
1P3M |
1P3B |
1P3F |
1P3G |
1P3I |
1P3O |
1P3P |
| *Mutants identified in the original screen by Kruger et al (1995). Coordinate errors for all the mutants lay in the range 0.34–0.37, which is very close to that reported for the original model 1AOI (Davey et al, 2002). | |||||||||||
Class II mutations target H3 R116. This residue, which is positioned to form two salt bridges with a nearby phosphate, instead bends back and makes two salt bridges with H3 D123 (Figure 1B). This arrangement is protected from solvent by a hydrophobic ‘van der Waals cup' located directly above it. It is formed by H3 residues M120, P121, K122, and holds a well-localized chloride ion (Davey et al, 2002, see Figure 4A). This feature has been observed in all previously determined nucleosome structures from Xenopus laevis (Luger et al, 1997a; Suto et al, 2000; Davey et al, 2002; Suto et al, 2003), and is also conserved in nucleosomes from other species (Harp et al, 2000; White et al, 2001; S Chakravarthy and K Luger, unpublished results). The original genetic screen identified a histidine substitution of this residue. We also analyzed an alanine control mutant of H3 R116 (Table I).
Figure 4.
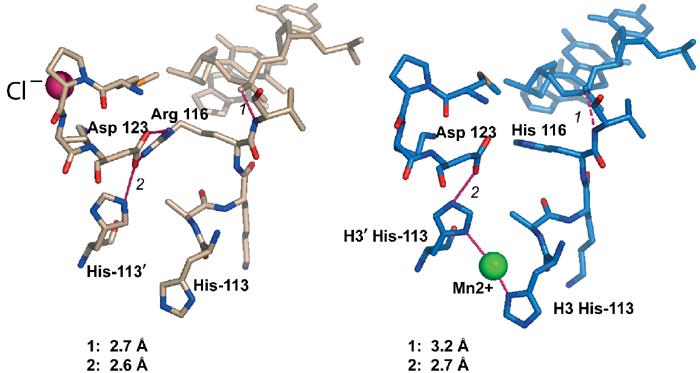
Changes in local interactions in the H3 Arg-116-His mutant. (A) Wild-type structure. Hydrogen bonds are shown in magenta, and their respective bond length is given in the inset. (B) NCPs containing H3 Arg-116-His shown in the same view. Note the loss of the chloride ion (magenta in A), and the appearance of a well-defined manganese ion (green) chelating H3 His-113 and H3′ His-113.
Class III mutations affect H4 V43, which is part of a hydrophobic core that stabilizes the conformation of the L1L2 loop arrangement (Figure 1B and C). The L1L2 loops closely approach the DNA phosphate backbone to allow hydrogen bond formation between the main-chain amide and phosphate oxygen, and also orient H4 R45 and H3 T118 (the two residues affected by class I mutations; Figure 1B and C; Luger and Richmond, 1998). Mutation of this residue to isoleucine (as observed in the original genetic screen) might disturb this arrangement, resulting in loss of interactions between main chains or side chains and the DNA. We also investigated the effect of an alanine substitution to understand the consequences of introducing a less bulky residue into this tightly packed interface.
Class IV residues involve H3 E105, whose mutation to lysine results in a relatively weak Sin phenotype (Kruger et al, 1995). This mutation is not likely to affect the protein–DNA interface, since it is located on the surface of the histone octamer (Figure 1B). However, mono-nucleosomes reconstituted with this mutant histone do exhibit slightly faster sliding rates in vitro. It is also possible that the resulting changes in the surface landscape lead to changes in higher-order structure, or changed interactions with unidentified protein factors. None of these features would have been obvious in the context of a mono-nucleosome (apart from perhaps changes in crystal packing), and the mutation was therefore not further characterized in this study.
The overall structure of mutant nucleosomes remains unchanged
Recombinant NCPs reconstituted with histones bearing the single point mutations listed in Table I were prepared as described previously (Dyer et al, 2003). We chose to introduce the mutations into the coding region of X. laevis histones (Luger et al, 1997a) rather than into the yeast sequences. This choice is justified by the fact that the entire region in H3 and H4, including the affected residues, is highly conserved between yeast and X. laevis (and other species; Sullivan et al, 2002) and warranted by our observation that nucleosomes prepared from yeast histones diffract only to 3.2 Å resolution (White et al, 2001), as opposed to ∼2.0 Å for X. laevis nucleosomes (Davey and Richmond, 2002). In addition, yeast histones H3 and H4 show 84 and 92% identity to X. laevis histones. The yeast nucleosome structure published previously shows an RMSD of only 0.47 with respect to the Xenopus nucleosome structure (White et al, 2001). The mutations had no adverse effects on protein expression, histone octamer refolding, or nucleosome reconstitution and purification.
Crystals for 11 mutant nucleosomes were obtained under similar conditions as reported earlier (Luger et al, 1997a). The structures were solved by molecular replacement from data collected either in-house or at the synchrotron. The details of data collection and refinement are listed in Table I. The presence of each Sin mutation and the conformation of mutated and surrounding residues were confirmed by calculating 7 Å radius SA-omit maps around each of the affected residues (Supplementary Figure 1). In each case, the mutation was also unambiguously apparent in the first maps after molecular replacement (not shown). All 11 structures have been deposited with the Protein Data Bank, and the pdb access codes are given in Table I.
All mutant NCP structures superimpose onto the wild-type structure (pdb entry code 1AOI) with RMSDs of less than 0.6 Å, demonstrating that the Sin mutations do not result in drastic changes in nucleosome structure. The overall path of the DNA remains undisturbed over its entire length, as indicated by the relatively small RMSDs for DNA phosphates between wild-type and mutant NCP structures that had been aligned based on the histone octamer (Table II). Since we used the same 146 base pair DNA fragment as in our previous studies, formation of diffracting crystals required ‘stretching' and twisting of the DNA by one base pair in one-half of the palindrome (Muthurajan et al, 2003; Suto et al, 2003). The precise location in nucleosomal DNA where this twist defect is dissipated is identical in all Sin mutant NCPs, and corresponds to the originally identified location (Luger et al, 1997a).
Table 2.
Structural alignment between Sin mutant and wild-type nucleosomes
| H3 mutants |
H4 mutants |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R116A | R116Ha | T118A | T118H | T118Ia | R45A | R45Ca | R45E | R45Ha | V43A | V43Ia | |
| Overall (protein and DNA) | 0.55 | 0.53 | 0.58 | 0.54 | 0.6 | 0.60 | 0.59 | 0.57 | 0.58 | 0.465 | 0.45 |
| Overall protein | 0.44 | 0.48 | 0.55 | 0.43 | 0.51 | 0.54 | 0.56 | 0.45 | 0.48 | 0.36 | 0.35 |
| Cα | 0.31 | 0.35 | 0.50 | 0.28 | 0.32 | 0.3 | 0.36 | 0.29 | 0.29 | 0.22 | 0.23 |
| DNA–phosphates |
0.60 |
0.50 |
0.53 |
0.58 |
0.62 |
0.57 |
0.51 |
0.56 |
0.58 |
0.5 |
0.51 |
| Numbers are RMSDs in Å. Histone octamers were aligned with Lsqman. | |||||||||||
| aMutants identified in the original screen by Kruger et al (1995). | |||||||||||
Overall, histone main-chain and side-chain conformations, apart from the mutated regions, were also highly similar between all structures (Table II). None of the mutations led to increases in local crystallographic B-factors for either DNA or protein, indicating that no local disorder is introduced as a consequence of the Sin mutations. As in previous structures, the histone N-terminal tails were mostly disordered (Luger et al, 1997a) (Table I), and the visible histone tail regions exhibit random coil structure.
Class I mutations result in a loss of selected protein–DNA interactions
Class I mutants (affecting residues H4 R45 and H3 T118) displayed the most severe Sin phenotype, next to H4 V43I (Kruger et al, 1995). Surprisingly, structural distortions in NCPs bearing class I mutations were minimal. Figure 2 shows a superposition of the corresponding DNA region between wild-type (wheat) and various H4 R45 mutant NCP structures (green), based on a structural alignment between the histone octamer main chains. Mutation of H4 R45 to cysteine or alanine resulted in an ‘empty' minor groove, with no additional distortions in the DNA (Figure 2B and E). This also holds true when H4 R45 is replaced with bulky or negatively charged side chains, as observed in the structure of NCP R45H, and in H4 R45E (Figure 2C and D). These side chains explicitly avoid the minor groove by adopting rotamer conformations that bend away from the minor groove. H4 R45E has a water molecule mediating an interaction with the DNA base in the minor groove (Figure 2D). We conclude that the side chain of H4 R45, despite the fact that no direct hydrogen bonds are being made by this residue, contributes significantly to this particular protein–DNA interaction interface by making van der Waals contacts with the minor groove. A loss of these interactions results in the Sin phenotype.
Figure 2.
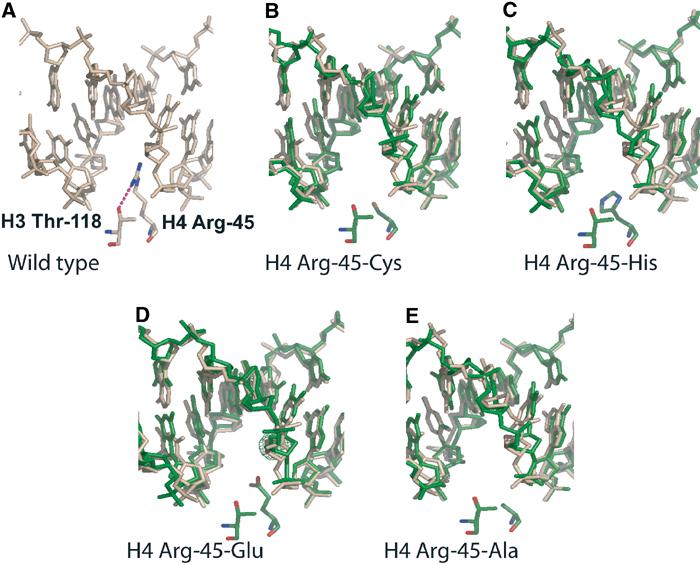
Close-up view of NCP structures reconstituted with H4 Arg-45 mutants. (A) Wild-type NCP, showing the minor groove at SHL+0.5, and H3 Arg-45 and H3 Thr-118 side chains. Hydrogen bonds are indicated by a dotted line. (B–E) Equivalent region from NCPs reconstituted with Arg-45-Cys, H4 Arg-45-His, H4 Arg-45-Glu, and H4 Arg-45-Ala, respectively (shown in green). DNA from the wild-type structure (based on an alignment of the entire histone octamer main chain) is shown in wheat to emphasize that no structural rearrangements occur as a consequence of the mutations.
The replacement of H3 T118 with the much bulkier isoleucine or histidine, and the inability of these side chains to hydrogen bond with the guanidinium group of H4 R45, pushes H4 R45 further back into the minor groove (Figure 3B and C). This results in a 1.6 Å shift in the position of a phosphate that is contacted by the main chain of H3 αN in NCPs reconstituted with H3 T118H, and a 1.2 Å shift in NCPs containing H3 T118I, as seen in Figure 3B and C (shown in wheat and blue, respectively). As a consequence, all hydrogen bonds between this particular phosphate atom and the protein main chain at H3 αN are lost. The distortions are most extreme in the H3 T118H substitution (Figure 3C). The alanine substitution has only very subtle effects, since it introduces no steric clash with H4 R45 (Figure 3D). Figure 3E shows the different positions that are assumed by H4 R45 in the different mutant structures. The more extreme positions of H4 R45 (as observed in the structures of NCP H3 T118H and NCP H3 T118I) potentially allow for base-specific interactions with an adenine.
Figure 3.
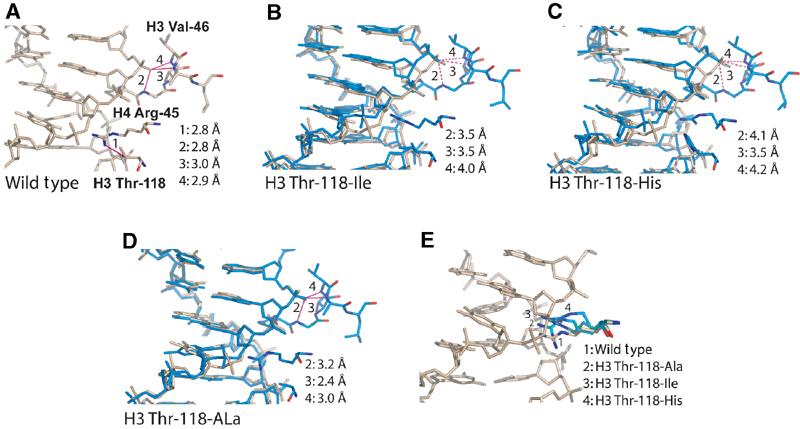
Close-up view of NCP structures reconstituted with H3 Thr-118 mutants. (A) Wild-type structure. Hydrogen bonds between the phosphate and the protein main chain (H3 αN) are indicated by solid lines, and are numbered from 2 to 4. Stretched or broken bonds are indicated by dotted lines. Bond lengths are listed in the inset. (B–E) NCP H3 Thr-118-Ile, H3 Thr-118-His, and H3 Thr-118-Ala are shown in blue in the same view. DNA from wild-type NCP (superimposed as described in Figure 2) is shown in wheat. (E) Superposition of Arg-45 side chains from wild type and H3 Thr-118 mutants. Note that the bulkier the substitution for H3 Thr-118, the further H4 Arg-45 is pushed into the minor groove.
Class II mutations subtly alter the conformation of the histone octamer main chain, and change solvent structure
H3 R116 forms a salt bridge with H3 D123 rather than interacting with the nearby DNA backbone (Figure 1B). This arrangement places the main chain around H3 R116 in perfect position to make several hydrogen bonds with the phosphate backbone of the DNA. Mutation of H3 R116 to histidine (Kruger et al, 1995) or alanine, results in the loss of this salt bridge, and in subtle changes in main-chain conformation of the H3 L2 loop. Several of the hydrogen bonds between the protein main chain and phosphates, and also between the side chains and phosphates, are weakened by less favorable distances and angles between the two binding partners (Figure 4). This result is significant since, as with all other mutants, it is independently observed for both H3 chains (H3 and H3′).
A novel feature of NCPs reconstituted with H3 R116H is the presence of a highly occupied manganese ion that is trapped between the nitrogen atoms (ND1) of H3 H113 and H3′ H113 (Figure 4B). These two residues are buried deep within the H3–H3′ four-helix bundle structure that holds together the (H3–H4)2 tetramer, and are essential for maintaining the stability of this interface (Luger et al, 1997a). In the wild-type structure, H3 H113 is held in a particular rotamer conformation that allows ND1 to make a hydrogen bond with H3 D123 (Figure 4A). The subtle structural changes invoked by the mutation from H3 R116 to histidine allows H113 to flip and juxtapose its ND1 with that of H3′ H113, while maintaining structurally equivalent hydrogen bonds between its NE2 and H3 D123 (Figure 4B). Given the vital role of these two histidine residues in organizing the central (H3–H4)2 tetramer, it is intriguing to speculate that the ability of these two side chains to bind divalent ions in response to subtle structural changes elsewhere may affect nucleosome stability and/or sliding behavior.
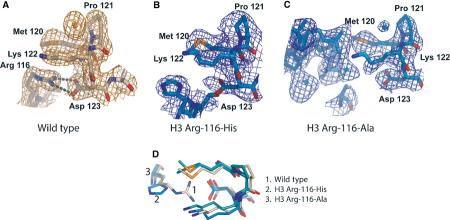
In wild-type NCP, a ‘van der Waals cup' holding an exceptionally well-localized chloride ion is located directly above this salt bridge (Figures 4A and 5A). This chloride ion is a constant feature in all nucleosome structures reported so far, and is also present in nine of the 11 structures described here. It is however absent in NCPs reconstituted with H3 R116H (Figure 5B), even though the conformation of the van der Waals cup is maintained in both of these mutant nucleosomes (Figure 5D). Its occupancy is greatly reduced in H3 R116A (Figure 5C). This demonstrates that the underlying salt bridge, although ∼7 Å distant from the ion, has a stabilizing effect on this particular ion.
Figure 5.
Changed solvent structure in H3 Arg-116 mutants. (A) The ‘van der Waals cup' formed by H3 Met-120, Pro-121, and Lys-122, holding the chloride ion. 2Fo-Fc electron density, contoured at 1 sigma, is shown in gold. (B) The same region is shown for H3 Arg-116-His (blue). Note the missing density for the chloride ion. (C) In H3 Arg-116-Ala, minor density is observed for the chloride ion, indicating only partial occupancy. (D) Superposition of the three structures, viewed from above with respect to the views shown in (A–C). Wild type is shown in wheat, and mutants in blue. The chloride ion is omitted for clarity.
Class III mutations result in weaker interactions between octamer and DNA
Mutations in H4 V43 resulted in only minor structural changes. The hydrophobic core around H4 V43 in the wild type (Figure 1C) is completely maintained upon the addition of a methyl group as a consequence of the H4 V43I mutation. However, minor reorganization of the main chain in response to the mutation leads to subtle increases in hydrogen bond lengths between the protein and eight of the DNA phosphates (not shown). The control mutation of H4 V43A has virtually no effect on the structure. In accordance, both H4 V43 mutations exhibit the smallest RMSDs for all parameters (Table II).
Sin mutations lead to increased nucleosome-sliding rates
We were interested in determining whether the observed local disruptions of protein–DNA interactions had an effect on the dynamic properties of the nucleosome. To this end, we exploited the fact that salt-gradient reconstitution of nucleosomes at 4°C onto either 5S DNA (Flaus et al, 1996), α-satellite DNA (Luger et al, 1999), or DNA derived from the mouse MMTV promoter (Flaus and Richmond, 1998) gives rise to NCPs that are positioned in at least three different translational positions offset by 10 bp with respect to each other (indicated schematically in Figures 6 and 7). For a 146 bp DNA fragment, only the central position utilizes the full range of protein–DNA interactions, making it thermodynamically more stable than others. The different nucleosome species are readily resolved by native gel electrophoresis (Luger et al, 1999), and the octamer can be mobilized in cis by the input of thermal energy to arrive at the lowest energy state (Flaus et al, 1996; Flaus and Owen-Hughes, 2001). The temperature and time required for repositioning is a function of the DNA sequence (Flaus and Richmond, 1998), and is likely to be dependent on the strength of interactions between the protein and the DNA.
Figure 6.
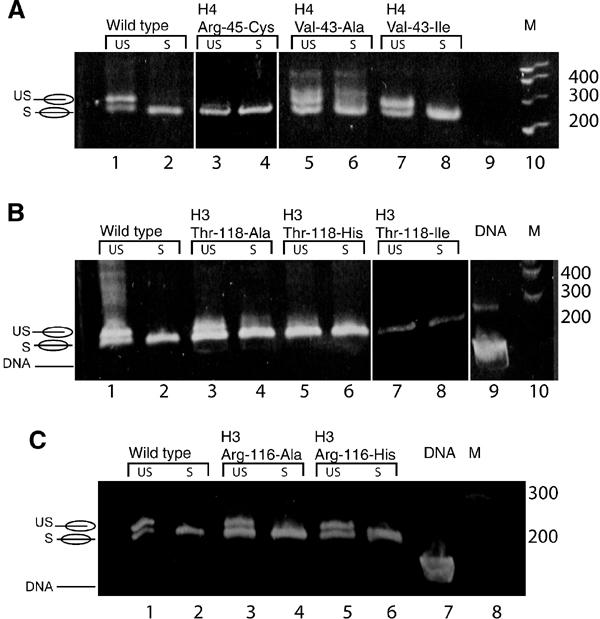
Temperature-induced repositioning of nucleosomes. Samples were kept on ice (‘unshifted'-US), or incubated for 2 h at 37°C (‘shifted'-S), and analyzed by 5% native PAGE. (A) Selected H4 mutant nucleosomes together with wild-type NCP, and (B, C) histone H3 mutant nucleosomes. Gels were stained with ethidium bromide. The position of unshifted and shifted NCPs is indicated schematically.
Figure 7.
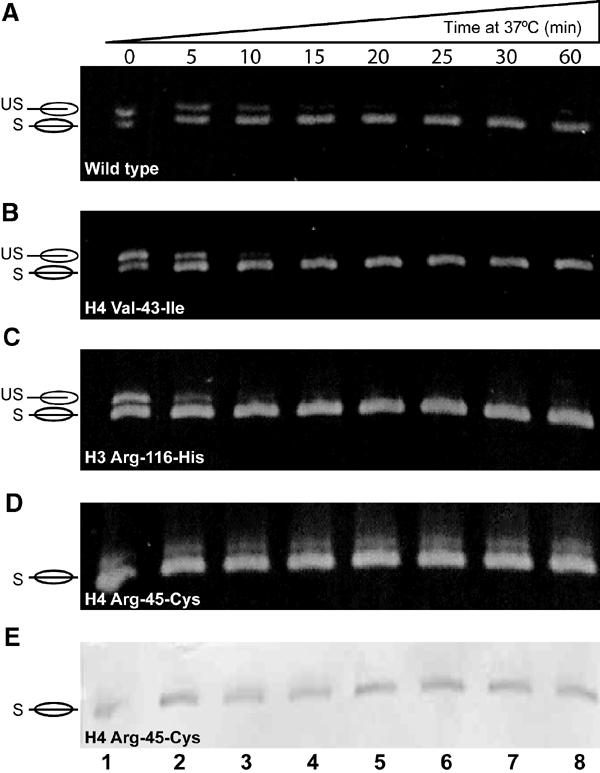
Time course of shifting for NCPs containing Sin mutants. Various NCP samples (as indicated) were incubated at 37°C for the indicated times, and analyzed by 5% native PAGE. (A–D) Gels stained with ethidim bromide. (E) The same gel as in (D), subsequently stained with Coomassie blue.
We observed that all of the H4 R45 mutants, and all of the H3 T118 mutants with the exception of T118A, reconstituted only in the thermodynamically stable position, and did not require heat shifting (Figure 6A, lanes 3 and 4; Figure 6B, lanes 5–8). In contrast, class II and class III mutant nucleosomes did require heat shifting (Figure 6C, lanes 3–6, and Figure 6A, Lanes 5–8), and we investigated the possibility that shifting in these mutants occurred at a faster rate compared to wild type.
As representatives of each class, we analyzed nucleosomes containing H4 R45C (class I, Figure 7D and E), H3 R116H (class II, Figure 7C), and H4 V43I (class III, Figure 7B) by measuring the time required for complete shifting at a fixed temperature (37°C). Wild-type nucleosomes are shown as controls (Figure 7A). NCPs containing H3 R116H or H4 V43I required less time for complete shifting than wild type. ImageQuant analysis of two sets of experiments showed that 100% shifting requires 15, 20, and 30 min, respectively, for NCPs reconstituted with H3 R116H, H4 V43I, and wild-type octamers at 37°C (Figure 7). Note that there is no change in the migration of H4 R45C nucleosomes during the time course, since these NCPs are already in the thermodynamically most stable position (Figure 7D). The faint diffuse upper band does not stain with Coomassie blue (Figure 7E), and is likely a noncanonical nucleosome that we occasionally observe. In some instances, it was possible to obtain samples of H4 R45C that did require shifting, and these samples shifted in under 5 min (not shown). Such samples could be obtained if extreme care was taken to control the temperature during reconstitution and sample handling (e.g. Figure 8A, lane 6), again suggesting that the energy barrier of these mutants towards repositioning is significantly lowered.
Figure 8.
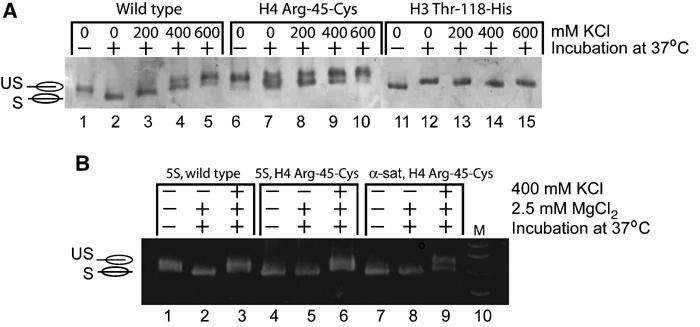
Back-shifting of wild-type and mutant nucleosomes under different conditions. (A) NCPs (wild type and mutant) reconstituted with palindromic α-satellite DNA were treated as indicated above each lane, and analyzed by native PAGE. Gels were stained with either ethidium bromide (bottom) or Coomassie blue (top). Lanes 3–5, 8–10, and 13–14 use completely shifted sample. Back-shifting is induced upon the addition of salt. (B) Shifting and back-shifting of wild-type and H4 Arg-45-Cys nucleosomes (assembled on either 5S DNA or α-satellite DNA) under conditions as indicated above each lane. Note that shifting and back-shifting are independent of the DNA sequence used, and are not inhibited by 2.5 mM magnesium chloride (compare lanes 1 and 2 in (A) with (B), lanes 1 and 2).
To exclude that the observed tendency of the Sin mutant NCPs for faster repositioning is only a peculiarity of the strongly positioning palindromic sequence derived from α-satellite DNA, we repeated these experiments using a 146 bp DNA fragment from the Lytechinus variegatus 5S rRNA gene (Richmond et al, 1988), with identical results (not shown).
The heat shifting process is reversible
We have established that salt-induced dissociation of NCPs is a multistate process. At approximately 600 mM NaCl, the ends of the DNA lose contact with the (H2A–H2B) dimer, whereas dimer–tetramer and tetramer–DNA contacts are still maintained (Park and Luger in preparation). Under these conditions, the process of heat shifting is reversible, and nucleosomes again occupy the two off-centered positions in addition to centrally positioned nucleosomes (‘back-shifting'). Figure 8A shows this phenomenon for wild-type NCP, and for mutant NCP reconstituted with either H4 R45C or H3 T118H. KCl 200, 400, or 600 mM was added to a completely shifted nucleosome sample, followed by incubation at 37°C for 90 min. For wild-type NCP, back-shifting initiated at 400 mM KCl and was complete at 600 mM KCl (Figure 8A, lanes 4 and 5). In contrast, NCPs reconstituted with H4 R45C initiated back-shifting at 200 mM KCl (Figure 8A, lane 8). These data lend further support to the hypothesis that the wild-type H4 R45 may prevent excessive nucleosome sliding. NCPs reconstituted with H3 T118H were consistently ‘pre-shifted' (Figures 6A and 8A), and this process could not be reversed upon addition of salt. These results indicate that, in these mutants, the shifted state may be more stabilized compared to the unshifted state.
The rates of shifting and back-shifting are unchanged at magnesium concentrations that mediate chromatin fiber folding
The magnesium-mediated collapse of a defined array of 12 nucleosomes into more compact ‘higher-order' structures is completely inhibited in arrays containing the H4 R45C mutant (Horn et al, 2002). In light of our finding that mono-nucleosomes harboring H4 R45C exhibit virtually no structural differences compared to wild type, but displayed a lowered energy barrier towards repositioning, we asked whether the observed increased sliding rates of mutant nucleosomes may be responsible for this behavior. We therefore repeated shifting and back-shifting experiments using a 146 bp DNA fragment derived from the 5S rRNA gene (Richmond et al, 1988) and α-satellite 146 bp DNA in the absence and presence of various concentrations of Mg2+. The 5S DNA fragment is the positioning region contained within the 12-208-mer repeats that have traditionally been used to study higher-order chromatin structure (Simpson et al, 1985), and was also used in Horn et al (2002).
As with α-satellite DNA, class I mutants are either ‘pre-shifted' (Figure 8B) or exhibit faster sliding rates (not shown). On either DNA fragment, magnesium concentrations of 1.75 and 2.5 mM (identical to those used to induce fiber formation in Horn et al, 2002) did not affect sliding or back-shifting (Figure 8B). Magnesium concentrations of >35 mM were required for inhibition of temperature-induced nucleosome sliding on 146 bp DNA fragments (not shown).
Mutant nucleosomes dissociate at lower salt concentrations
We next tested whether Sin mutations result in decreased nucleosome stability, monitored by salt-induced dissociation of nucleosomes. In a folded nucleosome, histone tyrosine fluorescence is quenched by the presence of the nearby DNA bases, and the loss of quenching is concomitant with nucleosome dissociation (Oohara and Wada, 1987; Luger et al, 1997b). We monitored tyrosine fluorescence in response to increased ionic strength, and confirmed dissociation of wild-type NCP at a midpoint of 1.1 M NaCl (Figure 9). Nucleosomes reconstituted with H4 R45C, H3 R116H, and H4 V43I all exhibited midpoints that were shifted to a significantly lower ionic strength (0.89, 0.85, and 0.91 M, respectively; Figure 9). As reported earlier (Oohara and Wada, 1987; Luger et al, 1997b), dissociation of wild-type NCPs monitored with this assay occurs in three transitions, with approximate midpoints of 0.9, 1.1, and 1.5 M NaCl. These may be attributed to increased ‘breathing' of the ends of the DNA, dissociation of the (H2A–H2B) dimer, and dissociation of the (H3–H4)2 tetramer, respectively. Only one transition was observed for H4 R45C and H4 R116H mutants, indicating that dissociation of the mutant (H3–H4)2 tetramer is now concomitant with increase in DNA end dissociation, whereas the two lower transitions remained in H4 V43I NCPs. These results are in agreement with our conclusions that the Sin mutants lead to subtle destabilization of the DNA–(H3–H4)2 tetramer interactions, and correlate well with the effects of the mutations on nucleosome sliding.
Figure 9.
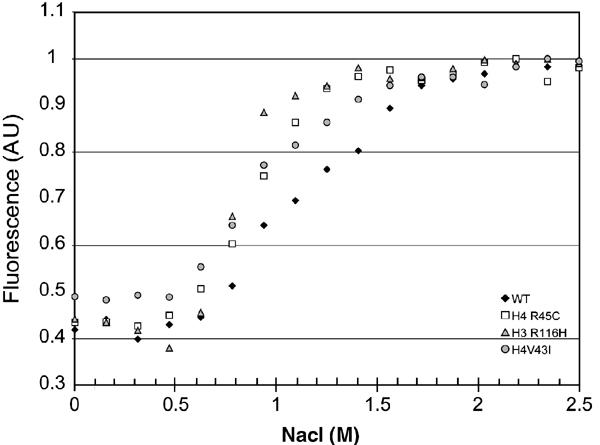
Salt-induced nucleosome dissociation. Loss of tyrosine quenching as a consequence of salt-induced nucleosome dissociation was monitored by following fluorescence emission at 306 nM (excitation 275 nm). Normalized curves are shown. Black diamonds: WT; white squares: NCP H4 Arg-45-Cys; gray triangles: NCP H3 Arg-116-His; gray circles: NCP H4-Val-43-Ile.
Discussion
The nucleosome is characterized by tight interactions between the histone octamer complex and the DNA over its entire length. In the wild-type nucleosome structure, 44 phosphates are hydrogen-bonded with a total of 60 main-chain amides over all 14 SHLs. Three phosphates (two in one DNA strand, one in the other strand) are engaged in such interactions at SHLs±5.5, 4.5, and 3.5. Interactions are weaker at SHLs±6.5 and 1.5, where only one phosphate is bound (Luger and Richmond, 1998). At SHL±0.5 (the interfaces that organize the central 20 base pairs of nucleosomal DNA), five phosphates are bound by a total of nine amides due to additional interactions with H3 αN, making this region the most tightly bound of all SHLs. Despite a wealth of structural information, the relative contribution of individual bonds to binding at each site, and the contribution of each site to the overall stability of the nucleosome is not known in quantitative terms (Davey et al, 2002).
Here we present the structural and biophysical analysis of 11 NCPs containing individual point mutations in the structured regions of histones H3 and H4 that organize SHL±0.5. These encompass five of the six original histone Sin mutants (Kruger et al, 1995), and several less-intrusive control mutations. The affected residues are highly conserved among species, and are found in a similar constellation at other SHLs within the nucleosome (see below). We find that introduction of these mutant histones into nucleosomes results in the localized loss or weakening of selected protein–DNA interactions in the vicinity of the site of mutation, rather than in profound changes in the path of the DNA, or in structural rearrangements within the histone octamer. As a result of destabilized (H3–H4)2–DNA interactions, mutant nucleosomes dissociate at significantly lower ionic strength compared to wild-type nucleosomes. One consequence of potential biological relevance is the observation that the energy barrier towards temperature-induced nucleosome repositioning (‘sliding') in mutant nucleosomes is reduced, despite the fact that only a small percentage of the direct protein–DNA interactions are lost in each of the mutant nucleosomes. This effect is independent of the DNA sequence, as shown by our comparison between nucleosomes reconstituted with α-satellite DNA and 5S DNA. It occurs both at low salt concentrations in the process of attaining the energetically favored ‘symmetric' position of the 146 bp of DNA on the histone octamer (heat shifting), as well as at elevated salt concentrations, where positioning effects are governed by the (H3–H4)2 tetramer (back-shifting).
Sin mutants exert their effect on nucleosome structure and stability in a variety of ways. In NCPs reconstituted with H3 T118I and H3 T118H, all three of the main-chain hydrogen bonds to one particular phosphate are lost at SHL±0.5, due to a rearrangement of the phosphate position. However, the two flanking phosphates are still held firmly. In other H3 mutants, and in H4 V43I, bonds are simply stretched to varying degrees. The effect of mutating H4 R45 is exerted without affecting any of the protein–phosphate hydrogen bonds, but is due to the loss of stabilizing interactions from the aliphatic side chain that reaches into the minor groove. Mutation of this residue has the most pronounced in vitro and in vivo effect. The number of interactions lost between protein and DNA that are observed in our structures is well correlated with increased sliding rates, with the stability towards salt-induced dissociation, with the severity of the Sin phenotype (Kruger et al, 1995), and with previously observed in vivo and in vitro effects (Kurumizaka and Wolffe, 1997; Wechser et al, 1997). In particular, H3 R116H and H3 T118I were more accessible towards digestion by micrococcal nuclease, and lost the ability for precise rotational positioning.
Earlier studies showed that one particular Sin mutant (H4 R45C) completely inhibits intramolecular folding of a regular 12-nucleosome array, thus providing an attractive hypothesis for a mechanism by which Sin mutants may alleviate the need for SWI/SNF (Horn et al, 2002). While these results shed new light on the possible in vivo function and substrate of SWI/SNF, the structural basis by which this mutation disrupts higher-order structure formation remained speculative. Our results suggest the possibility that the increased propensity of the histone octamer to reposition itself in cis along the DNA, as observed with H4 R45C (and other Sin mutants), inhibits the formation of higher-order chromatin structures. We have shown that nucleosome-sliding rates are unaffected at magnesium concentrations that are necessary to induce fiber formation in vitro. It will be of interest to examine whether inhibition of higher-order structure formation is a general effect of all Sin mutants.
Due to the structural similarity of the (H3–H4) and (H2A–H2B) histone fold dimers, and due to the internal two-fold symmetry of each histone fold dimer, very similar main-chain and side-chain arrangements are found at 12 of the 14 minor grooves that face the histone octamer (Luger, 2003). A recent genetic study identified regions in the structured domains of histones H3 and H4 that affect silencing in yeast (Park et al, 2002). Several of the affected residues (the lrs mutants) are the exact structural equivalents of the Sin mutations, located at the other end of the (H3–H4) histone fold dimer (SHL±2.5; Figure 1A, and Luger, 2003). While the in vitro consequences of these mutations remain to be established, the correlation in the affected residues must be more than coincidental. For example, it is tempting to speculate that arginine residues that protrude into 12 of the 14 minor grooves facing the histone octamer (Luger, 2003) may increase the resistance towards nucleosome sliding, much like cogs protruding from a gear-wheel.
The present study illustrates the role of some of the key histone residues in maintaining the structural and functional integrity of the nucleosome. We find that disruption of selected interactions between octamer and DNA has no impact on the overall structure of the nucleosome, but facilitates repositioning of the histone octamer with respect to the DNA. Thus, these mutants offer an explanation for how SWI/SNF may facilitate the access of transcription factors to chromatin in vivo. Histone Sin mutations and ATP-dependent remodeling factors both lower the energy barrier for nucleosome sliding by an as yet undetermined mechanism. The observation that Sin mutants only partially relieve the requirement for Swi/Snf (Kruger et al, 1995), and the identification of other Sin mutants that are not located at the histone–DNA interface (e.g. Hirschhorn et al, 1995; Santisteban et al, 1997; Recht and Osley, 1999) suggests additional in vivo mechanisms for making chromatin accessible to the transcription machinery. There could be additional remodeling events involving other remodeling factors, histone modifications, or the action of histone chaperones.
Our results demonstrate clearly the two conflicting functions of the histone octamer in DNA organization. Interactions between protein and DNA must be strong and numerous enough to achieve compaction and bending of nucleosomal DNA; on the other hand, the interactions must also be readily reversible to allow chromatin fluidity that appears to be required for the function of DNA processing enzymes. Single point mutations can upset this delicate balance, providing one additional reason (besides the need for maintaining structural integrity) for the extreme conservation of histone sequence throughout evolution.
Materials and methods
Nucleosome core particle assembly and structure determination
Previously reported protocols were used to express, purify, and reconstitute the Sin mutant histones in a recombinant X. laevis background with a 146 bp palindromic DNA fragment derived from human α-satellite DNA (Luger et al, 1997a; Luger et al, 1999; Dyer et al, 2003). Crystals for all mutants were obtained by vapor diffusion in the previously reported range of MnCl2 and KCl concentration (Luger et al, 1997a). Diffraction-quality crystals were routinely obtained from sitting drops, occasionally using macroseeding, and were flash cooled after stepwise soaks in MPD for cryoprotection.
Data were either collected at beam line 5.0.2 at the Advanced Light Source (Berkeley), or on a Rigaku RU-H3R rotating anode generator equipped with an Osmic Confocal multilayer optics system, and an R-Axis IV image plate detector (Table I). Data were processed using Denzo and Scalepack (Otwinowski and Minor, 1997). Molecular replacement was carried out using the previously published nucleosome structure (pdb access code 1AOI). Refinement and model building were carried out using CNS (Brunger et al, 1997) and ‘O' (Jones et al, 1991), respectively. The structures were checked using Simulated annealing-OMIT map calculations (radius of 7 Å around the mutated residue) and validated using PROCHECK (Laskowski et al, 1993) and MODELSTATS (Brunger et al, 1997). The figures in this paper were made with Pymol (DeLano, 2002).
Temperature-dependent shifting assays
Heat shifting was carried out as for wild-type nucleosomes (Luger et al, 1999) by heating the sample at 37°C for 2 h. Shifting time courses were carried out by incubating aliquots for the time points indicated (Figure 7) in a PCR thermal cycler set at 37°C. Shifting was arrested by rapid cooling to 0°C, and 20% (w/v) sucrose was added to a final concentration of 5% (w/v). A measure of 3 μl (0.2–2.0 μg) of sample was loaded on a 5% polyacrylamide gel (acryl amide:bis-acrylamide 59:1, 0.2 × TBE) (Luger et al, 1999). Gels were run at 4°C. The gels were first stained with ethidium bromide and then with Coomassie brilliant blue. Samples were adjusted to 1.75 or 2.5 mM magnesium chloride by adding appropriate volumes of 20 mM magnesium chloride stock solutions to the unshifted nucleosome samples and shifted as described above.
For back-shifting experiments, completely shifted nucleosomes were brought to 200, 400, or 600 mM KCl by the addition of 4 M KCl. The samples were incubated at 37°C for 90 min. Aliquots were electrophoresed and stained as above.
NCP dissociation assay
In all, 5 μl aliquots of a 1.5 mg/ml nucleosome preparation (using α-satellite DNA and the indicated mutants) in 20 mM Tris 7.5, 1 mM EDTA, and 1 mM DTT were brought to the indicated salt concentration with 5 M NaCl (final volume of 125 μl; final nucleosome concentration constant at 2.93 × 10−7 M) and equilibrated at 25°C. Tyrosine fluorescence was measured at an emission of 306 nm (excitation 275 nm, band width=2 nm). An automated titrating differential/ratio Aviv spectrofluorometer model ATF 105 was used for measurements.
Supplementary Material
Supplementary Fig. 1. SA- omit maps for H3 and H4 Sin mutant NCPs. All maps are contoured at 1 sigma. Close-ups from the mutant structure (shown in green for H4 mutations, and in blue for H3 mutations) are shown in views that best emphasize each mutant, with the wild type residue (white) superimposed on top of the mutated residue.
Acknowledgments
We thank Andrew Flaus and Tom Owen-Hughes for helpful suggestions and critical reading of the manuscript, Srinivas Chakravarthy and Stephanie Ebbesen for help with histone and DNA purification, and staff at the ALS (beam line 5.0.2) for technical support. We thank Craig Peterson, Kim Crowley, and Andrew Flaus for donation of some of the histone mutant expression plasmids. This work was funded by grant GM61909.
References
- Aoyagi S, Narlikar G, Zheng C, Sif S, Kingston RE, Hayes JJ (2002) Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone–DNA interactions. Mol Cell Biol 22: 3653–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB (2002) Nucleosome sliding: facts and fiction. EMBO J 21: 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Rice LM (1997) New applications of simulated annealing in X-ray crystallography and solution NMR. Structure 5: 325–336 [DOI] [PubMed] [Google Scholar]
- Davey CA, Richmond TJ (2002) DNA-dependent divalent cation binding in the nucleosome core particle. Proc Natl Acad Sci USA 99: 11169–11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ (2002) Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol 319: 1097–1113 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL User's Manual. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Dyer PN, Edayathumangalam RS, White C, Mutharajan UM, Bao Y, Chakravarthy S, Luger K (2004) Methods for reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol, in press [DOI] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ (2003) Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Flaus A, Luger K, Tan S, Richmond TJ (1996) Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc Natl Acad Sci USA 93: 1370–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2001) Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev 11: 148–154 [DOI] [PubMed] [Google Scholar]
- Flaus A, Richmond TJ (1998) Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J Mol Biol 275: 427–441 [DOI] [PubMed] [Google Scholar]
- Harp JM, Hanson BL, Timm DE, Bunick GJ (2000) Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Crystallogr D Biol Crystallogr 56 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Bortvin AL, Ricupero Hovasse SL, Winston F (1995) A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol 15: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Crowley KA, Carruthers LM, Hansen JC, Peterson CL (2002) The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat Struct Biol 9: 167–171 [DOI] [PubMed] [Google Scholar]
- Imbalzano AN (1998) Energy-dependent chromatin remodelers: complex complexes and their components. Crit Rev Eukaryot Gene Exp 8: 225–255 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjelgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B (2002) High-resolution mapping of changes in histone–DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol 22: 7524–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, Herskowitz I (1991) A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol 11: 4135–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I (1995) Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev 9: 2770–2779 [DOI] [PubMed] [Google Scholar]
- Kurumizaka H, Wolffe AP (1997) Sin mutations of histone H3: influence on nucleosome core structure and function. Mol Cell Biol 17: 6953–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R, MacArthur M, Moss D, Thornton J (1993) PROCHECK: a program to evaluate stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lorch Y, Cairns BR, Zhang M, Kornberg RD (1998) Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell 94: 29–34 [DOI] [PubMed] [Google Scholar]
- Luger K (2003) Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev 13: 127–135 [DOI] [PubMed] [Google Scholar]
- Luger K, Maeder AW, Richmond RK, Sargent DF, Richmond TJ (1997a) X-ray structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–259 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ (1997b) Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol 272: 301–311 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Luger K, Richmond TJ (1998) DNA binding within the nucleosome core. Curr Opin Struct Biol 8: 33–40 [DOI] [PubMed] [Google Scholar]
- Muthurajan UM, Park YJ, Edayathumangalam RS, Suto RK, Chakravarthy S, Dyer PN, Luger K (2003) Structure and dynamics of nucleosomal DNA. Biopolymers 68: 547–556 [DOI] [PubMed] [Google Scholar]
- Oohara I, Wada A (1987) Spectroscopic studies on histone–DNA interactions. II. Three transitions in nucleosomes resolved by salt-titration. J Mol Biol 196: 399–411 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode, Volume 276, Macromolecular Crystallography, Part A. Carter CW, Sweet RM (eds) New York: Academic Press [DOI] [PubMed]
- Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD (2002) A core nucleosome surface crucial for transcriptional silencing. Nat Genet 16: 16. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Schnitzler GR, Kingston RE (2000) Octamer transfer and creation of stably remodeled nucleosomes by human SWI–SNF and its isolated ATPases. Mol Cell Biol 20: 6380–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Osley MA (1999) Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi–Snf in yeast. EMBO J 18: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TJ, Searles MA, Simpson RT (1988) Crystals of a nucleosome core particle containing defined sequence DNA. J Mol Biol 199: 161–170 [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Arents G, Moudrianakis EN, Smith MM (1997) Histone octamer function in vivo: mutations in the dimer–tetramer interfaces disrupt both gene activation and repression. EMBO J 16: 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT, Thoma F, Brubaker JM (1985) Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42: 799–808 [DOI] [PubMed] [Google Scholar]
- Sullivan S, Sink DW, Trout KL, Makalowska I, Taylor PM, Baxevanis AD, Landsman D (2002) The histone database. Nucleic Acids Res 30: 341–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A. Z Nat Struct Biol 7: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K (2003) Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J Mol Biol 326: 371–380 [DOI] [PubMed] [Google Scholar]
- Utley RT, Cote J, Owen Hughes T, Workman JL (1997) SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem 272: 12642–12649 [DOI] [PubMed] [Google Scholar]
- Wechser MA, Kladde MP, Alfieri JA, Peterson CL (1997) Effects of Sin-versions of histone H4 on yeast chromatin structure and function. EMBO J 16: 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. SA- omit maps for H3 and H4 Sin mutant NCPs. All maps are contoured at 1 sigma. Close-ups from the mutant structure (shown in green for H4 mutations, and in blue for H3 mutations) are shown in views that best emphasize each mutant, with the wild type residue (white) superimposed on top of the mutated residue.